Prediction and Visualization of Non-Enhancing Tumor in Glioblastoma via T1w/T2w-Ratio Map
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Selection
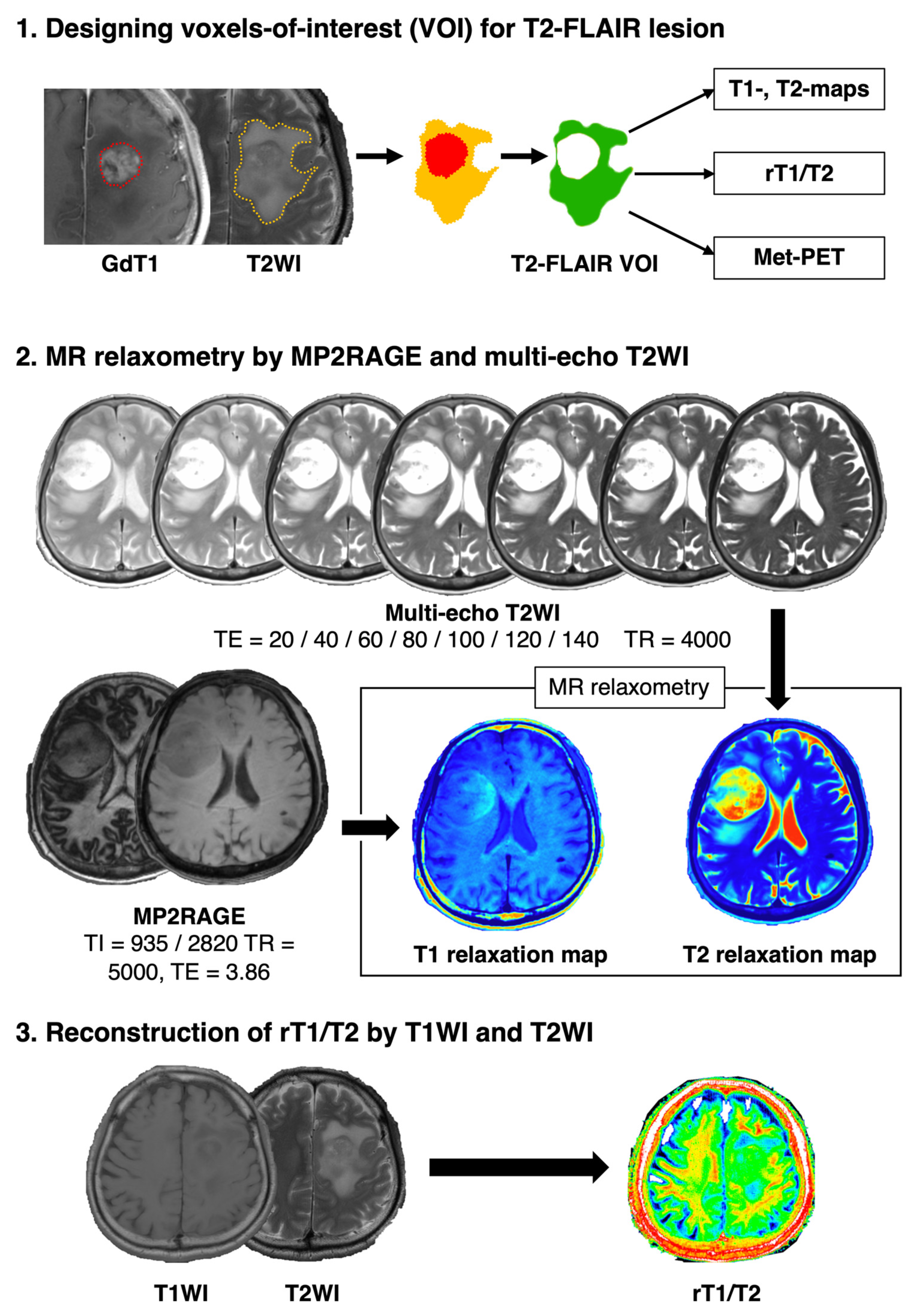
2.2. Image Co-Registration and Voxel-Of-Interest (VOI) Definition
2.3. T1- and T2-Relaxometry by MP2RAGE and Multi-Echo T2WIs
2.4. 11C-Methionine Positron Emission Tomography (Met-PET)
2.5. T1w/T2w-Ratio Map (rT1/T2) Reconstruction
2.6. Analysis of the Relationship between T1- and T2-Relaxation Maps and rT1/T2
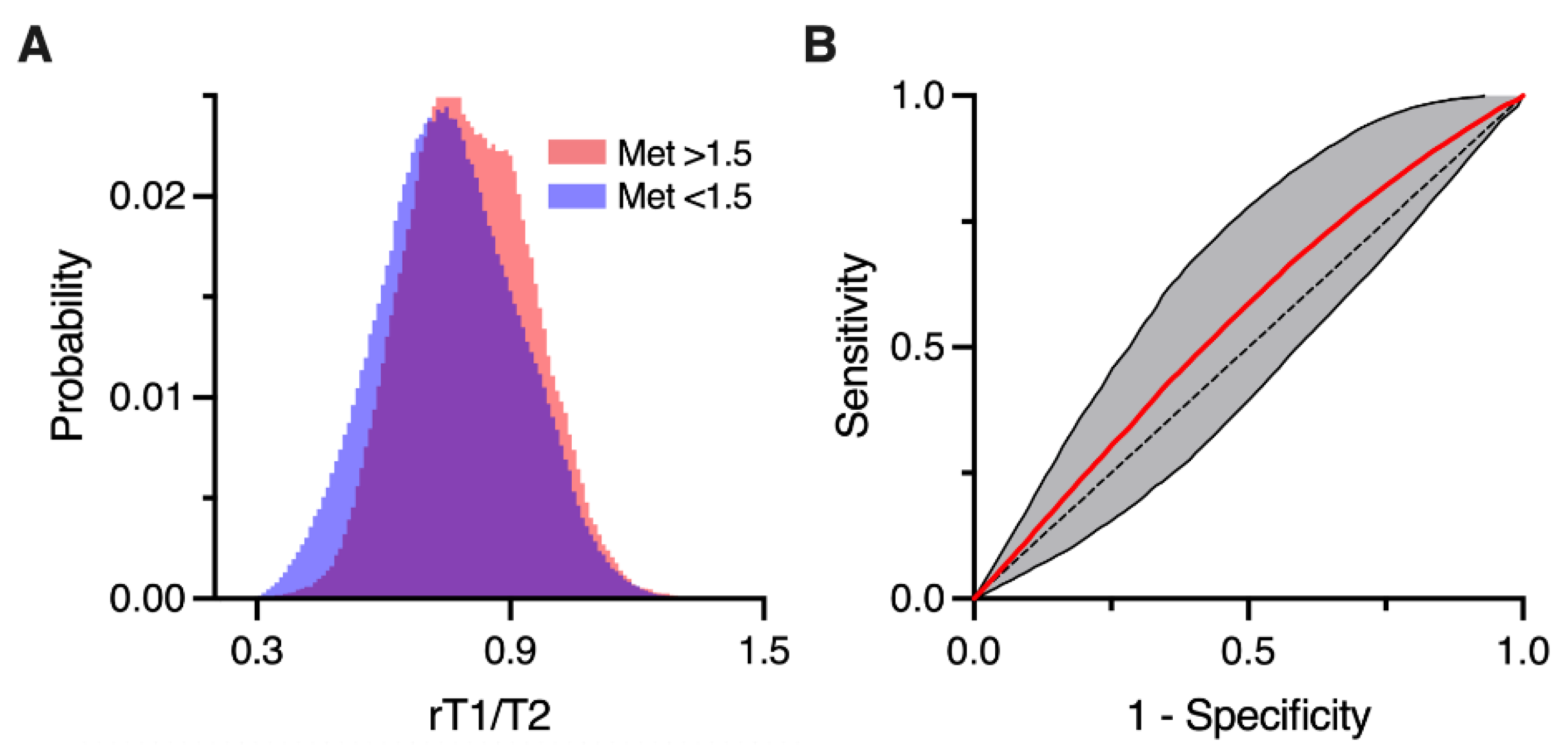
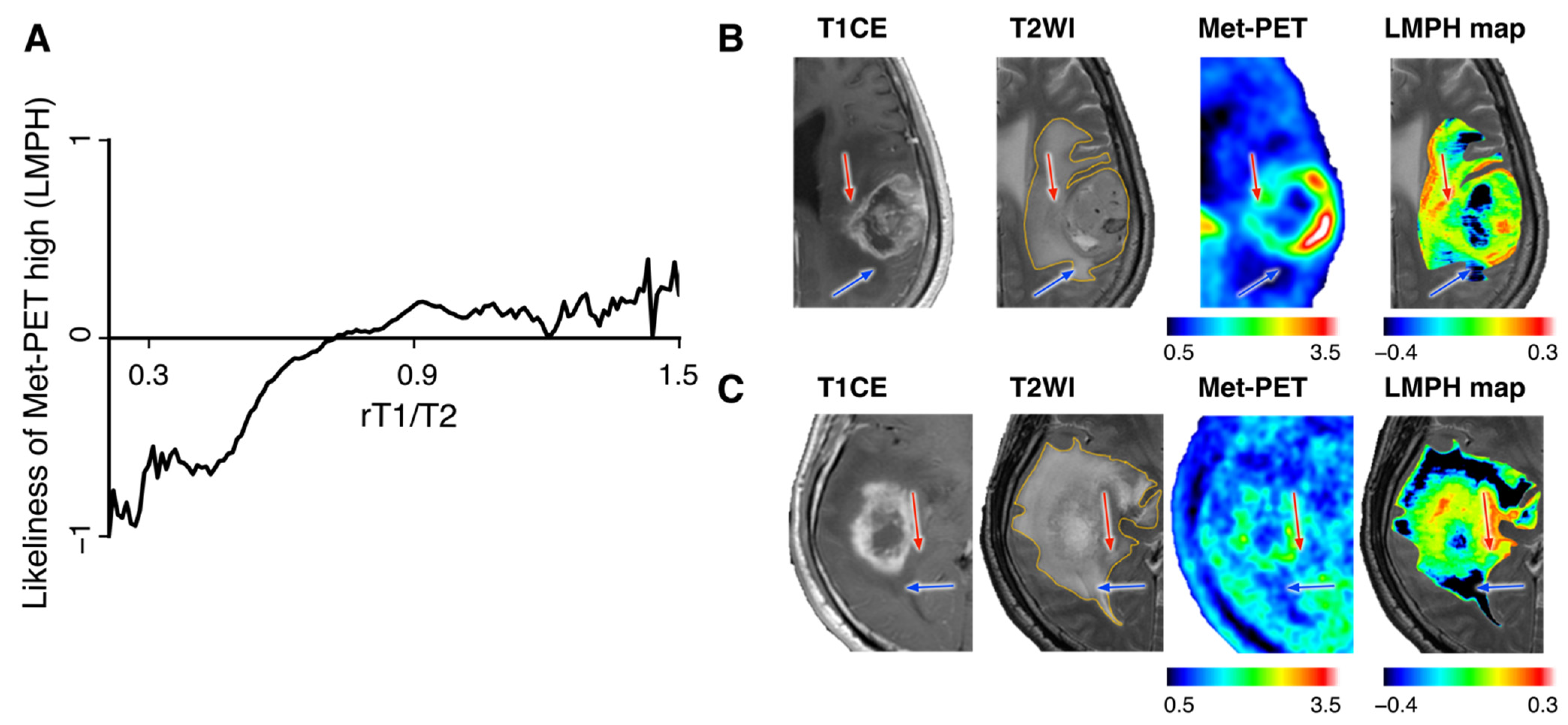
2.7. Comparing rT1/T2 and Met-PET and Defining the “Likeliness of Met-PET High”
2.8. Prediction Accuracy Estimation Analysis of LMPH Deriving from rT1/T2
2.9. Statistical Analysis
3. Results
3.1. rT1/T2 Logarithmically Correlated with T1- and T2-Relaxation Time
3.2. rT1/T2 Distributions of Met-PET High and Low T2-FLAIR Hyperintense Lesions
3.3. Likeliness of Met-PET High Derived from rT1/T2 (LMPH) Helped Classify Met-PET High and Low T2-FLAIR Hyperintense Lesions
3.4. LMPH Map for Visualizing Met-PET High NET
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Lacroix, M.; Abi-Said, D.; Fourney, D.R.; Gokaslan, Z.L.; Shi, W.; DeMonte, F.; Lang, F.F.; McCutcheon, I.E.; Hassenbusch, S.J.; Holland, E.; et al. A Multivariate Analysis of 416 Patients with Glioblastoma Multiforme: Prognosis, Extent of Resection, and Survival. J. Neurosurg. 2001, 95, 190–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.M.; Suki, D.; Hess, K.; Sawaya, R. The Influence of Maximum Safe Resection of Glioblastoma on Survival in 1229 Patients: Can We Do Better than Gross-Total Resection? J. Neurosurg. 2016, 124, 977–988. [Google Scholar] [CrossRef] [Green Version]
- Sanai, N.; Polley, M.-Y.; McDermott, M.W.; Parsa, A.T.; Berger, M.S. An Extent of Resection Threshold for Newly Diagnosed Glioblastomas: Clinical Article. J. Neurosurg. 2011, 115, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Molinaro, A.M.; Hervey-Jumper, S.; Morshed, R.A.; Young, J.; Han, S.J.; Chunduru, P.; Zhang, Y.; Phillips, J.J.; Shai, A.; Lafontaine, M.; et al. Association of Maximal Extent of Resection of Contrast-Enhanced and Non–Contrast-Enhanced Tumor With Survival Within Molecular Subgroups of Patients With Newly Diagnosed Glioblastoma. JAMA Oncol. 2020, 6, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Kracht, L.W.; Miletic, H.; Busch, S.; Jacobs, A.H.; Voges, J.; Hoevels, M.; Klein, J.C.; Herholz, K.; Heiss, W.-D. Delineation of Brain Tumor Extent with [11C]l-Methionine Positron Emission Tomography. Clin. Cancer Res. 2004, 10, 7163–7170. [Google Scholar] [CrossRef] [Green Version]
- Nariai, T.; Tanaka, Y.; Wakimoto, H.; Aoyagi, M.; Tamaki, M.; Ishiwata, K.; Senda, M.; Ishii, K.; Hirakawa, K.; Ohno, K. Usefulness of l-[Methyl-11C] Methionine—Positron Emission Tomography as a Biological Monitoring Tool in the Treatment of Glioma. J. Neurosurg. 2005, 103, 498–507. [Google Scholar] [CrossRef] [PubMed]
- Okita, Y.; Kinoshita, M.; Goto, T.; Kagawa, N.; Kishima, H.; Shimosegawa, E.; Hatazawa, J.; Hashimoto, N.; Yoshimine, T. 11C-Methionine Uptake Correlates with Tumor Cell Density Rather than with Microvessel Density in Glioma: A Stereotactic Image-Histology Comparison. Neuroimage 2010, 49, 2977–2982. [Google Scholar] [CrossRef]
- Ikeda, H.; Abe, T.; Watanabe, K. Usefulness of Composite Methionine–Positron Emission Tomography/3.0-Tesla Magnetic Resonance Imaging to Detect the Localization and Extent of Early-Stage Cushing Adenoma: Clinical Article. J. Neurosurg. 2010, 112, 750–755. [Google Scholar] [CrossRef]
- Galldiks, N.; Ullrich, R.; Schroeter, M.; Fink, G.R.; Jacobs, A.H.; Kracht, L.W. Volumetry of [11C]-Methionine PET Uptake and MRI Contrast Enhancement in Patients with Recurrent Glioblastoma Multiforme. Eur. J. Nucl. Med. Mol. Imaging 2010, 37, 84–92. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, M.; Arita, H.; Goto, T.; Okita, Y.; Isohashi, K.; Watabe, T.; Kagawa, N.; Fujimoto, Y.; Kishima, H.; Shimosegawa, E.; et al. A Novel PET Index, 18F-FDG–11C-Methionine Uptake Decoupling Score, Reflects Glioma Cell Infiltration. J. Nucl. Med. 2012, 53, 1701–1708. [Google Scholar] [CrossRef] [Green Version]
- Arita, H.; Kinoshita, M.; Kagawa, N.; Fujimoto, Y.; Kishima, H.; Hashimoto, N.; Yoshimine, T. 11C-methionine Uptake and Intraoperative 5-aminolevulinic Acid-induced Fluorescence as Separate Index Markers of Cell Density in Glioma. Cancer 2011, 118, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, M.; Uchikoshi, M.; Tateishi, S.; Miyazaki, S.; Sakai, M.; Ozaki, T.; Asai, K.; Fujita, Y.; Matsuhashi, T.; Kanemura, Y.; et al. Magnetic Resonance Relaxometry for Tumor Cell Density Imaging for Glioma: An Exploratory Study via 11C-Methionine PET and Its Validation via Stereotactic Tissue Sampling. Cancers 2021, 13, 4067. [Google Scholar] [CrossRef] [PubMed]
- Glasser, M.F.; Essen, D.C.V. Mapping Human Cortical Areas In Vivo Based on Myelin Content as Revealed by T1- and T2-Weighted MRI. J. Neurosci. 2011, 31, 11597–11616. [Google Scholar] [CrossRef] [Green Version]
- Ganzetti, M.; Wenderoth, N.; Mantini, D. Whole Brain Myelin Mapping Using T1- and T2-Weighted MR Imaging Data. Front. Hum. Neurosci. 2014, 8, 671. [Google Scholar] [CrossRef] [PubMed]
- Pareto, D.; Garcia-Vidal, A.; Alberich, M.; Auger, C.; Montalban, X.; Tintoré, M.; Sastre-Garriga, J.; Rovira, À. Ratio of T1-Weighted to T2-Weighted Signal Intensity as a Measure of Tissue Integrity: Comparison with Magnetization Transfer Ratio in Patients with Multiple Sclerosis. Am. J. Neuroradiol. 2020, 41, 461–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preziosa, P.; Bouman, P.M.; Kiljan, S.; Steenwijk, M.D.; Meani, A.; Pouwels, P.J.; Rocca, M.A.; Filippi, M.; Geurts, J.J.G.; Jonkman, L.E. Neurite Density Explains Cortical T1-Weighted/T2-Weighted Ratio in Multiple Sclerosis. J. Neurol. Neurosurg. Psychiatry 2021, 92, 790–792. [Google Scholar] [CrossRef]
- Hatakeyama, T.; Kawai, N.; Nishiyama, Y.; Yamamoto, Y.; Sasakawa, Y.; Ichikawa, T.; Tamiya, T. 11C-Methionine (MET) and 18F-Fluorothymidine (FLT) PET in Patients with Newly Diagnosed Glioma. Eur. J. Nucl. Med. Mol. Imaging 2008, 35, 2009–2017. [Google Scholar] [CrossRef]
- Kinoshita, M.; Arita, H.; Okita, Y.; Kagawa, N.; Kishima, H.; Hashimoto, N.; Tanaka, H.; Watanabe, Y.; Shimosegawa, E.; Hatazawa, J.; et al. Comparison of Diffusion Tensor Imaging and 11C-Methionine Positron Emission Tomography for Reliable Prediction of Tumor Cell Density in Gliomas. J. Neurosurg. 2016, 125, 1136–1142. [Google Scholar] [CrossRef] [Green Version]
- Hirata, T.; Kinoshita, M.; Tamari, K.; Seo, Y.; Suzuki, O.; Wakai, N.; Achiha, T.; Umehara, T.; Arita, H.; Kagawa, N.; et al. 11C-Methionine-18F-FDG Dual-PET-Tracer–Based Target Delineation of Malignant Glioma: Evaluation of Its Geometrical and Clinical Features for Planning Radiation Therapy. J. Neurosurg. 2018, 131, 676–686. [Google Scholar] [CrossRef]
- Verburg, N.; Hoefnagels, F.W.A.; Barkhof, F.; Boellaard, R.; Goldman, S.; Guo, J.; Heimans, J.J.; Hoekstra, O.S.; Jain, R.; Kinoshita, M.; et al. Diagnostic Accuracy of Neuroimaging to Delineate Diffuse Gliomas within the Brain: A Meta-Analysis. Am. J. Neuroradiol. 2017, 38, 1884–1891. [Google Scholar] [CrossRef] [Green Version]
- Verburg, N.; Koopman, T.; Yaqub, M.M.; Hoekstra, O.S.; Lammertsma, A.A.; Barkhof, F.; Pouwels, P.J.W.; Reijneveld, J.C.; Heimans, J.J.; Rozemuller, A.J.M.; et al. Improved Detection of Diffuse Glioma Infiltration with Imaging Combinations: A Diagnostic Accuracy Study. Neuro-Oncology 2019, 22, 412–422. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Kazerooni, A.F.; Ware, J.B.; Mamourian, E.; Anderson, H.; Guiry, S.; Sako, C.; Raymond, C.; Yao, J.; Brem, S.; et al. Quantification of Tumor Microenvironment Acidity in Glioblastoma Using Principal Component Analysis of Dynamic Susceptibility Contrast Enhanced MR Imaging. Sci. Rep. 2021, 11, 15011. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, S.; Sanada, T.; Sakai, M.; Arisawa, A.; Kagawa, N.; Shimosegawa, E.; Nakanishi, K.; Kanemura, Y.; Kinoshita, M.; Kishima, H. Prediction and Visualization of Non-Enhancing Tumor in Glioblastoma via T1w/T2w-Ratio Map. Brain Sci. 2022, 12, 99. https://doi.org/10.3390/brainsci12010099
Yamamoto S, Sanada T, Sakai M, Arisawa A, Kagawa N, Shimosegawa E, Nakanishi K, Kanemura Y, Kinoshita M, Kishima H. Prediction and Visualization of Non-Enhancing Tumor in Glioblastoma via T1w/T2w-Ratio Map. Brain Sciences. 2022; 12(1):99. https://doi.org/10.3390/brainsci12010099
Chicago/Turabian StyleYamamoto, Shota, Takahiro Sanada, Mio Sakai, Atsuko Arisawa, Naoki Kagawa, Eku Shimosegawa, Katsuyuki Nakanishi, Yonehiro Kanemura, Manabu Kinoshita, and Haruhiko Kishima. 2022. "Prediction and Visualization of Non-Enhancing Tumor in Glioblastoma via T1w/T2w-Ratio Map" Brain Sciences 12, no. 1: 99. https://doi.org/10.3390/brainsci12010099
APA StyleYamamoto, S., Sanada, T., Sakai, M., Arisawa, A., Kagawa, N., Shimosegawa, E., Nakanishi, K., Kanemura, Y., Kinoshita, M., & Kishima, H. (2022). Prediction and Visualization of Non-Enhancing Tumor in Glioblastoma via T1w/T2w-Ratio Map. Brain Sciences, 12(1), 99. https://doi.org/10.3390/brainsci12010099







