A Microfluidic In Vitro Three-Dimensional Dynamic Model of the Blood–Brain Barrier to Study the Transmigration of Immune Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Design and Fabrication
2.2. Flow Simulation Using Computational Fluid Dynamics
2.3. Cell Culture of BBB Models
2.4. TEER Measurement
2.5. FITC–Dextran Permeability Assay
2.6. Immunoflourescence Imaging
2.7. Flow Cytometric Analysis
2.8. RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.9. Migration Assay
2.10. Statistical Analyses
3. Results
3.1. Laminar Flow Is Maintained in TripleB Slides
3.2. The Endothelial Cell Layer of the BBB Aligns with the Direction of Flow in the Dynamic In Vitro Model Resulting in Pronounced Cell-to-Cell Tight Junction Formation
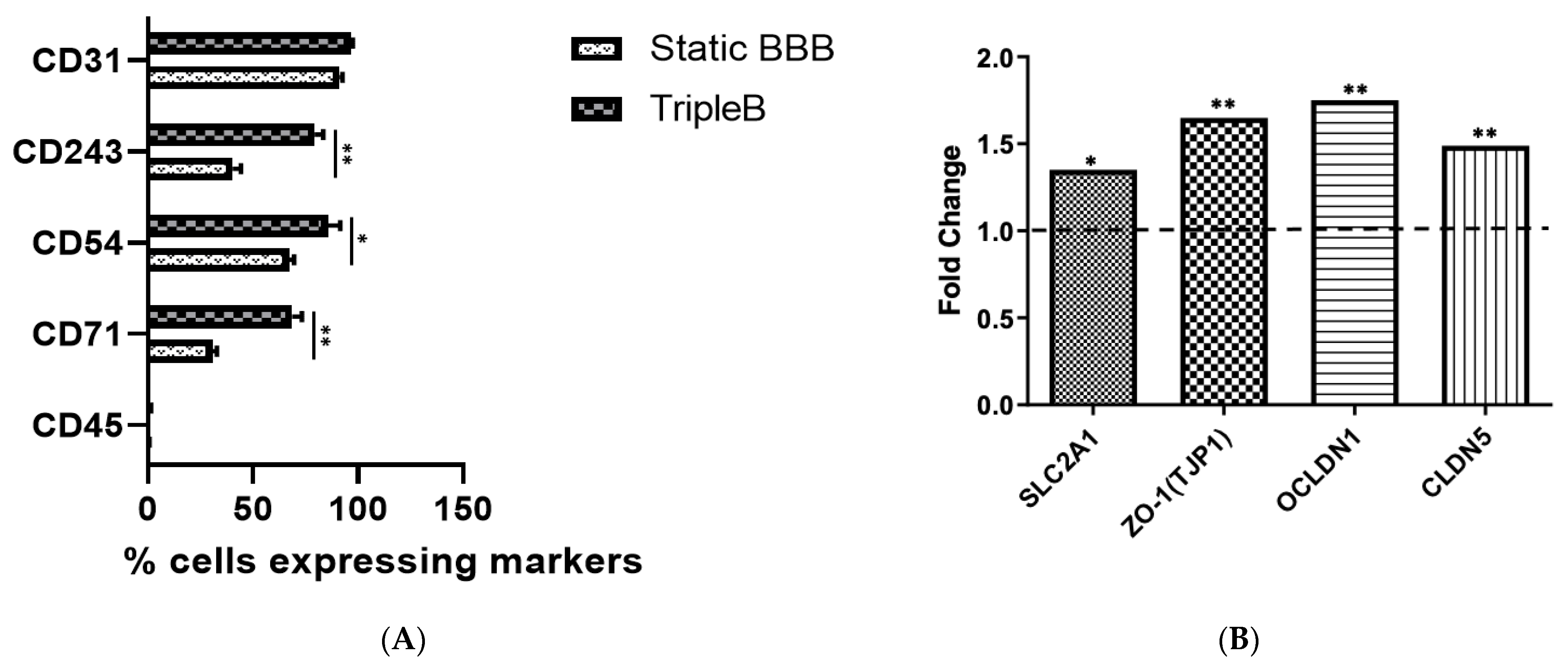
3.3. A Significantly More Stringent and Impermeable Barrier Was Formed in the Dynamic TripleB Model
3.4. Upregulated Expression of Different Tight Junction Proteins as Well as of the Hallmark BBB Protein, Pg-p, Was Found in the Dynamic TripleB Model
4. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Deuschl, G.; Beghi, E.; Fazekas, F.; Varga, T.; Christoforidi, K.A.; Sipido, E.; Bassetti, C.L.; Vos, T.; Feigin, V.L. The Burden of Neurological Diseases in Europe: An Analysis for the Global Burden of Disease Study 2017. Lancet Public Health 2020, 5, e551–e567. [Google Scholar] [CrossRef]
- Feigin, V.L.; Vos, T.; Alahdab, F.; Amit, A.M.L.; Bärnighausen, T.W.; Beghi, E.; Beheshti, M.; Chavan, P.P.; Criqui, M.H.; Desai, R. Burden of Neurological Disorders across the US from 1990-2017: A Global Burden of Disease Study. JAMA Neurol. 2021, 78, 165–176. [Google Scholar] [PubMed]
- Feigin, V.L.; Abajobir, A.A.; Abate, K.H.; Abd-Allah, F.; Abdulle, A.M.; Abera, S.F.; Abyu, G.Y.; Ahmed, M.B.; Aichour, A.N.; Aichour, I. Global, Regional, and National Burden of Neurological Disorders during 1990–2015: A Systematic Analysis for the Global Burden of Disease Study 2015. Lancet Neurol. 2017, 16, 877–897. [Google Scholar] [CrossRef]
- Chen, Y.; Dalwadi, G.; Benson, H.A.E. Drug Delivery across the Blood-Brain Barrier. Curr. Drug Deliv. 2004, 1, 361–376. [Google Scholar] [CrossRef]
- Mulvihill, J.J.E.; Cunnane, E.M.; Ross, A.M.; Duskey, J.T.; Tosi, G.; Grabrucker, A.M. Drug Delivery across the Blood–Brain Barrier: Recent Advances in the Use of Nanocarriers. Nanomedicine 2020, 15, 205–214. [Google Scholar] [CrossRef]
- Abbott, N.J.; Patabendige, A.A.K.; Dolman, D.E.M.; Yusof, S.R.; Begley, D.J. Structure and Function of the Blood--Brain Barrier. Neurobiol. Dis. 2010, 37, 13–25. [Google Scholar] [CrossRef]
- Abbott, N.J. Physiology of the Blood–Brain Barrier and Its Consequences for Drug Transport to the Brain. In International Congress Series; Elsevier: Amsterdam, The Netherlands, 2005; Volume 1277, pp. 3–18. [Google Scholar]
- Pandit, R.; Chen, L.; Götz, J. The Blood-Brain Barrier: Physiology and Strategies for Drug Delivery. Adv. Drug Deliv. Rev. 2020, 165, 1–14. [Google Scholar] [CrossRef]
- Daneman, R.; Prat, A. The Blood–Brain Barrier. Cold Spring Harb. Perspect Biol. 2015, 7, a020412. [Google Scholar] [CrossRef]
- Haseloff, R.F.; Dithmer, S.; Winkler, L.; Wolburg, H.; Blasig, I.E. Transmembrane Proteins of the Tight Junctions at the Blood–Brain Barrier: Structural and Functional Aspects. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2015; Volume 38, pp. 16–25. [Google Scholar]
- Jiang, X.; Andjelkovic, A.v.; Zhu, L.; Yang, T.; Bennett, M.V.L.; Chen, J.; Keep, R.F.; Shi, Y. Blood-Brain Barrier Dysfunction and Recovery after Ischemic Stroke. Prog. Neurobiol. 2018, 163, 144–171. [Google Scholar] [CrossRef]
- Alavijeh, M.S.; Chishty, M.; Qaiser, M.Z.; Palmer, A.M. Drug Metabolism and Pharmacokinetics, the Blood-Brain Barrier, and Central Nervous System Drug Discovery. NeuroRx 2005, 2, 554–571. [Google Scholar] [CrossRef]
- Cardoso, F.L.; Brites, D.; Brito, M.A. Looking at the Blood–Brain Barrier: Molecular Anatomy and Possible Investigation Approaches. Brain Res. Rev. 2010, 64, 328–363. [Google Scholar] [CrossRef] [PubMed]
- Luissint, A.-C.; Artus, C.; Glacial, F.; Ganeshamoorthy, K.; Couraud, P.-O. Tight Junctions at the Blood Brain Barrier: Physiological Architecture and Disease-Associated Dysregulation. Fluids Barriers CNS 2012, 9, 23. [Google Scholar] [CrossRef] [PubMed]
- Nair, M.; Jayant, R.D.; Kaushik, A.; Sagar, V. Getting into the Brain: Potential of Nanotechnology in the Management of NeuroAIDS. Adv. Drug Deliv. Rev. 2016, 103, 202–217. [Google Scholar] [CrossRef]
- Gajdács, M. The Concept of an Ideal Antibiotic: Implications for Drug Design. Molecules 2019, 24, 892. [Google Scholar] [CrossRef]
- Prinz, M.; Mildner, A. Microglia in the CNS: Immigrants from Another World. Glia 2011, 59, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Aday, S.; Cecchelli, R.; Hallier-Vanuxeem, D.; Dehouck, M.P.; Ferreira, L. Stem Cell-Based Human Blood–Brain Barrier Models for Drug Discovery and Delivery. Trends Biotechnol. 2016, 34, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Ballabh, P.; Braun, A.; Nedergaard, M. The Blood--Brain Barrier: An Overview: Structure, Regulation, and Clinical Implications. Neurobiol. Dis. 2004, 16, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Helms, H.C.; Abbott, N.J.; Burek, M.; Cecchelli, R.; Couraud, P.-O.; Deli, M.A.; Förster, C.; Galla, H.J.; Romero, I.A.; Shusta, E.V.; et al. In Vitro Models of the Blood–Brain Barrier: An Overview of Commonly Used Brain Endothelial Cell Culture Models and Guidelines for Their Use. J. Cereb. Blood Flow Metab. 2016, 36, 862–890. [Google Scholar] [CrossRef] [PubMed]
- Löscher, W.; Potschka, H. Blood-Brain Barrier Active Efflux Transporters: ATP-Binding Cassette Gene Family. NeuroRx 2005, 2, 86–98. [Google Scholar] [CrossRef]
- Idriss, H.T.; Hannun, Y.A.; Boulpaep, E.; Basavappa, S. Regulation of Volume-Activated Chloride Channels by P-Glycoprotein: Phosphorylation Has the Final Say! J. Physiol. 2000, 524, 629. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Pastan, I. Biochemistry of Multidrug Resistance Mediated by the Multidrug Transporter. Annu. Rev. Biochem. 1993, 62, 385–427. [Google Scholar] [CrossRef] [PubMed]
- Virgintino, D.; Robertson, D.; Errede, M.; Benagiano, V.; Girolamo, F.; Maiorano, E.; Roncali, L.; Bertossi, M. Expression of P-Glycoprotein in Human Cerebral Cortex Microvessels. J. Histochem. Cytochem. 2002, 50, 1671–1676. [Google Scholar] [CrossRef] [PubMed]
- Schinkel, A.H. P-Glycoprotein, a Gatekeeper in the Blood–Brain Barrier. Adv. Drug Deliv. Rev. 1999, 36, 179–194. [Google Scholar] [CrossRef]
- Ramakrishnan, P. The Role of P-Glycoprotein in the Blood-Brain Barrier. Einstein QJ Biol. Med. 2003, 19, 160–165. [Google Scholar]
- Van Assema, D.M.E.; Lubberink, M.; Boellaard, R.; Schuit, R.C.; Windhorst, A.D.; Scheltens, P.; Lammertsma, A.A.; van Berckel, B.N.M. P-Glycoprotein Function at the Blood–Brain Barrier: Effects of Age and Gender. Mol. Imaging Biol. 2012, 14, 771–776. [Google Scholar] [CrossRef] [PubMed]
- de Lange, E.C.M.; vd Berg, D.J.; Bellanti, F.; Voskuyl, R.A.; Syvänen, S. P-Glycoprotein Protein Expression versus Functionality at the Blood-Brain Barrier Using Immunohistochemistry, Microdialysis and Mathematical Modeling. Eur. J. Pharm. Sci. 2018, 124, 61–70. [Google Scholar] [CrossRef]
- Bauer, M.; Tournier, N.; Langer, O. Imaging P-Glycoprotein Function at the Blood–Brain Barrier as a Determinant of the Variability in Response to Central Nervous System Drugs. Clin. Pharmacol. Ther. 2019, 105, 1061. [Google Scholar] [CrossRef]
- Fromm, M.F. P-Glycoprotein: A Defense Mechanism Limiting Oral Bioavailability and CNS Accumulation of Drugs. Int. J. Clin. Pharmacol. Ther. 2000, 38, 69–74. [Google Scholar] [CrossRef]
- Wilhelm, I.; Fazakas, C.; Krizbai, I.A. In Vitro Models of the Blood-Brain Barrier. Acta Neurobiol. Exp. 2011, 71, 113–128. [Google Scholar]
- Gomes, M.J.; Mendes, B.; Martins, S.; Sarmento, B. Cell-Based In Vitro Models for Studying Blood–Brain Barrier (BBB) Permeability. In Concepts and Models for Drug Permeability Studies; Elsevier: Amsterdam, The Netherlands, 2016; pp. 169–188. [Google Scholar]
- Lecuyer, M.-A.; Kebir, H.; Prat, A. Glial Influences on BBB Functions and Molecular Players in Immune Cell Trafficking. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2016, 1862, 472–482. [Google Scholar] [CrossRef]
- Abbott, N.J. Blood–Brain Barrier Structure and Function and the Challenges for CNS Drug Delivery. J. Inherit. Metab. Dis. 2013, 36, 437–449. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER Measurement Techniques for In Vitro Barrier Model Systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Cucullo, L.; Hossain, M.; Puvenna, V.; Marchi, N.; Janigro, D. The Role of Shear Stress in Blood-Brain Barrier Endothelial Physiology. BMC Neurosci. 2011, 12, 40. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, C.F.; Verbridge, S.S.; Vlachos, P.P.; Rylander, M.N. Flow Shear Stress Regulates Endothelial Barrier Function and Expression of Angiogenic Factors in a 3D Microfluidic Tumor Vascular Model. Cell Adh. Migr. 2014, 8, 517–524. [Google Scholar] [CrossRef]
- Wong, A.; Ye, M.; Levy, A.; Rothstein, J.; Bergles, D.; Searson, P.C. The Blood-Brain Barrier: An Engineering Perspective. Front. Neuroeng. 2013, 6, 7. [Google Scholar] [CrossRef]
- Elbakary, B.; Badhan, R.K.S. A Dynamic Perfusion Based Blood-Brain Barrier Model for Cytotoxicity Testing and Drug Permeation. Sci. Rep. 2020, 10, 3788. [Google Scholar] [CrossRef]
- Wang, X.; Xu, B.; Xiang, M.; Yang, X.; Liu, Y.; Liu, X.; Shen, Y. Advances on Fluid Shear Stress Regulating Blood-Brain Barrier. Microvasc. Res. 2020, 128, 103930. [Google Scholar] [CrossRef]
- Choublier, N.; Müller, Y.; Gomez Baisac, L.; Laedermann, J.; de Rham, C.; Declèves, X.; Roux, A. Blood–Brain Barrier Dynamic Device with Uniform Shear Stress Distribution for Microscopy and Permeability Measurements. Appl. Sci. 2021, 11, 5584. [Google Scholar] [CrossRef]
- Garcia-Polite, F.; Martorell, J.; del Rey-Puech, P.; Melgar-Lesmes, P.; O’Brien, C.C.; Roquer, J.; Ois, A.; Principe, A.; Edelman, E.R.; Balcells, M. Pulsatility and High Shear Stress Deteriorate Barrier Phenotype in Brain Microvascular Endothelium. J. Cereb. Blood Flow Metab. 2017, 37, 2614–2625. [Google Scholar] [CrossRef]
- de Laere, M.; Sousa, C.; Meena, M.; Buckinx, R.; Timmermans, J.-P.; Berneman, Z.; Cools, N. Increased Transendothelial Transport of CCL3 Is Insufficient to Drive Immune Cell Transmigration through the Blood--Brain Barrier under Inflammatory Conditions In Vitro. Mediators Inflamm. 2017, 2017, 6752756. [Google Scholar] [CrossRef]
- Meena, M.; van Delen, M.; de Laere, M.; Sterkens, A.; Costas Romero, C.; Berneman, Z.; Cools, N. Transmigration across a Steady-State Blood–Brain Barrie Induces Activation of Circulating Dendritic Cells Partly Mediated by Actin Cytoskeletal Reorganization. Membranes 2021, 11, 700. [Google Scholar] [CrossRef] [PubMed]
- Bischel, L.L.; Coneski, P.N.; Lundin, J.G.; Wu, P.K.; Giller, C.B.; Wynne, J.; Ringeisen, B.R.; Pirlo, R.K. Electrospun Gelatin Biopapers as Substrate for In Vitro Bilayer Models of Blood− Brain Barrier Tissue. J. Biomed. Mater. Res. A 2016, 104, 901–909. [Google Scholar] [CrossRef]
- Palumbo, P.; Picchini, U.; Beck, B.; van Gelder, J.; Delbar, N.; DeGaetano, A. A General Approach to the Apparent Permeability Index. J. Pharmacokinet. Pharmacodyn. 2008, 35, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, A.B.; Calpena, A.C.; Mallandrich, M.; Clares, B. Validation of an Ex Vivo Permeation Method for the Intestinal Permeability of Different BCS Drugs and Its Correlation with Caco-2 In Vitro Experiments. Pharmaceutics 2019, 11, 638. [Google Scholar] [CrossRef] [PubMed]
- Ozeki, K.; Kato, M.; Sakurai, Y.; Ishigai, M.; Kudo, T.; Ito, K. Evaluation of the Appropriate Time Range for Estimating the Apparent Permeability Coefficient (Papp) in a Transcellular Transport Study. Int. J. Pharm. 2015, 495, 963–971. [Google Scholar] [CrossRef] [PubMed]
- Malek, A.M.; Izumo, S. Mechanism of Endothelial Cell Shape Change and Cytoskeletal Remodeling in Response to Fluid Shear Stress. J. Cell Sci. 1996, 109, 713–726. [Google Scholar] [CrossRef]
- Fisher, A.B.; Chien, S.; Barakat, A.I.; Nerem, R.M. Endothelial Cellular Response to Altered Shear Stress. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2001, 281, L529–L533. [Google Scholar] [CrossRef]
- Ballermann, B.J.; Dardik, A.; Eng, E.; Liu, A. Shear Stress and the Endothelium. Kidney Int. 1998, 54, S100–S108. [Google Scholar] [CrossRef]
- Tornavaca, O.; Chia, M.; Dufton, N.; Almagro, L.O.; Conway, D.E.; Randi, A.M.; Schwartz, M.A.; Matter, K.; Balda, M.S. ZO-1 Controls Endothelial Adherens Junctions, Cell–Cell Tension, Angiogenesis, and Barrier Formation. J. Cell Biol. 2015, 208, 821–838. [Google Scholar] [CrossRef]
- Bauer, H.-C.; Krizbai, I.A.; Bauer, H.; Traweger, A. “You Shall Not Pass”—Tight Junctions of the Blood Brain Barrier. Front. Neurosci. 2014, 8, 392. [Google Scholar] [CrossRef]
- Balda, M.S.; Matter, K. Tight Junctions and the Regulation of Gene Expression. Biochim. Biophys. Acta (BBA)-Biomembr. 2009, 1788, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Bauer, H.; Zweimueller-Mayer, J.; Steinbacher, P.; Lametschwandtner, A.; Bauer, H.-C. The Dual Role of Zonula Occludens (ZO) Proteins. J. Biomed. Biotechnol. 2010, 2010, 402593. [Google Scholar] [CrossRef] [PubMed]
- Gericke, B.; Borsdorf, S.; Wienböker, I.; Noack, A.; Noack, S.; Löscher, W. Similarities and Differences in the Localization, Trafficking, and Function of P-Glycoprotein in MDR1-EGFP-Transduced Rat versus Human Brain Capillary Endothelial Cell Lines. Fluids Barriers CNS 2021, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- Grant, G.A.; Abbott, N.J.; Janigro, D. Understanding the Physiology of the Blood-Brain Barrier: In Vitro Models. Physiology 1998, 13, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Kadry, H.; Noorani, B.; Cucullo, L. A Blood–Brain Barrier Overview on Structure, Function, Impairment, and Biomarkers of Integrity. Fluids Barriers CNS 2020, 17, 69. [Google Scholar] [CrossRef]
- Jamieson, J.J.; Searson, P.C.; Gerecht, S. Engineering the Human Blood-Brain Barrier In Vitro. J. Biol. Eng. 2017, 11, 37. [Google Scholar] [CrossRef]
- Garcia, C.M.; Darland, D.C.; Massingham, L.J.; D’Amore, P.A. Endothelial Cell–Astrocyte Interactions and TGFβ Are Required for Induction of Blood–Neural Barrier Properties. Dev. Brain Res. 2004, 152, 25–38. [Google Scholar] [CrossRef]
- Abbott, N.J. Astrocyte–Endothelial Interactions and Blood–Brain Barrier Permeability. J. Anat. 2002, 200, 523–534. [Google Scholar] [CrossRef]
- Gaillard, P.J.; Voorwinden, L.H.; Nielsen, J.L.; Ivanov, A.; Atsumi, R.; Engman, H.; Ringbom, C.; de Boer, A.G.; Breimer, D.D. Establishment and Functional Characterization of an In Vitro Model of the Blood–Brain Barrier, Comprising a Co-Culture of Brain Capillary Endothelial Cells and Astrocytes. Eur. J. Pharm. Sci. 2001, 12, 215–222. [Google Scholar] [CrossRef]
- Kulczar, C.; Lubin, K.E.; Lefebvre, S.; Miller, D.W.; Knipp, G.T. Development of a Direct Contact Astrocyte-Human Cerebral Microvessel Endothelial Cells Blood–Brain Barrier Coculture Model. J. Pharm. Pharmacol. 2017, 69, 1684–1696. [Google Scholar] [CrossRef]
- Siddharthan, V.; Kim, Y.v.; Liu, S.; Kim, K.S. Human Astrocytes/Astrocyte-Conditioned Medium and Shear Stress Enhance the Barrier Properties of Human Brain Microvascular Endothelial Cells. Brain Res. 2007, 1147, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Weksler, B.B.; Subileau, E.A.; Perriere, N.; Charneau, P.; Holloway, K.; Leveque, M.; Tricoire-Leignel, H.; Nicotra, A.; Bourdoulous, S.; Turowski, P. Blood-brain Barrier-specific Properties of a Human Adult Brain Endothelial Cell Line. FASEB J. 2005, 19, 1872–1874. [Google Scholar] [CrossRef] [PubMed]
- Raub, T.J.; Kuentzel, S.L.; Sawada, G.A. Permeability of Bovine Brain Microvessel Endothelial Cells In Vitro: Barrier Tightening by a Factor Released from Astroglioma Cells. Exp. Cell Res. 1992, 199, 330–340. [Google Scholar] [CrossRef]
- Wang, C.; Baker, B.M.; Chen, C.S.; Schwartz, M.A. Endothelial Cell Sensing of Flow Direction. Arter. Thromb. Vasc. Biol. 2013, 33, 2130–2136. [Google Scholar] [CrossRef] [PubMed]
- Islas, S.; Vega, J.; Ponce, L.; González-Mariscal, L. Nuclear Localization of the Tight Junction Protein ZO-2 in Epithelial Cells. Exp. Cell Res. 2002, 274, 138–148. [Google Scholar] [CrossRef]
- Traweger, A.; Fuchs, R.; Krizbai, I.A.; Weiger, T.M.; Bauer, H.-C.; Bauer, H. The Tight Junction Protein ZO-2 Localizes to the Nucleus and Interacts with the Heterogeneous Nuclear Ribonucleoprotein Scaffold Attachment Factor-B. J. Biol. Chem. 2003, 278, 2692–2700. [Google Scholar] [CrossRef]
- Jaramillo, B.E.; Ponce, A.; Moreno, J.; Betanzos, A.; Huerta, M.; Lopez-Bayghen, E.; Gonzalez-Mariscal, L. Characterization of the Tight Junction Protein ZO-2 Localized at the Nucleus of Epithelial Cells. Exp. Cell Res. 2004, 297, 247–258. [Google Scholar] [CrossRef]
- Wu, L.-W.; Yin, F.; Peng, J.; Wang, W.-D.; Gan, N. The Tight Junction Proteins ZO-1, Occludin and Actin Participate in the Permeability Increasing of Blood-Brain Barrier Induced by Hypoxia-Ischemia. Zhongguo Dang Dai Er Ke Za Zhi 2008, 10, 513–516. [Google Scholar]
- Hashimoto, Y.; Campbell, M. Tight Junction Modulation at the Blood-Brain Barrier: Current and Future Perspectives. Biochim. Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183298. [Google Scholar] [CrossRef]
- Lochhead, J.J.; Yang, J.; Ronaldson, P.T.; Davis, T.P. Structure, Function, and Regulation of the Blood-Brain Barrier Tight Junction in Central Nervous System Disorders. Front. Physiol. 2020, 11, 914. [Google Scholar] [CrossRef]
- Liu, W.; Wang, Z.; Zhang, L.; Wei, X.; Li, L. Tight Junction in Blood-brain Barrier: An Overview of Structure, Regulation, and Regulator Substances. CNS Neurosci. Ther. 2012, 18, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Wolburg, H.; Lippoldt, A. Tight Junctions of the Blood–Brain Barrier: Development, Composition and Regulation. Vascul. Pharmacol. 2002, 38, 323–337. [Google Scholar] [CrossRef]
- Huber, J.D.; Egleton, R.D.; Davis, T.P. Molecular Physiology and Pathophysiology of Tight Junctions in the Blood–Brain Barrier. Trends Neurosci. 2001, 24, 719–725. [Google Scholar] [CrossRef]
- Jóźwik, A.; Frymus, T. Comparison of the Immunofluorescence Assay with RT-PCR and Nested PCR in the Diagnosis of Canine Distemper. Vet. Res. Commun. 2005, 29, 347–359. [Google Scholar] [CrossRef]
- Reis, A.D.; Fink, M.C.D.; Machado, C.M.; Paz, J.d.P., Jr.; Oliveira, R.R.; Tateno, A.F.; Machado, A.F.; Cardoso, M.R.; Pannuti, C.S. Comparison of Direct Immunofluorescence, Conventional Cell Culture and Polymerase Chain Reaction Techniques for Detecting Respiratory Syncytial Virus in Nasopharyngeal Aspirates from Infants. Rev. Inst. Med. Trop. Sao Paulo 2008, 50, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, V.M.; Malik, S.V.S.; Kaur, S.; Kumar, S.; Barbuddhe, S.B. Comparison of PCR, Immunofluorescence Assay, and Pathogen Isolation for Diagnosis of Q Fever in Humans with Spontaneous Abortions. J. Clin. Microbiol. 2008, 46, 2038–2044. [Google Scholar] [CrossRef]
- Chotiprasitsakul, D.; Pewloungsawat, P.; Setthaudom, C.; Santanirand, P.; Pornsuriyasak, P. Performance of Real-Time PCR and Immunofluorescence Assay for Diagnosis of Pneumocystis Pneumonia in Real-World Clinical Practice. PLoS ONE 2020, 15, e0244023. [Google Scholar] [CrossRef]
- Urich, E.; Lazic, S.E.; Molnos, J.; Wells, I.; Freskgård, P.-O. Transcriptional Profiling of Human Brain Endothelial Cells Reveals Key Properties Crucial for Predictive In Vitro Blood-Brain Barrier Models. PLoS ONE 2012, 7, e38149. [Google Scholar] [CrossRef]
- Lutzky, V.P.; Carnevale, R.P.; Alvarez, M.J.; Maffia, P.C.; Zittermann, S.I.; Podhajcer, O.L.; Issekutz, A.C.; Chuluyan, H.E. Platelet-endothelial Cell Adhesion Molecule-1 (CD31) Recycles and Induces Cell Growth Inhibition on Human Tumor Cell Lines. J. Cell Biochem. 2006, 98, 1334–1350. [Google Scholar] [CrossRef]
- Clark, P.R.; Manes, T.D.; Pober, J.S.; Kluger, M.S. Increased ICAM-1 Expression Causes Endothelial Cell Leakiness, Cytoskeletal Reorganization and Junctional Alterations. J. Investig. Dermatol. 2007, 127, 762–774. [Google Scholar] [CrossRef]
- Cucullo, L.; Couraud, P.-O.; Weksler, B.; Romero, I.-A.; Hossain, M.; Rapp, E.; Janigro, D. Immortalized Human Brain Endothelial Cells and Flow-Based Vascular Modeling: A Marriage of Convenience for Rational Neurovascular Studies. J. Cereb. Blood Flow Metab. 2008, 28, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Arisaka, T.; Mitsumata, M.; Kawasumi, M.; Tohjima, T.; Hirose, S.; Yoshida, Y. Effects of Shear Stress on Glycosaminoglycan Synthesis in Vascular Endothelial Cells. Ann. N. Y. Acad. Sci. 1995, 748, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Santaguida, S.; Janigro, D.; Hossain, M.; Oby, E.; Rapp, E.; Cucullo, L. Side by Side Comparison between Dynamic versus Static Models of Blood–Brain Barrier In Vitro: A Permeability Study. Brain Res. 2006, 1109, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Butt, A.M.; Jones, H.C.; Abbott, N.J. Electrical Resistance across the Blood-brain Barrier in Anaesthetized Rats: A Developmental Study. J. Physiol. 1990, 429, 47–62. [Google Scholar] [CrossRef]
- Crone, C.; Olesen, S.P. Electrical Resistance of Brain Microvascular Endothelium. Brain Res. 1982, 241, 49–55. [Google Scholar] [CrossRef]
- Xu, H.; Li, Z.; Yu, Y.; Sizdahkhani, S.; Ho, W.S.; Yin, F.; Wang, L.; Zhu, G.; Zhang, M.; Jiang, L. A Dynamic In Vivo-like Organotypic Blood-Brain Barrier Model to Probe Metastatic Brain Tumors. Sci. Rep. 2016, 6, 36670. [Google Scholar] [CrossRef]
- Cucullo, L.; Marchi, N.; Hossain, M.; Janigro, D. A Dynamic In Vitro BBB Model for the Study of Immune Cell Trafficking into the Central Nervous System. J. Cereb. Blood Flow Metab. 2011, 31, 767–777. [Google Scholar] [CrossRef]
- Tsuji, A.; Tamai, I. Blood-Brain Barrier Function of P-Glycoprotein. Adv. Drug Deliv. Rev. 1997, 25, 287–298. [Google Scholar] [CrossRef]
- Noack, A.; Noack, S.; Buettner, M.; Naim, H.Y.; Löscher, W. Intercellular Transfer of P-Glycoprotein in Human Blood-Brain Barrier Endothelial Cells Is Increased by Histone Deacetylase Inhibitors. Sci. Rep. 2016, 6, 29253. [Google Scholar] [CrossRef]
- Roninson, I.B. The Role of the MDR1 (P-Glycoprotein) Gene in Multidrug Resistance In Vitro and In Vivo. Biochem. Pharmacol. 1992, 43, 95–102. [Google Scholar] [CrossRef]
- Schinkel, A.H.; Smit, J.J.M.; van Tellingen, M.; Beijnen, J.H.; Wagenaar, E.; van Deemter, L.; Mol, C.; van der Valk, M.A.; Robanus-Maandag, E.C.; te Riele, H.P.J. Disruption of the Mouse Mdr1a P-Glycoprotein Gene Leads to a Deficiency in the Blood-Brain Barrier and to Increased Sensitivity to Drugs. Cell 1994, 77, 491–502. [Google Scholar] [CrossRef]
- Bart, J.; Willemsen, A.T.M.; Groen, H.J.M.; van der Graaf, W.T.A.; Wegman, T.D.; Vaalburg, W.; de Vries, E.G.E.; Hendrikse, N.H. Quantitative Assessment of P-Glycoprotein Function in the Rat Blood–Brain Barrier by Distribution Volume of [11C] Verapamil Measured with PET. Neuroimage 2003, 20, 1775–1782. [Google Scholar] [CrossRef]
- Gaillard, P.J.; van der Sandt, I.C.J.; Voorwinden, L.H.; Vu, D.; Nielsen, J.L.; de Boer, A.G.; Breimer, D.D. Astrocytes Increase the Functional Expression of P-Glycoprotein in an In Vitro Model of the Blood-Brain Barrier. Pharm. Res. 2000, 17, 1198–1205. [Google Scholar] [CrossRef] [PubMed]
- Eisenblätter, T.; Hüwel, S.; Galla, H.-J. Characterisation of the Brain Multidrug Resistance Protein (BMDP/ABCG2/BCRP) Expressed at the Blood–Brain Barrier. Brain Res. 2003, 971, 221–231. [Google Scholar] [CrossRef]
- Austin Doyle, L.; Ross, D.D. Multidrug Resistance Mediated by the Breast Cancer Resistance Protein BCRP (ABCG2). Oncogene 2003, 22, 7340–7358. [Google Scholar] [CrossRef]
- Johnsen, K.B.; Burkhart, A.; Melander, F.; Kempen, P.J.; Vejlebo, J.B.; Siupka, P.; Nielsen, M.S.; Andresen, T.L.; Moos, T. Targeting Transferrin Receptors at the Blood-Brain Barrier Improves the Uptake of Immunoliposomes and Subsequent Cargo Transport into the Brain Parenchyma. Sci. Rep. 2017, 7, 10396. [Google Scholar] [CrossRef]
- Jefferies, W.A.; Brandon, M.R.; Hunt, S.v.; Williams, A.F.; Gatter, K.C.; Mason, D.Y. Transferrin Receptor on Endothelium of Brain Capillaries. Nature 1984, 312, 162–163. [Google Scholar] [CrossRef]
- Santa-Maria, A.R.; Walter, F.R.; Figueiredo, R.; Kincses, A.; Vigh, J.P.; Heymans, M.; Culot, M.; Winter, P.; Gosselet, F.; Dér, A. Flow Induces Barrier and Glycocalyx-Related Genes and Negative Surface Charge in a Lab-on-a-Chip Human Blood-Brain Barrier Model. J. Cereb. Blood Flow Metab. 2021, 41, 2201–2215. [Google Scholar] [CrossRef]
- Zakharova, M.; do Carmo, M.A.P.; van der Helm, M.W.; de Graaf, M.N.S.; Orlova, V.; van den Berg, A.; van der Meer, A.D.; Broersen, K.; Segerink, L.I. Multiplexed Blood–Brain Barrier Organ-on-Chip. Lab Chip 2020, 20, 3132–3143. [Google Scholar] [CrossRef]
- Raimondi, I.; Izzo, L.; Tunesi, M.; Comar, M.; Albani, D.; Giordano, C. Organ-on-a-Chip In Vitro Models of the Brain and the Blood-Brain Barrier and Their Value to Study the Microbiota-Gut-Brain Axis in Neurodegeneration. Front. Bioeng. Biotechnol. 2020, 7, 435. [Google Scholar] [CrossRef]
- Williams-Medina, A.; Deblock, M.; Janigro, D. In Vitro Models of the Blood–Brain Barrier: Tools in Translational Medicine. Front. Med. Technol. 2021, 30, 623950. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Yoon, J.-Y. In Situ Sensors for Blood-Brain Barrier (BBB) on a Chip. Sens. Actuators Rep. 2021, 3, 100031. [Google Scholar] [CrossRef]
- Linville, R.M.; Searson, P.C. Next-Generation In Vitro Blood–Brain Barrier Models: Benchmarking and Improving Model Accuracy. Fluids Barriers CNS 2021, 18, 56. [Google Scholar] [CrossRef] [PubMed]
- van der Helm, M.W.; van der Meer, A.D.; Eijkel, J.C.T.; van den Berg, A.; Segerink, L.I. Microfluidic Organ-on-Chip Technology for Blood-Brain Barrier Research. Tissue Barriers 2016, 4, e1142493. [Google Scholar] [CrossRef] [PubMed]
- Cabezas, R.; Ávila, M.; Gonzalez, J.; El-Bachá, R.S.; Báez, E.; García-Segura, L.M.; Jurado Coronel, J.C.; Capani, F.; Cardona-Gomez, G.P.; Barreto, G.E. Astrocytic Modulation of Blood Brain Barrier: Perspectives on Parkinson’s Disease. Front. Cell Neurosci. 2014, 8, 211. [Google Scholar] [CrossRef] [PubMed]
- Török, O.; Schreiner, B.; Schaffenrath, J.; Tsai, H.-C.; Maheshwari, U.; Stifter, S.A.; Welsh, C.; Amorim, A.; Sridhar, S.; Utz, S.G. Pericytes Regulate Vascular Immune Homeostasis in the CNS. Proc. Natl. Acad. Sci. USA 2021, 118, e2016587118. [Google Scholar] [CrossRef]
- Zhang, S.; Wan, Z.; Kamm, R.D. Vascularized Organoids on a Chip: Strategies for Engineering Organoids with Functional Vasculature. Lab Chip 2021, 21, 473–488. [Google Scholar] [CrossRef]
- di Lullo, E.; Kriegstein, A.R. The Use of Brain Organoids to Investigate Neural Development and Disease. Nat. Rev. Neurosci. 2017, 18, 573–584. [Google Scholar] [CrossRef]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced Pluripotent Stem Cell Technology: A Decade of Progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef]
- Workman, M.J.; Svendsen, C.N. Recent Advances in Human IPSC-Derived Models of the Blood–Brain Barrier. Fluids Barriers CNS 2020, 17, 30. [Google Scholar] [CrossRef]
- Delsing, L.; Herland, A.; Falk, A.; Hicks, R.; Synnergren, J.; Zetterberg, H. Models of the Blood-Brain Barrier Using IPSC-Derived Cells. Mol. Cell. Neurosci. 2020, 107, 103533. [Google Scholar] [CrossRef] [PubMed]
- Vatine, G.D.; Barrile, R.; Workman, M.J.; Sances, S.; Barriga, B.K.; Rahnama, M.; Barthakur, S.; Kasendra, M.; Lucchesi, C.; Kerns, J. Human IPSC-Derived Blood-Brain Barrier Chips Enable Disease Modeling and Personalized Medicine Applications. Cell Stem Cell 2019, 24, 995–1005. [Google Scholar] [CrossRef] [PubMed]
- Linville, R.M.; DeStefano, J.G.; Sklar, M.B.; Xu, Z.; Farrell, A.M.; Bogorad, M.I.; Chu, C.; Walczak, P.; Cheng, L.; Mahairaki, V. Human IPSC-Derived Blood-Brain Barrier Microvessels: Validation of Barrier Function and Endothelial Cell Behavior. Biomaterials 2019, 190, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Nishihara, H.; Gastfriend, B.D.; Soldati, S.; Perriot, S.; Mathias, A.; Sano, Y.; Shimizu, F.; Gosselet, F.; Kanda, T.; Palecek, S.P. Advancing Human Induced Pluripotent Stem Cell-derived Blood-brain Barrier Models for Studying Immune Cell Interactions. FASEB J. 2020, 34, 16693–16715. [Google Scholar] [CrossRef]
- Mossu, A.; Rosito, M.; Khire, T.; Li Chung, H.; Nishihara, H.; Gruber, I.; Luke, E.; Dehouck, L.; Sallusto, F.; Gosselet, F. A Silicon Nanomembrane Platform for the Visualization of Immune Cell Trafficking across the Human Blood–Brain Barrier under Flow. J. Cereb. Blood Flow Metab. 2019, 39, 395–410. [Google Scholar] [CrossRef]
- Cecchelli, R.; Aday, S.; Sevin, E.; Almeida, C.; Culot, M.; Dehouck, L.; Coisne, C.; Engelhardt, B.; Dehouck, M.-P.; Ferreira, L. A Stable and Reproducible Human Blood-Brain Barrier Model Derived from Hematopoietic Stem Cells. PLoS ONE 2014, 9, e99733. [Google Scholar] [CrossRef]
- Qian, T.; Maguire, S.E.; Canfield, S.G.; Bao, X.; Olson, W.R.; Shusta, E.v.; Palecek, S.P. Directed Differentiation of Human Pluripotent Stem Cells to Blood-Brain Barrier Endothelial Cells. Sci. Adv. 2017, 3, e1701679. [Google Scholar] [CrossRef]
- Jones, H.C.; Keep, R.F.; Butt, A.M. The Development of Ion Regulation at the Blood-Brain Barrier. Prog. Brain Res. 1992, 91, 123–131. [Google Scholar]
- Gendelman, H.E.; Ding, S.; Gong, N.; Liu, J.; Ramirez, S.H.; Persidsky, Y.; Mosley, R.L.; Wang, T.; Volsky, D.J.; Xiong, H. Monocyte Chemotactic Protein-1 Regulates Voltage-Gated K+ Channels and Macrophage Transmigration. J. Neuroimmune Pharmacol. 2009, 4, 47–59. [Google Scholar] [CrossRef]
- Breschi, G.L.; Cametti, M.; Mastropietro, A.; Librizzi, L.; Baselli, G.; Resnati, G.; Metrangolo, P.; de Curtis, M. Different Permeability of Potassium Salts across the Blood-Brain Barrier Follows the Hofmeister Series. PLoS ONE 2013, 8, e78553. [Google Scholar] [CrossRef]
- Miah, M.K.; Chowdhury, E.A.; Bickel, U.; Mehvar, R. Evaluation of [14C] and [13C] Sucrose as Blood–Brain Barrier Permeability Markers. J. Pharm. Sci. 2017, 106, 1659–1669. [Google Scholar] [CrossRef] [PubMed]
- Paulson, J.R.; Roder, K.E.; McAfee, G.; Allen, D.D.; van der Schyf, C.J.; Abbruscato, T.J. Tobacco Smoke Chemicals Attenuate Brain-to-Blood Potassium Transport Mediated by the Na, K, 2Cl-Cotransporter during Hypoxia-Reoxygenation. J. Pharmacol. Exp. Ther. 2006, 316, 248–254. [Google Scholar] [CrossRef] [PubMed]
- Behrens, M.; Hüwel, S.; Galla, H.-J.; Humpf, H.-U. Blood-Brain Barrier Effects of the Fusarium Mycotoxins Deoxynivalenol, 3 Acetyldeoxynivalenol, and Moniliformin and Their Transfer to the Brain. PLoS ONE 2015, 10, e0143640. [Google Scholar] [CrossRef] [PubMed]
- Oppenheim, H.A.; Lucero, J.; Guyot, A.-C.; Herbert, L.M.; McDonald, J.D.; Mabondzo, A.; Lund, A.K. Exposure to Vehicle Emissions Results in Altered Blood Brain Barrier Permeability and Expression of Matrix Metalloproteinases and Tight Junction Proteins in Mice. Part. Fibre Toxicol. 2013, 10, 62. [Google Scholar] [CrossRef] [PubMed]
- Huber, J.D.; Hau, V.S.; Borg, L.; Campos, C.R.; Egleton, R.D.; Davis, T.P. Blood-Brain Barrier Tight Junctions Are Altered during a 72-h Exposure to λ-Carrageenan-Induced Inflammatory Pain. Am. J. Physiol.-Heart Circ. Physiol. 2002, 283, H1531–H1537. [Google Scholar] [CrossRef]
- Hawkins, B.T.; Sykes, D.B.; Miller, D.S. Rapid, Reversible Modulation of Blood–Brain BarrierP-Glycoprotein Transport Activity by Vascular Endothelial Growth Factor. J. Neurosci. 2010, 30, 1417–1425. [Google Scholar] [CrossRef]
- Ronaldson, P.T.; DeMarco, K.M.; Sanchez-Covarrubias, L.; Solinsky, C.M.; Davis, T.P. Transforming Growth Factor-β Signaling Alters Substrate Permeability and Tight Junction Protein Expression at the Blood-Brain Barrier during Inflammatory Pain. J. Cereb. Blood Flow Metab. 2009, 29, 1084–1098. [Google Scholar] [CrossRef]
- Bickel, U.; Schumacher, O.P.; Kang, Y.-S.; Voigt, K. Poor Permeability of Morphine 3-Glucuronide and Morphine 6-Glucuronide through the Blood-Brain Barrier in the Rat. J. Pharmacol. Exp. Ther. 1996, 278, 107–113. [Google Scholar]
- Ziylan, Y.Z.; LeFauconnier, J.M.; Bernard, G.; Bourre, J.M. Effect of Dexamethasone on Transport of A-Aminoisobutyric Acid and Sucrose Across the Blood-Brain Barrier. J. Neurochem. 1988, 51, 1338–1342. [Google Scholar] [CrossRef]
- Yin, D.; Wang, X.; Konda, B.M.; Ong, J.M.; Hu, J.; Sacapano, M.R.; Ko, M.K.; Espinoza, A.J.; Irvin, D.K.; Shu, Y. Increase in Brain Tumor Permeability in Glioma-Bearing Rats with Nitric Oxide Donors. Clin. Cancer Res. 2008, 14, 4002–4009. [Google Scholar] [CrossRef]
- Jin, L.; Nation, R.L.; Li, J.; Nicolazzo, J.A. Species-Dependent Blood-Brain Barrier Disruption of Lipopolysaccharide: Amelioration by Colistin In Vitro and In Vivo. Antimicrob. Agents Chemother. 2013, 57, 4336–4342. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meena, M.; Vandormael, R.; De Laere, M.; Pintelon, I.; Berneman, Z.; Watts, R.; Cools, N. A Microfluidic In Vitro Three-Dimensional Dynamic Model of the Blood–Brain Barrier to Study the Transmigration of Immune Cells. Brain Sci. 2022, 12, 1293. https://doi.org/10.3390/brainsci12101293
Meena M, Vandormael R, De Laere M, Pintelon I, Berneman Z, Watts R, Cools N. A Microfluidic In Vitro Three-Dimensional Dynamic Model of the Blood–Brain Barrier to Study the Transmigration of Immune Cells. Brain Sciences. 2022; 12(10):1293. https://doi.org/10.3390/brainsci12101293
Chicago/Turabian StyleMeena, Megha, Robin Vandormael, Maxime De Laere, Isabel Pintelon, Zwi Berneman, Regan Watts, and Nathalie Cools. 2022. "A Microfluidic In Vitro Three-Dimensional Dynamic Model of the Blood–Brain Barrier to Study the Transmigration of Immune Cells" Brain Sciences 12, no. 10: 1293. https://doi.org/10.3390/brainsci12101293
APA StyleMeena, M., Vandormael, R., De Laere, M., Pintelon, I., Berneman, Z., Watts, R., & Cools, N. (2022). A Microfluidic In Vitro Three-Dimensional Dynamic Model of the Blood–Brain Barrier to Study the Transmigration of Immune Cells. Brain Sciences, 12(10), 1293. https://doi.org/10.3390/brainsci12101293




