Node Centrality Measures Identify Relevant Structural MRI Features of Subjects with Autism
Abstract
1. Introduction
2. Materials and Methods
2.1. ABIDE Dataset and Sample Selection Criteria
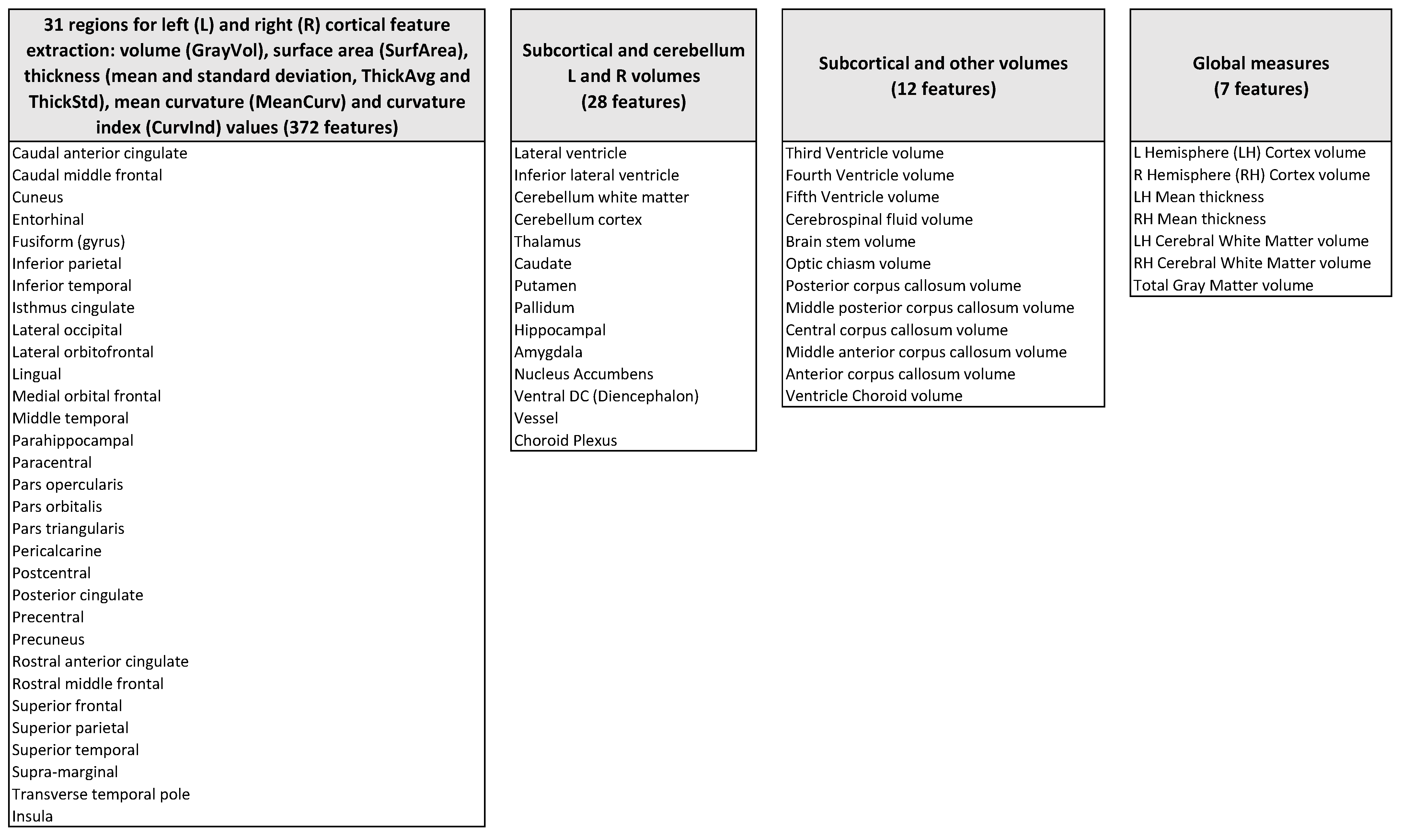
2.2. Extraction of Descriptive Features
- Volume, surface area, thickness (mean and standard deviation), mean curvature, and curvature index of 31 bilateral cortical structures (372 features);
- Volumes of bilateral sub-cortical structures and cerebellum (28 features);
- Volumes of other subcortical structures, including the corpus callosum and brainstem (12 features);
- Global measures–i.e. bilateral average global cortex volumes and thicknesses, bilateral white matter average volumes, and total gray matter volume (seven features).
- Volumetric features were divided by the global brain volume, considering the FreeSurfer feature corresponding to brain segmented volume without ventricles (BrainSegVolNotVent);
- Cortical thicknesses were divided by the mean cortical thickness (computed as the average between the left and right hemisphere average thickness values);
- Cortical areas were divided by the sum of surface area values of all cortical structures.
2.3. Weighted Correlation Networks (WCN)
- Each feature was identified as a node: ;
- For each group, the Spearman’s correlation matrix was computed;
- Node pairs with exceeding the 0.85-th quantile of the whole Spearman’s coefficients distribution were considered as linked (self-loops were discarded);
- After thresholding, we generated both a binarized and a weighted version of each network, with the weights assigned via the formula , using the chordal distance and setting .
2.4. Network Matrices and Node Centrality Measures
- The node degree and node strength , defined as
- The average degree and strength of each node’s nearest neighbors, denoted and , respectively;
- The closeness centrality and weighted closeness centrality , defined aswhere and are the topological and weighted distance between nodes i and j, respectively;
- The betweenness centrality and weighted betweenness centrality , defined aswhere, for both the topological and the weighted network, is the number of shortest paths between nodes i and j, and is the number of paths passing through node i;
- The clustering coefficient , defined as the fraction of pairs of a node’s adjacent nodes that are also adjacent to each other (and thus not defined for i s.t. ), computed as
- Spectral Centrality and Weighted Spectral Centrality, defined as:where the superscript “” is used to specify that the spectral centrality is considered, and and denote the first nontrivial eigenvector of the Laplacian for the topological and the weighted networks, respectively [20].
2.5. Network Subsampling
- The large difference in samples in and (, ) can cause a different amount of fluctuations in sample correlation ;
- By reconstructing a single network instance for each group, only two values would be produced (one for and one for ), making a statistical comparison impossible between the two groups.
- Whole-network statistics: For each network, a single centrality value was calculated as the average over all node centrality measures (obtaining a distribution of values for each centrality measure in the TD group to be compared with a single value for the AUT group);
- Single-node statistics: We compared the centrality values separately for each node (thus obtaining a table of centrality values for the TD subsampled group, to be compared with the 818 values for the single instance of the AUT group).
2.6. Group Comparison
3. Results
3.1. Age Bias in TD and AUT Groups
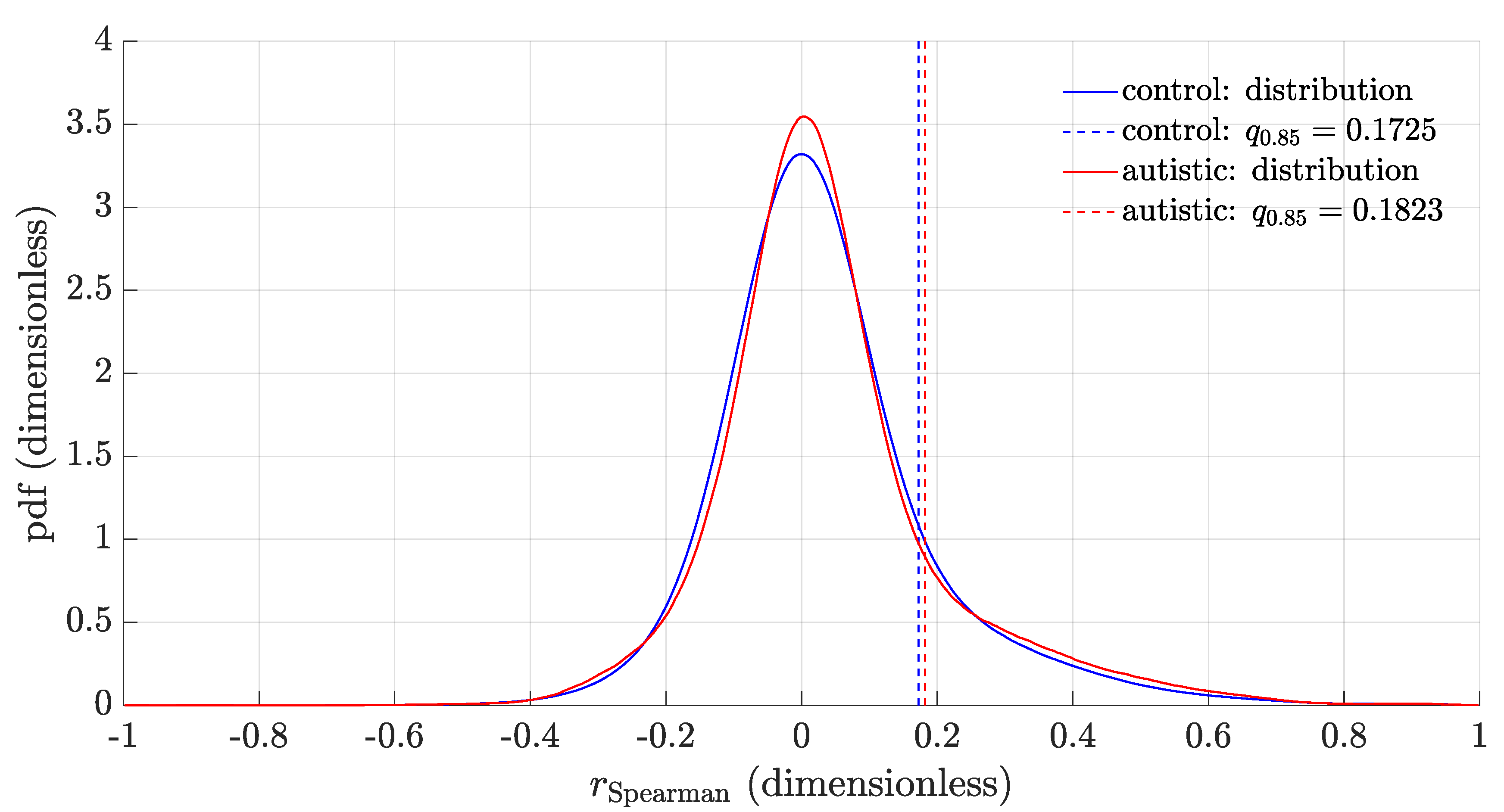
3.2. Distribution of the Feature Correlation Coefficients
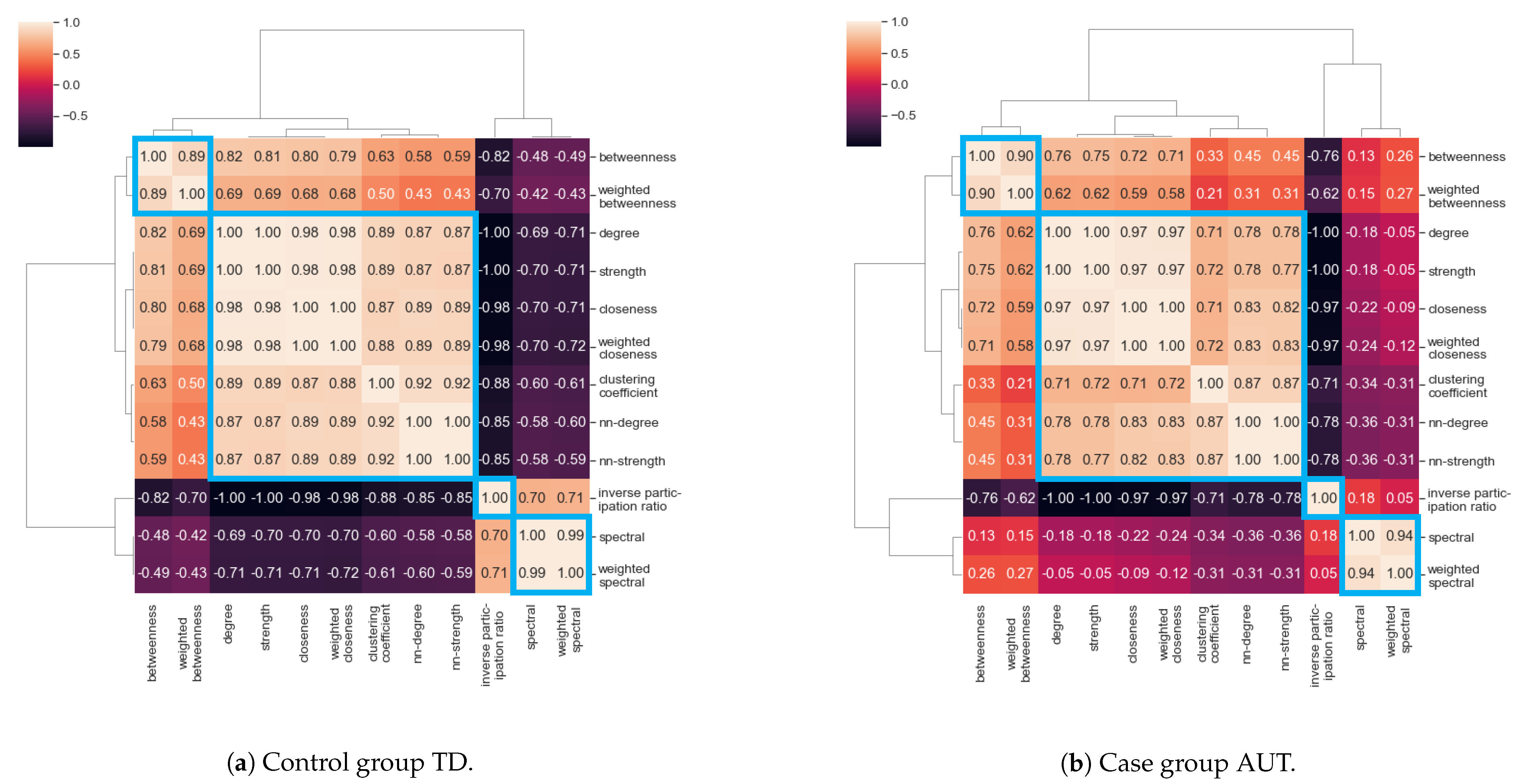
3.3. Correlation between Centrality Measures
- Cluster I: Betweenness centrality and weighted betweenness centrality;
- Cluster II: Degree, strength, nearest-neighbor degree, nearest-neighbor strength, closeness centrality, weighted closeness centrality, clustering coefficient;
- Cluster III: Inverse participation ratio;
- Cluster IV: Spectral centrality and weighted spectral centrality.
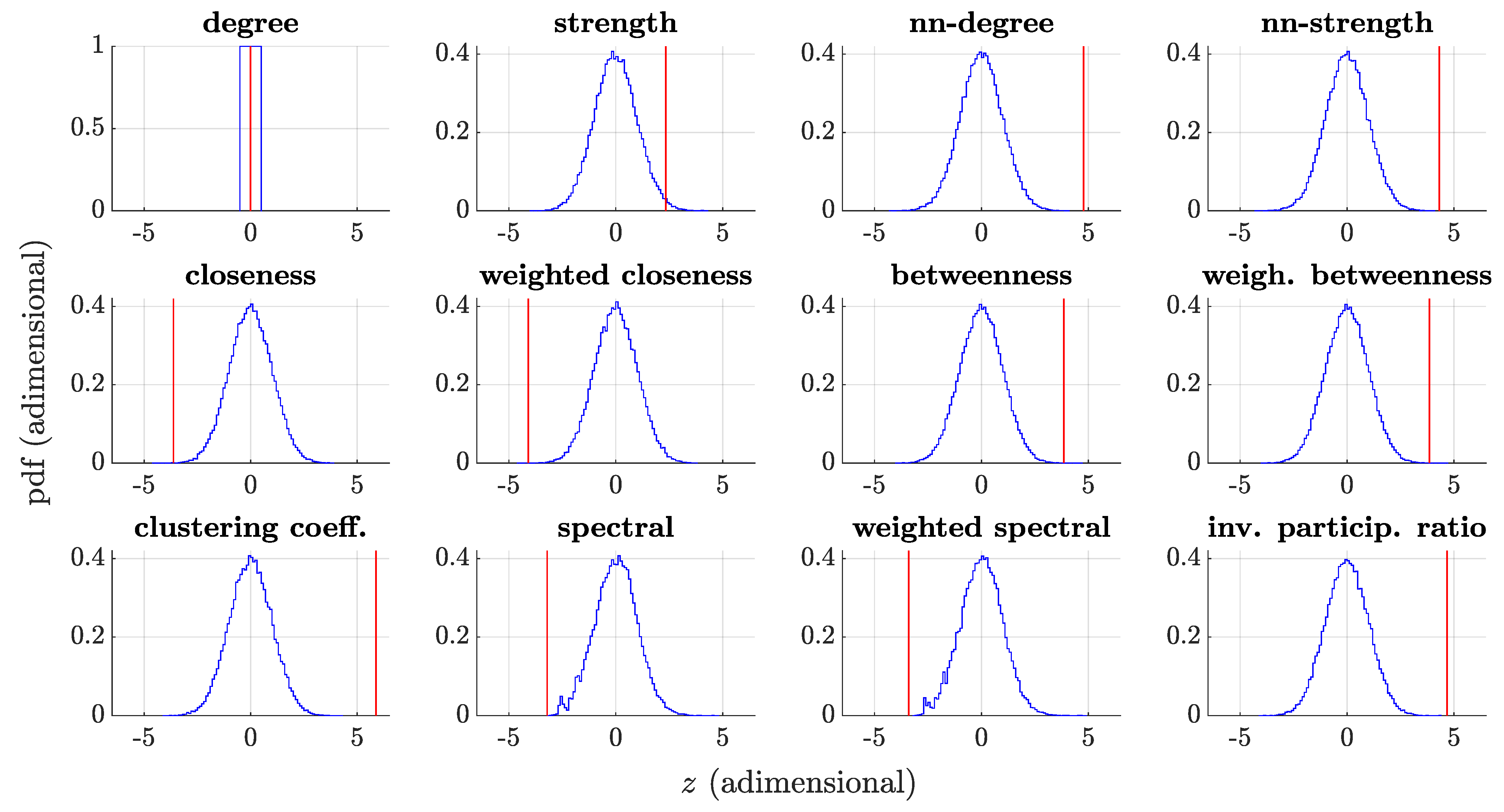
3.4. Whole-Network Comparison
3.5. Single-Node Comparison
3.6. Network Visualization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TD | Typically-developing subjects |
| AUT | Autistic disorder/autistic subjects |
| M | Number of subjects |
| N | Number of features/nodes (equivalent) |
| Spearman correlation coefficient | |
| G | Network (G standing for graph) |
| A | Adjacency matrix |
| W | Weights matrix |
| (topo) | Relative to the topological network |
| (weighted) | Relative to the weighted network |
| K | Degree |
| S | Strength |
| Average degree of nearest neighbors | |
| Average strength of nearest neighbors | |
| CC | Closeness centrality |
| WCC | Weighted closeness centrality |
| BC | Betweenness centrality |
| WBC | Weighted betweenness centrality |
| C, Clust | Clustering coefficient |
| SC | Spectral centrality |
| WSC | Weighted spectral centrality |
| IPR | Inverse participation ratio |
References
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (DSM-5®); American Psychiatric Publishing: Washington, DC, USA, 2013. [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders-IV Text Revision; American Psychiatric Publishing: Washington, DC, USA, 2000. [Google Scholar]
- Lord, C.; Bishop, S.L. Recent advances in autism research as reflected in DSM-5 criteria for autism spectrum disorder. Annu. Rev. Clin. Psychol. 2015, 11, 53–70. [Google Scholar] [CrossRef] [PubMed]
- Mottron, L.; Bzdok, D. Autism spectrum heterogeneity: Fact or artifact? Mol. Psychiatry 2020, 25, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Grove, J.; Ripke, S.; Als, T.D.; Mattheisen, M.; Walters, R.K.; Won, H.; Pallesen, J.; Agerbo, E.; Andreassen, O.A.; Anney, R.; et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 2019, 51, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Green, J. Editorial Perspective: Delivering autism intervention through development. J. Child Psychol. Psychiatry 2019, 60, 1353–1356. [Google Scholar] [CrossRef] [PubMed]
- Rødgaard, E.M.; Jensen, K.; Vergnes, J.N.; Soulières, I.; Mottron, L. Temporal changes in effect sizes of studies comparing individuals with and without autism: A meta-analysis. JAMA Psychiatry 2019, 76, 1124–1132. [Google Scholar] [CrossRef]
- McAlonan, G.M.; Suckling, J.; Wong, N.; Cheung, V.; Lienenkaemper, N.; Cheung, C.; Chua, S.E. Distinct patterns of grey matter abnormality in high-functioning autism and Asperger’s syndrome. J. Child Psychol. Psychiatry 2008, 49, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Toal, F.; Daly, E.; Page, L.; Deeley, Q.; Hallahan, B.; Bloemen, O.; Cutter, W.; Brammer, M.; Curran, S.; Robertson, D.; et al. Clinical and anatomical heterogeneity in autistic spectrum disorder: A structural MRI study. Psychol. Med. 2010, 40, 1171. [Google Scholar] [CrossRef]
- Kwon, H.; Ow, A.W.; Pedatella, K.E.; Lotspeich, L.J.; Reiss, A.L. Voxel-based morphometry elucidates structural neuroanatomy of high-functioning autism and Asperger syndrome. Dev. Med. Child Neurol. 2004, 46, 760–764. [Google Scholar] [CrossRef]
- Via, E.; Radua, J.; Cardoner, N.; Happé, F.; Mataix-Cols, D. Meta-analysis of gray matter abnormalities in autism spectrum disorder: Should Asperger disorder be subsumed under a broader umbrella of autistic spectrum disorder? Arch. Gen. Psychiatry 2011, 68, 409–418. [Google Scholar] [CrossRef]
- Qi, S.; Morris, R.; Turner, J.A.; Fu, Z.; Jiang, R.; Deramus, T.P.; Zhi, D.; Calhoun, V.D.; Sui, J. Common and unique multimodal covarying patterns in autism spectrum disorder subtypes. Mol. Autism 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Mazurek, M.O.; Lu, F.; Symecko, H.; Butter, E.; Bing, N.M.; Hundley, R.J.; Poulsen, M.; Kanne, S.M.; Macklin, E.A.; Handen, B.L. A prospective study of the concordance of DSM-IV and DSM-5 diagnostic criteria for autism spectrum disorder. J. Autism Dev. Disord. 2017, 47, 2783–2794. [Google Scholar] [CrossRef]
- Di Martino, A.; Yan, C.G.; Li, Q.; Denio, E.; Castellanos, F.X.; Alaerts, K.; Anderson, J.S.; Assaf, M.; Bookheimer, S.Y.; Dapretto, M.; et al. The autism brain imaging data exchange: Towards a large-scale evaluation of the intrinsic brain architecture in autism. Mol. Psychiatry 2014, 19, 659–667. [Google Scholar] [CrossRef] [PubMed]
- Dale, A.M.; Fischl, B.; Sereno, M.I. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage 1999, 9, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Fischl, B.; Sereno, M.I.; Dale, A.M. Cortical surface-based analysis: II. inflation, flattening, and a surface-based coordinate system. Neuroimage 1999, 9, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Fischl, B.; Salat, D.H.; Busa, E.; Albert, M.; Dieterich, M.; Haselgrove, C.; Van Der Kouwe, A.; Killiany, R.; Kennedy, D.; Klaveness, S.; et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 2002, 33, 341–355. [Google Scholar] [CrossRef]
- Fischl, B.; Van Der Kouwe, A.; Destrieux, C.; Halgren, E.; Ségonne, F.; Salat, D.H.; Busa, E.; Seidman, L.J.; Goldstein, J.; Kennedy, D.; et al. Automatically parcellating the human cerebral cortex. Cereb. Cortex 2004, 14, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Desikan, R.S.; Ségonne, F.; Fischl, B.; Quinn, B.T.; Dickerson, B.C.; Blacker, D.; Buckner, R.L.; Dale, A.M.; Maguire, R.P.; Hyman, B.T.; et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006, 31, 968–980. [Google Scholar] [CrossRef]
- Pauls, S.D.; Remondini, D. Measures of centrality based on the spectrum of the Laplacian. Phys. Rev. E 2012, 85, 066127. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Pagnozzi, A.M.; Conti, E.; Calderoni, S.; Fripp, J.; Rose, S.E. A systematic review of structural MRI biomarkers in autism spectrum disorder: A machine learning perspective. Int. J. Dev. Neurosci. 2018, 71, 68–82. [Google Scholar] [CrossRef]
- Schmaal, L.; Ching, C.R.; McMahon, A.B.; Jahanshad, N.; Thompson, P.M. Neuroimaging, genetics, and personalized psychiatry: Developments and opportunities from the ENIGMA consortium. In Personalized Psychiatry; Academic Press: Cambridge, MA, USA, 2020; pp. 483–497. [Google Scholar]
- Button, K.S.; Ioannidis, J.P.; Mokrysz, C.; Nosek, B.A.; Flint, J.; Robinson, E.S.; Munafò, M.R. Power failure: Why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 2013, 14, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Iacoboni, M.; Dapretto, M. The mirror neuron system and the consequences of its dysfunction. Nat. Rev. Neurosci. 2006, 7, 942–951. [Google Scholar] [CrossRef] [PubMed]
- Beer, J.S.; John, O.P.; Scabini, D.; Knight, R.T. Orbitofrontal cortex and social behavior: Integrating self-monitoring and emotion-cognition interactions. J. Cogn. Neurosci. 2006, 18, 871–879. [Google Scholar] [CrossRef]
- Hardan, A.Y.; Girgis, R.R.; Lacerda, A.L.; Yorbik, O.; Kilpatrick, M.; Keshavan, M.S.; Minshew, N.J. Magnetic resonance imaging study of the orbitofrontal cortex in autism. J. Child Neurol. 2006, 21, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Hill, E.L. Executive dysfunction in autism. Trends Cogn. Sci. 2004, 8, 26–32. [Google Scholar] [CrossRef]
- Cai, R.Y.; Richdale, A.L.; Uljarević, M.; Dissanayake, C.; Samson, A.C. Emotion regulation in autism spectrum disorder: Where we are and where we need to go. Autism Res. 2018, 11, 962–978. [Google Scholar] [CrossRef] [PubMed]
- Dubbelink, L.M.O.; Geurts, H.M. Planning skills in autism spectrum disorder across the lifespan: A meta-analysis and meta-regression. J. Autism Dev. Disord. 2017, 47, 1148–1165. [Google Scholar] [CrossRef]
- Leung, R.C.; Zakzanis, K.K. Brief report: Cognitive flexibility in autism spectrum disorders: A quantitative review. J. Autism Dev. Disord. 2014, 44, 2628–2645. [Google Scholar] [CrossRef] [PubMed]
- Bishop, D.V.; Norbury, C.F. Executive functions in children with communication impairments, in relation to autistic symptomatology: I: Generativity. Autism 2005, 9, 7–27. [Google Scholar] [CrossRef] [PubMed]
- Barendse, E.M.; Hendriks, M.P.; Jansen, J.F.; Backes, W.H.; Hofman, P.A.; Thoonen, G.; Kessels, R.P.; Aldenkamp, A.P. Working memory deficits in high-functioning adolescents with autism spectrum disorders: Neuropsychological and neuroimaging correlates. J. Neurodev. Disord. 2013, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Solomon, M.; Ozonoff, S.J.; Ursu, S.; Ravizza, S.; Cummings, N.; Ly, S.; Carter, C.S. The neural substrates of cognitive control deficits in autism spectrum disorders. Neuropsychologia 2009, 47, 2515–2526. [Google Scholar] [CrossRef]
- Amodio, D.M.; Frith, C.D. Meeting of minds: The medial frontal cortex and social cognition. Nat. Rev. Neurosci. 2006, 7, 268–277. [Google Scholar] [CrossRef]
- Xu, J.; Wang, C.; Xu, Z.; Li, T.; Chen, F.; Chen, K.; Gao, J.; Wang, J.; Hu, Q. Specific functional connectivity patterns of middle temporal gyrus subregions in children and adults with autism spectrum disorder. Autism Res. 2020, 13, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Narzisi, A.; Muratori, F.; Calderoni, S.; Fabbro, F.; Urgesi, C. Neuropsychological profile in high functioning autism spectrum disorders. J. Autism Dev. Disord. 2013, 43, 1895–1909. [Google Scholar] [CrossRef]
- Southwick, J.S.; Bigler, E.D.; Froehlich, A.; DuBray, M.B.; Alexander, A.L.; Lange, N.; Lainhart, J.E. Memory functioning in children and adolescents with autism. Neuropsychology 2011, 25, 702. [Google Scholar] [CrossRef]
- Van Rooij, D.; Anagnostou, E.; Arango, C.; Auzias, G.; Behrmann, M.; Busatto, G.F.; Calderoni, S.; Daly, E.; Deruelle, C.; Di Martino, A.; et al. Cortical and subcortical brain morphometry differences between patients with autism spectrum disorder and healthy individuals across the lifespan: Results from the ENIGMA ASD Working Group. Am. J. Psychiatry 2018, 175, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Rizzolatti, G.; Fabbri-Destro, M. Mirror neurons: From discovery to autism. Exp. Brain Res. 2010, 200, 223–237. [Google Scholar] [CrossRef]
- Carper, R.A.; Moses, P.; Tigue, Z.D.; Courchesne, E. Cerebral lobes in autism: Early hyperplasia and abnormal age effects. Neuroimage 2002, 16, 1038–1051. [Google Scholar] [CrossRef]
- Jou, R.J.; Minshew, N.J.; Keshavan, M.S.; Hardan, A.Y. Cortical gyrification in autistic and Asperger disorders: A preliminary magnetic resonance imaging study. J. Child Neurol. 2010, 25, 1462–1467. [Google Scholar] [CrossRef] [PubMed]
- Travers, B.G.; Kana, R.K.; Klinger, L.G.; Klein, C.L.; Klinger, M.R. Motor learning in individuals with autism spectrum disorder: Activation in superior parietal lobule related to learning and repetitive behaviors. Autism Res. 2015, 8, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Freitag, C.M.; Konrad, C.; Häberlen, M.; Kleser, C.; von Gontard, A.; Reith, W.; Troje, N.F.; Krick, C. Perception of biological motion in autism spectrum disorders. Neuropsychologia 2008, 46, 1480–1494. [Google Scholar] [CrossRef]
- Cavanna, A.E.; Trimble, M.R. The precuneus: A review of its functional anatomy and behavioural correlates. Brain 2006, 129, 564–583. [Google Scholar] [CrossRef] [PubMed]
- Schilbach, L.; Wohlschlaeger, A.M.; Kraemer, N.C.; Newen, A.; Shah, N.J.; Fink, G.R.; Vogeley, K. Being with virtual others: Neural correlates of social interaction. Neuropsychologia 2006, 44, 718–730. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Rolls, E.T.; Gu, H.; Zhang, J.; Feng, J. Autism: Reduced connectivity between cortical areas involved in face expression, theory of mind, and the sense of self. Brain 2015, 138, 1382–1393. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, M.V.; Chakrabarti, B.; Bullmore, E.T.; Sadek, S.A.; Pasco, G.; Wheelwright, S.J.; Suckling, J.; Consortium, M.A.; Baron-Cohen, S. Atypical neural self-representation in autism. Brain 2010, 133, 611–624. [Google Scholar] [CrossRef]
- Di Martino, A.; Ross, K.; Uddin, L.Q.; Sklar, A.B.; Castellanos, F.X.; Milham, M.P. Functional brain correlates of social and nonsocial processes in autism spectrum disorders: An activation likelihood estimation meta-analysis. Biol. Psychiatry 2009, 65, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Dickstein, D.P.; Pescosolido, M.F.; Reidy, B.L.; Galvan, T.; Kim, K.L.; Seymour, K.E.; Laird, A.R.; Di Martino, A.; Barrett, R.P. Developmental meta-analysis of the functional neural correlates of autism spectrum disorders. J. Am. Acad. Child Adolesc. Psychiatry 2013, 52, 279–289. [Google Scholar] [CrossRef]
- Di Martino, A.; O’connor, D.; Chen, B.; Alaerts, K.; Anderson, J.S.; Assaf, M.; Balsters, J.H.; Baxter, L.; Beggiato, A.; Bernaerts, S.; et al. Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Sci. Data 2017, 4, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Uddin, L.Q.; Menon, V. The anterior insula in autism: Under-connected and under-examined. Neurosci. Biobehav. Rev. 2009, 33, 1198–1203. [Google Scholar] [CrossRef] [PubMed]
- Schaaf, R.C.; Toth-Cohen, S.; Johnson, S.L.; Outten, G.; Benevides, T.W. The everyday routines of families of children with autism: Examining the impact of sensory processing difficulties on the family. Autism 2011, 15, 373–389. [Google Scholar] [CrossRef]
- Hyde, K.L.; Samson, F.; Evans, A.C.; Mottron, L. Neuroanatomical differences in brain areas implicated in perceptual and other core features of autism revealed by cortical thickness analysis and voxel-based morphometry. Hum. Brain Mapp. 2010, 31, 556–566. [Google Scholar] [CrossRef] [PubMed]
- Scheel, C.; Rotarska-Jagiela, A.; Schilbach, L.; Lehnhardt, F.G.; Krug, B.; Vogeley, K.; Tepest, R. Imaging derived cortical thickness reduction in high-functioning autism: Key regions and temporal slope. Neuroimage 2011, 58, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Grill-Spector, K.; Kourtzi, Z.; Kanwisher, N. The lateral occipital complex and its role in object recognition. Vis. Res. 2001, 41, 1409–1422. [Google Scholar] [CrossRef]
- Weigelt, S.; Koldewyn, K.; Kanwisher, N. Face recognition deficits in autism spectrum disorders are both domain specific and process specific. PLoS ONE 2013, 8, e74541. [Google Scholar] [CrossRef] [PubMed]
- Van der Hallen, R.; Evers, K.; Brewaeys, K.; Van den Noortgate, W.; Wagemans, J. Global processing takes time: A meta-analysis on local-global visual processing in ASD. Psychol. Bull. 2015, 141, 549. [Google Scholar] [CrossRef] [PubMed]

| p | all TD | all AUT | Right-Handed Male TD | Right-Handed Male AUT |
|---|---|---|---|---|
| all TD | 1 | 0.699 | 0.698 | 0.553 |
| all AUT | 1 | 0.408 | 0.993 | |
| right-handed male TD | 1 | 0.784 | ||
| right-handed male AUT | 1 |
| Cluster | Centrality Measure | z-Score | p-Value |
|---|---|---|---|
| I | Betweenness | 3.85 | |
| Weighted betweenness | 3.84 | ||
| II | Clustering coefficient | 5.88 | < |
| nn-degree | 4.77 | < | |
| nn-strength | 4.31 | < | |
| Weighted closeness | −4.09 | ||
| Closeness | −3.62 | 0.00014 | |
| Strength | 2.33 | 0.011 | |
| Degree | 0 by constr. | 1 by constr. | |
| III | Inverse participation ratio | 4.69 | < |
| IV | Weighted spectral | −3.40 | 0.0019 |
| Spectral | −3.21 | 0.0019 |
| Rank | Hemisphere | Brain Area | Measure | zBC |
|---|---|---|---|---|
| 1 | R | Postcentral | Gray volume | 2.808 |
| 2 | R | Medial orbitofrontal | Gray volume | 2.584 |
| 3 | R | Superior frontal | Average thickness | 2.310 |
| 4 | R | Rostral middle frontal | Average thickness | 2.214 |
| 5 | L | Rostral middle frontal | Gray volume | 2.207 |
| Rank | Hemisphere | Brain Area | Measure | zClust |
|---|---|---|---|---|
| 1 | R | Pars triangularis | Surface area | 6.705 |
| 2 | R | Caudal middle frontal | Surface area | 6.046 |
| 3 | R | Rostral middle frontal | Surface area | 5.893 |
| 4 | R | Caudal middle frontal | Gray volume | 5.686 |
| 5 | R | Postcentral | Mean curvature | 5.097 |
| Rank | Hemisphere | Brain Area | Measure | zIPR |
|---|---|---|---|---|
| 1 | R | Middle temporal | Surface area | 4.087 |
| 2 | R | Entorhinal | Standard thickness | 3.643 |
| 3 | L | Lateral occipital | Gray volume | 2.772 |
| 4 | L | Insula | Surface area | 2.735 |
| 5 | R | Insula | Surface area | 2.734 |
| Rank | Hemisphere | Brain Area | Measure | zWSC |
|---|---|---|---|---|
| 1 | L | Precentral | Mean curvature | −1.836 |
| 2 | L | Precuneus | Standard thickness | −1.708 |
| 3 | L | Postcentral | Curvature index | −1.705 |
| 4 | L | Superior parietal | Curvature index | −1.679 |
| 5 | L | Superior frontal | Standard thickness | −1.626 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zanghieri, M.; Menichetti, G.; Retico, A.; Calderoni, S.; Castellani, G.; Remondini, D. Node Centrality Measures Identify Relevant Structural MRI Features of Subjects with Autism. Brain Sci. 2021, 11, 498. https://doi.org/10.3390/brainsci11040498
Zanghieri M, Menichetti G, Retico A, Calderoni S, Castellani G, Remondini D. Node Centrality Measures Identify Relevant Structural MRI Features of Subjects with Autism. Brain Sciences. 2021; 11(4):498. https://doi.org/10.3390/brainsci11040498
Chicago/Turabian StyleZanghieri, Marcello, Giulia Menichetti, Alessandra Retico, Sara Calderoni, Gastone Castellani, and Daniel Remondini. 2021. "Node Centrality Measures Identify Relevant Structural MRI Features of Subjects with Autism" Brain Sciences 11, no. 4: 498. https://doi.org/10.3390/brainsci11040498
APA StyleZanghieri, M., Menichetti, G., Retico, A., Calderoni, S., Castellani, G., & Remondini, D. (2021). Node Centrality Measures Identify Relevant Structural MRI Features of Subjects with Autism. Brain Sciences, 11(4), 498. https://doi.org/10.3390/brainsci11040498