Time Course Transcriptome Analysis of Spina Bifida Progression in Fetal Rats
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Congenital Retinoic Acid (RA) Induced Spina Bifida Animal Model
2.3. Tissue Processing and RNA Extraction
2.4. cDNA Library Preparation and Sequencing
2.5. RNA-Seq Data Processing and Analysis
2.6. RT-qPCR Analysis
2.7. Statistical Analysis
3. Results
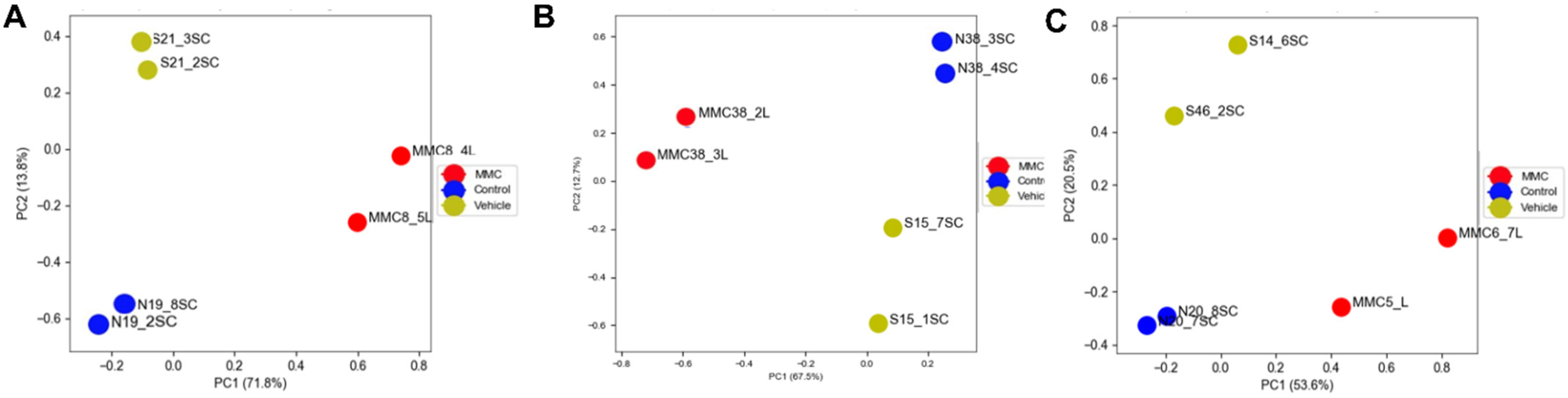
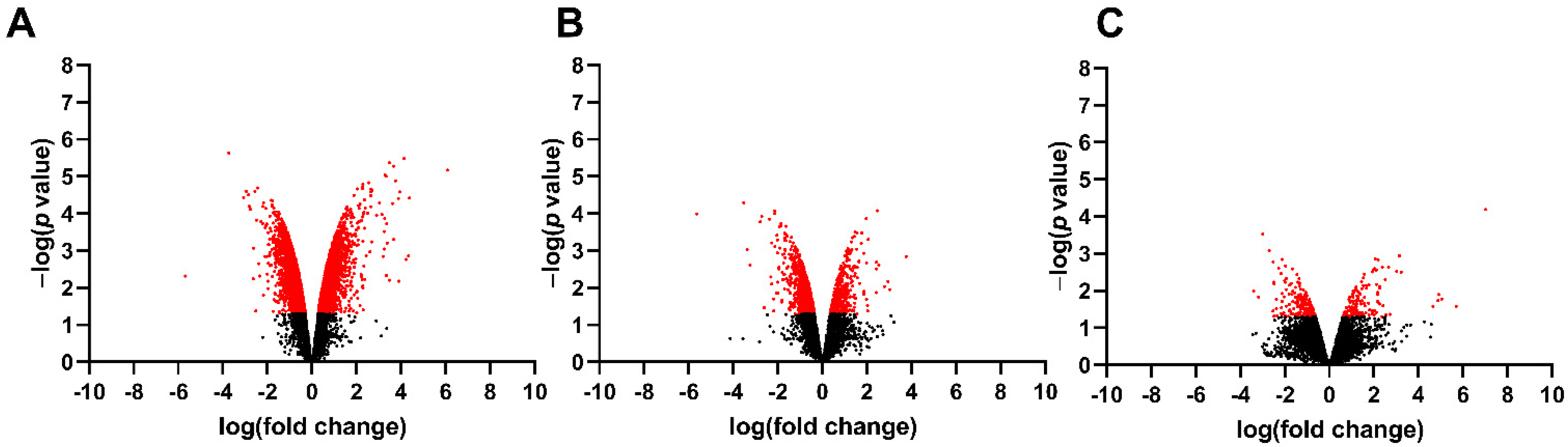
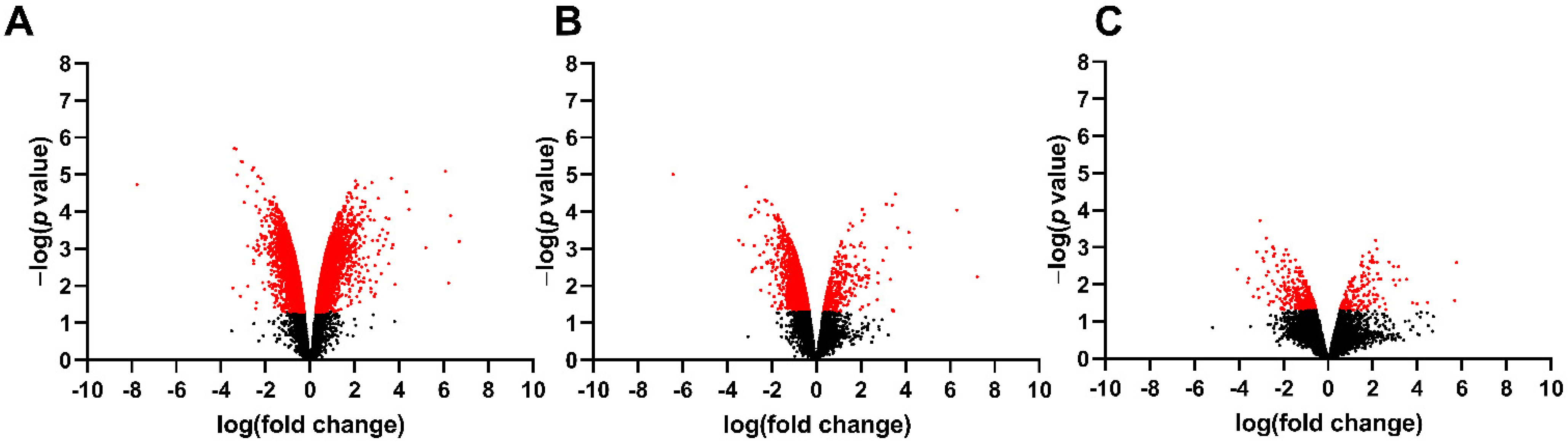
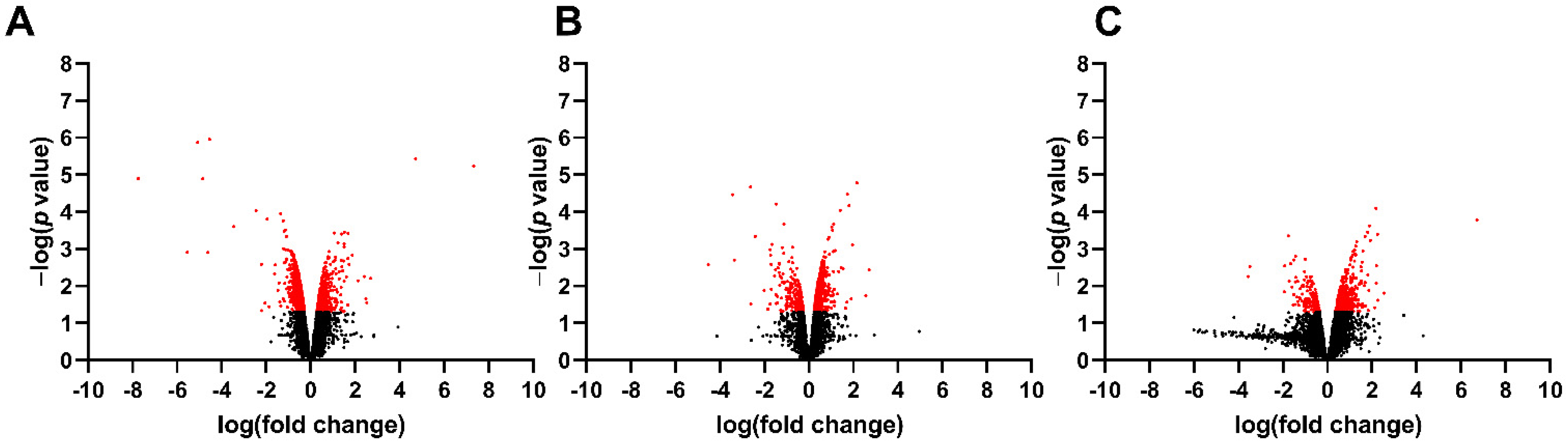
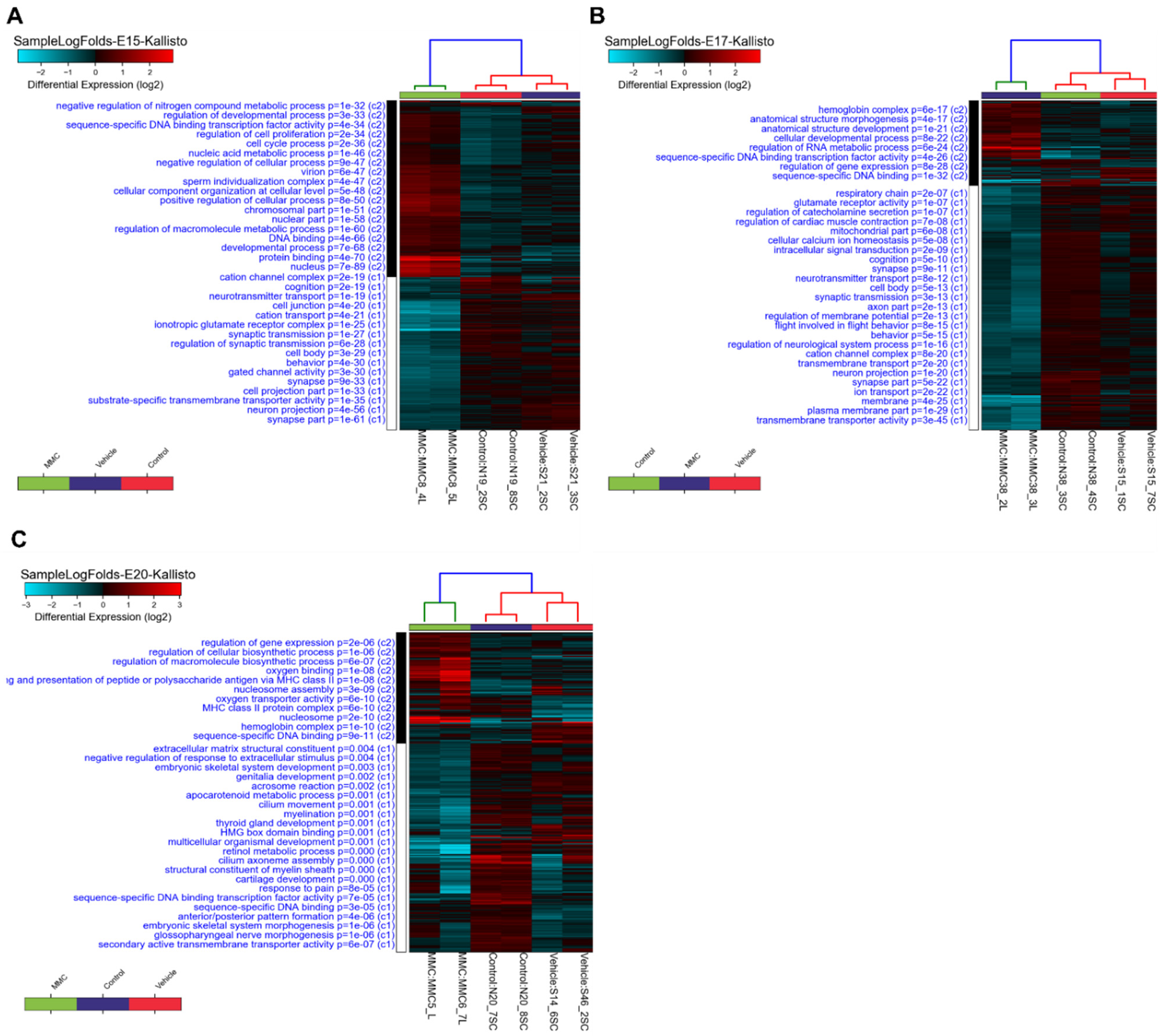
3.1. Differentially Expressed Genes in the Fetal Spinal Cord after MMC Occurs
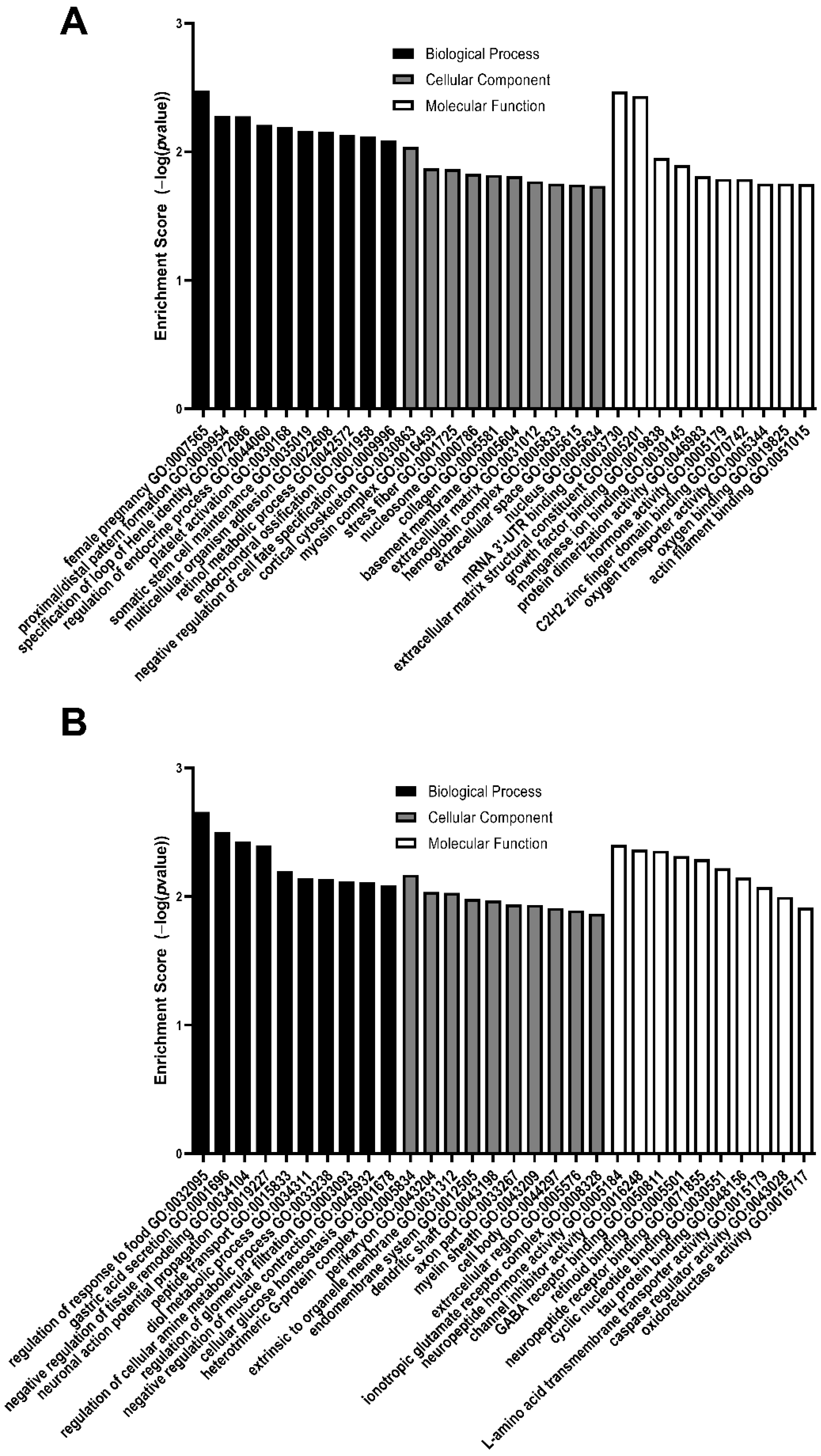
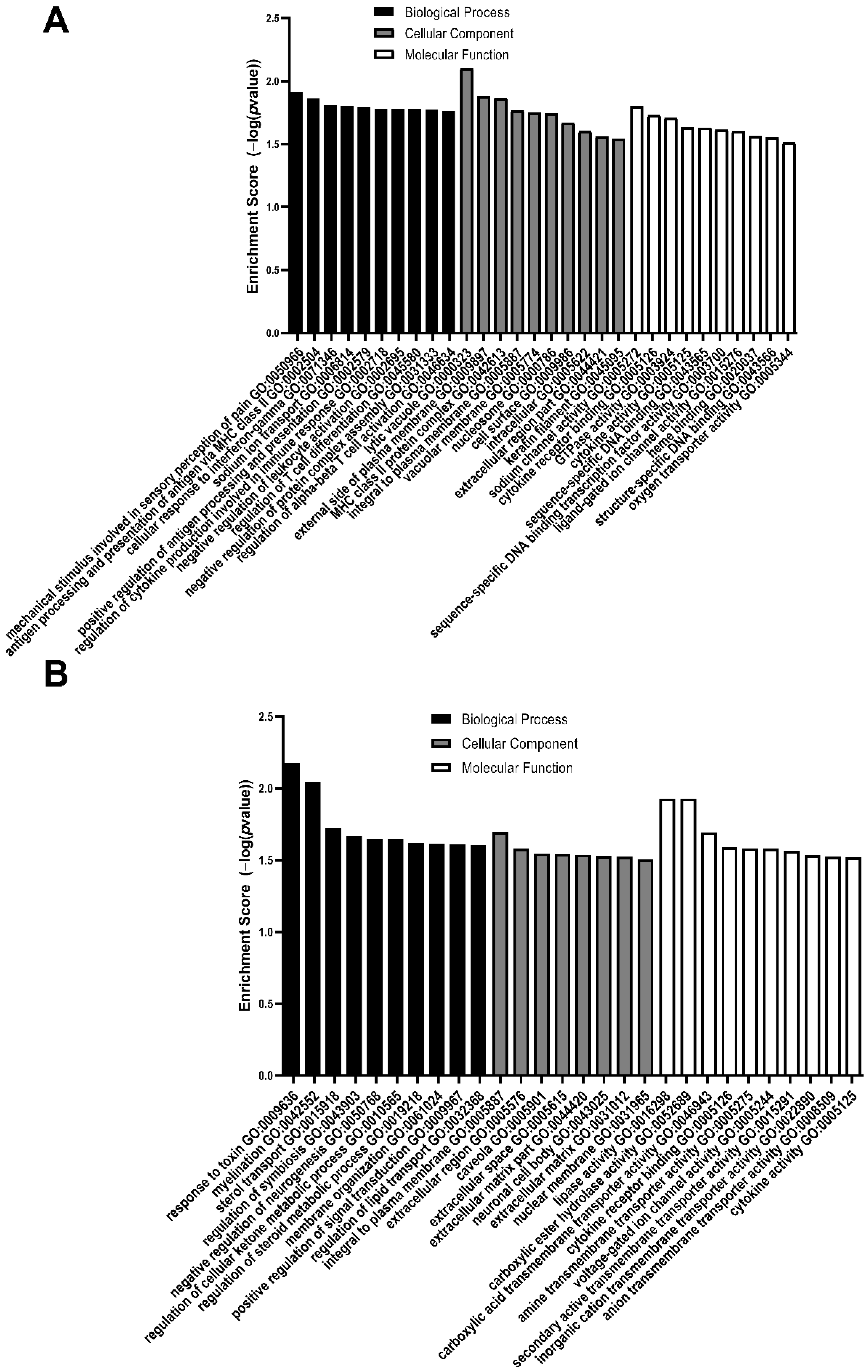
3.2. GO Analysis of Differentially Expressed Genes
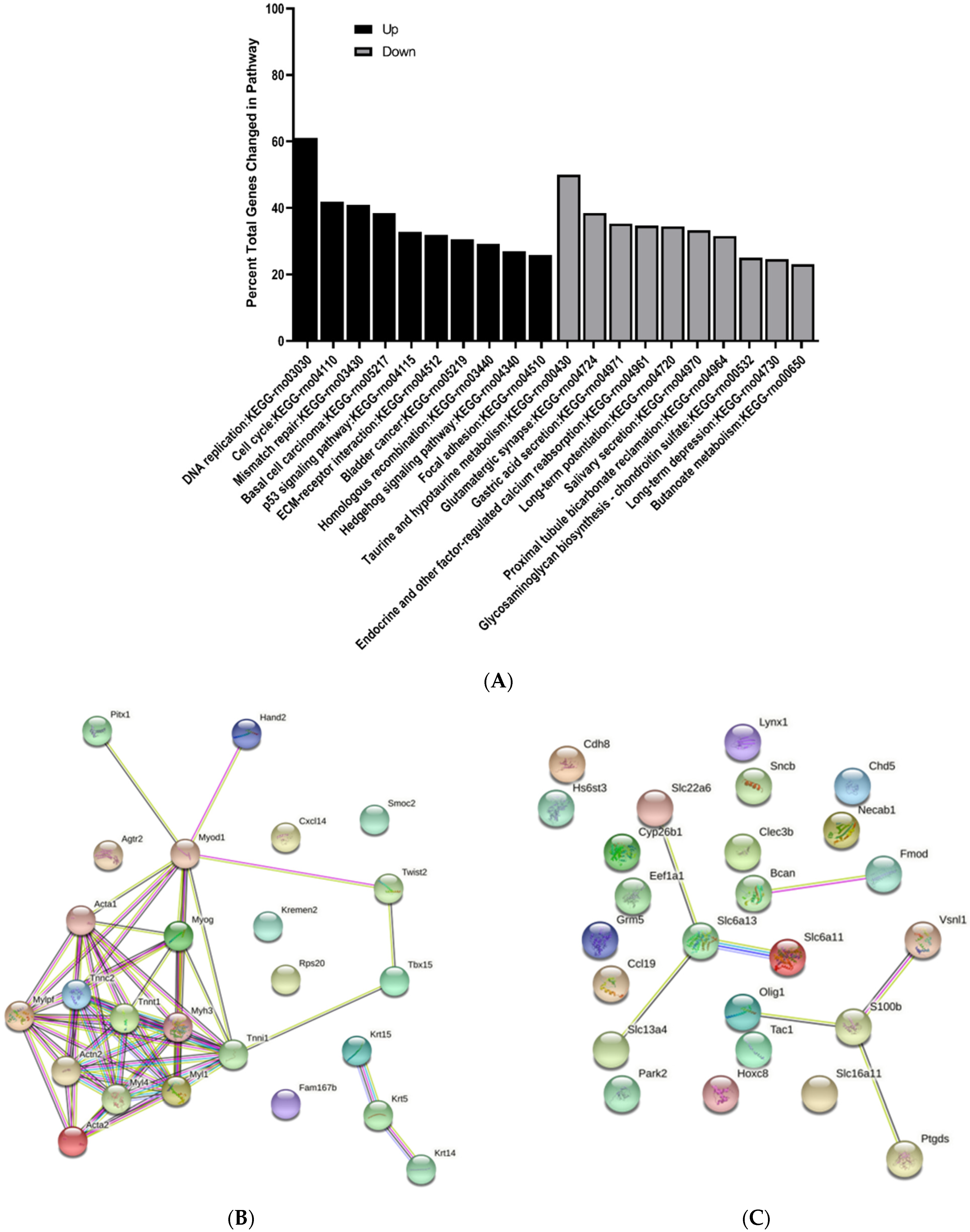
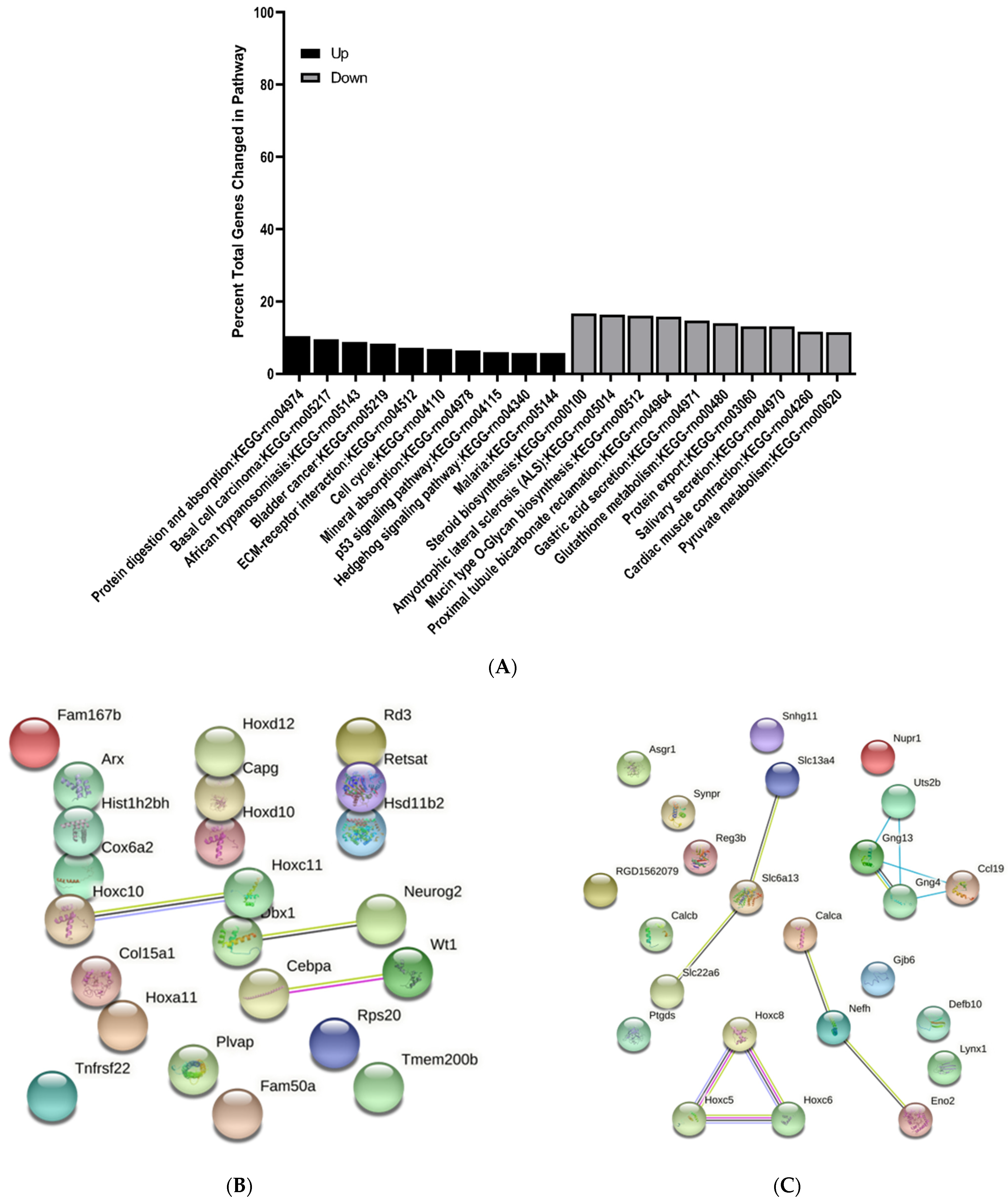
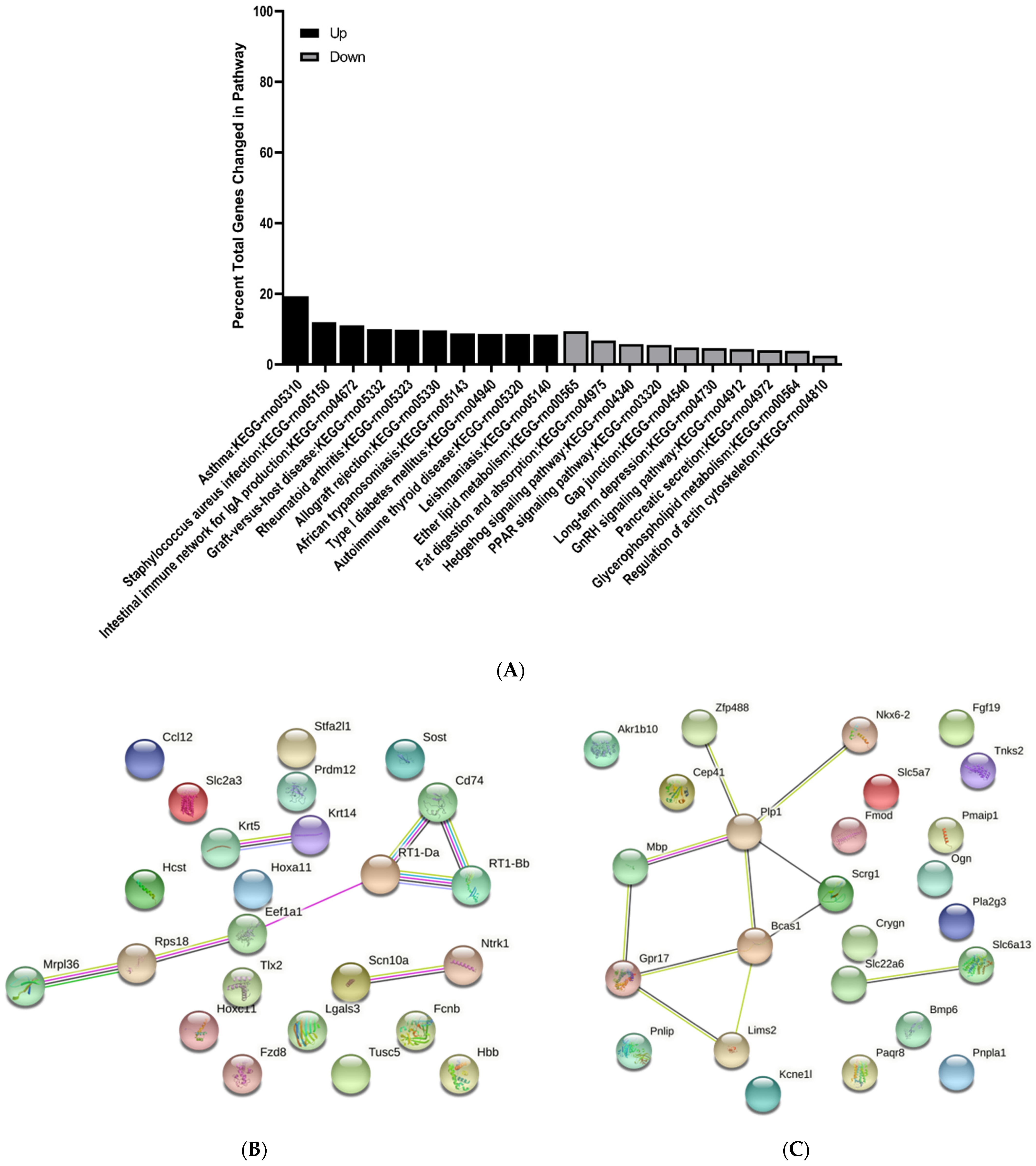
3.3. KEGG Pathway Enrichment Analysis of Differentially Expressed Genes
3.4. Protein–Protein Interaction Network Analysis of Differentially Expressed Genes
3.5. Validation by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Copp, A.J.; Adzick, N.S.; Chitty, L.S.; Fletcher, J.M.; Holmbeck, G.N.; Shaw, G.M. Spina bifida. Nat. Rev. Dis. Prim. 2015, 1, 15007. [Google Scholar] [CrossRef] [PubMed]
- Copp, A.J.; Greene, N.D.E. Neural tube defects—Disorders of neurulation and related embryonic processes. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavalli, P. Prevention of Neural Tube Defects and proper folate periconceptional supplementation. J. Prenat. Med. 2008, 2, 40. [Google Scholar] [PubMed]
- Alruwaili, A.A.; Das, J.M. Myelomeningocele. Encycl. Neurol. Sci. 2021, 246–249. [Google Scholar]
- Moldenhauer, J.S.; Adzick, N.S. Fetal surgery for myelomeningocele: After the Management of Myelomeningocele Study (MOMS). Semin. Fetal Neonatal Med. 2017, 22, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Maroto, A.; Illescas, T.; Meléndez, M.; Arévalo, S.; Rodó, C.; Peiró, J.L.; Belfort, M.; Cuxart, A.; Carreras, E. Ultrasound functional evaluation of fetuses with myelomeningocele: Study of the interpretation of results. J. Matern.-Fetal Neonatal Med. 2017, 30, 2301–2305. [Google Scholar] [CrossRef] [PubMed]
- Carreras, E.; Maroto, A.; Illescas, T.; Meléndez, M.; Arévalo, S.; Peiró, J.L.; García-Fontecha, C.G.; Belfort, M.; Cuxart, A. Prenatal ultrasound evaluation of segmental level of neurological lesion in fetuses with myelomeningocele: Development of a new technique. Ultrasound Obstet. Gynecol. 2016, 47, 162–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, C.M.; Mukherjee, S.; Wilburn, A.N.; Cates, C.; Lewkowich, I.P.; Deshmukh, H.; Zacharias, W.J.; Chougnet, C.A. Pulmonary Consequences of Prenatal Inflammatory Exposures: Clinical Perspective and Review of Basic Immunological Mechanisms. Front. Immunol. 2020, 11, 1285. [Google Scholar] [CrossRef] [PubMed]
- Reemst, K.; Noctor, S.C.; Lucassen, P.J.; Hol, E.M. The Indispensable Roles of Microglia and Astrocytes during Brain Development. Front. Hum. Neurosci. 2016, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayer, S.A.; Altman, J.; Russo, R.J.; Zhang, X. Timetables of Neurogenesis in the Human Brain Based on Experimentally Determ’ined Patterns in the Rat Deriving Principles of Central Nervous System Development from Experimental Studies in Rats. NeuroToxicology 1993, 14, 83–144. [Google Scholar] [PubMed]
- Oria, M.; Figueira, R.L.; Scorletti, F.; Sbragia, L.; Owens, K.; Li, Z.; Pathak, B.; Corona, M.U.; Marotta, M.; Encinas, J.L.; et al. CD200-CD200R imbalance correlates with microglia and pro-inflammatory activation in rat spinal cords exposed to amniotic fluid in retinoic acid-induced spina bifida. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Copp, A.J.; Stanier, P.; Greene, N.D.E. Neural tube defects—Recent advances, unsolved questions and controversies. Lancet Neurol. 2013, 12, 799. [Google Scholar] [CrossRef] [Green Version]
- Olsson, A.; Venkatasubramanian, M.; Chaudhri, V.K.; Aronow, B.J.; Salomonis, N.; Singh, H.; Grimes, H.L. Single-cell analysis of mixed-lineage states leading to a binary cell fate choice. Nature 2016, 537, 698–702. [Google Scholar] [CrossRef]
- Salomonis, N.; Schlieve, C.R.; Pereira, L.; Wahlquist, C.; Colas, A.; Zambon, A.C.; Vranizan, K.; Spindler, M.J.; Pico, A.R.; Cline, M.S.; et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc. Natl. Acad. Sci. USA 2010, 107, 10514–10519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emig, D.; Salomonis, N.; Baumbach, J.; Lengauer, T.; Conklin, B.R.; Albrecht, M. AltAnalyze and DomainGraph: Analyzing and visualizing exon expression data. Nucleic Acids Res. 2010, 38, W755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danzer, E.; Schwarz, U.; Wehrli, S.; Radu, A.; Adzick, N.S.; Flake, A.W. Retinoic acid induced myelomeningocele in fetal rats: Characterization by histopathological analysis and magnetic resonance imaging. Exp. Neurol. 2005, 194, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Pressler, R.; Auvin, S. Comparison of brain maturation among species: An example in translational research suggesting the possible use of bumetanide in newborn. Front. Neurol. 2013, 4, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Wei, X.; Fan, Y.; Miao, J.; Wang, L.; Zhang, Y.; Wu, D.; Yuan, Z. Altered Expression of 14-3-3ζ Protein in Spinal Cords of Rat Fetuses with Spina Bifida Aperta. PLoS ONE 2013, 8, e70457. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, W.B.; Kaplan, D.R.; Miller, F.D. The p53 family in nervous system development and disease. J. Neurochem. 2006, 97, 1571–1584. [Google Scholar] [CrossRef]
- Oppenheim, R.W.; Homma, S.; Marti, E.; Prevette, D.; Wang, S.; Yaginuma, H.; McMahon, A.P. Modulation of Early but Not Later Stages of Programmed Cell Death in Embryonic Avian Spinal Cord by Sonic Hedgehog. Mol. Cell. Neurosci. 1999, 13, 348–361. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, S.; Li, X.; Li, H.; Li, Y.; Zhang, C.; Lin, J. Effect of sonic hedgehog on motor neuron positioning in the spinal cord during chicken embryonic development. J. Cell. Mol. Med. 2019, 23, 3549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.-L.; Lin, Y.-W.; Nhieu, J.; Zhang, X.; Wei, L.-N. Sonic Hedgehog-Gli1 Signaling and Cellular Retinoic Acid Binding Protein 1 Gene Regulation in Motor Neuron Differentiation and Diseases. Int. J. Mol. Sci. 2020, 21, 4125. [Google Scholar] [CrossRef]
- Li, H.; Zhang, J.; Chen, S.; Wang, F.; Zhang, T.; Niswander, L. Genetic contribution of retinoid related genes to neural tube defects. Hum. Mutat. 2018, 39, 550. [Google Scholar] [CrossRef] [PubMed]
- Sequerra, E.B.; Goyal, R.; Castro, P.A.; Levin, J.B.; Borodinsky, L.N. NMDA Receptor Signaling Is Important for Neural Tube Formation and for Preventing Antiepileptic Drug-Induced Neural Tube Defects. J. Neurosci. 2018, 38, 4762–4773. [Google Scholar] [CrossRef]
- Fu, Y.; Wu, X.; Lu, J.; Huang, J. Presynaptic GABAB Receptor Regulates Activity-Dependent Maturation and Patterning of Inhibitory Synapses through Dynamic Allocation of Synaptic Vesicles. Front. Cell. Neurosci. 2012, 6, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finckenstein, F.G.; Davicioni, E.; Osborn, K.G.; Cavenee, W.K.; Arden, K.C.; Anderson, M.J. Transgenic mice expressing PAX3-FKHR have multiple defects in muscle development, including ectopic skeletal myogenesis in the developing neural tube. Transgenic Res. 2006, 15, 595–614. [Google Scholar] [CrossRef]
- Rolo, A.; Escuin, S.; Greene, N.D.E.; Copp, A.J. Rho GTPases in mammalian spinal neural tube closure. Small GTPases 2018, 9, 283. [Google Scholar] [CrossRef] [Green Version]
- Escuin, S.; Vernay, B.; Savery, D.; Gurniak, C.B.; Witke, W.; Greene, N.D.E.; Copp, A.J. Rho-kinase-dependent actin turnover and actomyosin disassembly are necessary for mouse spinal neural tube closure. J. Cell Sci. 2015, 128, 2468. [Google Scholar] [CrossRef] [Green Version]
- Dringen, R.; Pfeiffer, B.; Hamprecht, B. Synthesis of the Antioxidant Glutathione in Neurons: Supply by Astrocytes of CysGly as Precursor for Neuronal Glutathione. J. Neurosci. 1999, 19, 562–569. [Google Scholar] [CrossRef] [PubMed]
- Sedlak, T.W.; Paul, B.D.; Parker, G.M.; Hester, L.D.; Snowman, A.M.; Taniguchi, Y.; Kamiya, A.; Snyder, S.H.; Sawa, A. The glutathione cycle shapes synaptic glutamate activity. Proc. Natl. Acad. Sci. USA 2019, 116, 2701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rouleau, C.; Rakotoarivelo, C.; Petite, D.; Lambert, K.; Fabre, C.; Bonardet, A.; Mercier, J.; Baldet, P.; Privat, A.; Langley, K.; et al. Pyruvate modifies glycolytic and oxidative metabolism of rat embryonic spinal cord astrocyte cell lines and prevents their spontaneous transformation. J. Neurochem. 2007, 100, 1589–1598. [Google Scholar] [CrossRef]
- Díaz-García, C.M.; Yellen, G. Neurons rely on glucose rather than astrocytic lactate during stimulation. J. Neurosci. Res. 2019, 97, 883. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Wang, L.; Yi, D.; Jin, L.; Liu, J.; Zhang, Y.; Ren, A. Association between maternal single nucleotide polymorphisms in genes regulating glucose metabolism and risk for neural tube defects in offspring. Birth Defects Res. Part A Clin. Mol. Teratol. 2015, 103, 471–478. [Google Scholar] [CrossRef]
- Loeken, M.R. Current perspectives on the causes of neural tube defects resulting from diabetic pregnancy. Am. J. Med. Genet. Part C Semin. Med. Genet. 2005, 135C, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, S.; Gritti, L.; Crooks, D.; Dombrowski, Y. Oligodendrocytes in Development, Myelin Generation and Beyond. Cells 2019, 8, 1424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.-C.; Almazan, G. Role of Sonic Hedgehog Signaling in Oligodendrocyte Differentiation. Neurochem. Res. 2016 4112 2016, 41, 3289–3299. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Wang, F.; Yang, X.; Wang, H.; Niswander, L.; Zhang, T.; Li, H. Association between rare variants in specific functional pathways and human neural tube defects multiple subphenotypes. Neural Dev. 2020, 15, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Chaves-Filho, A.B.; Pinto, I.F.D.; Dantas, L.S.; Xavier, A.M.; Inague, A.; Faria, R.L.; Medeiros, M.H.G.; Glezer, I.; Yoshinaga, M.Y.; Miyamoto, S. Alterations in lipid metabolism of spinal cord linked to amyotrophic lateral sclerosis. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Scholl, C.; Weißmüller, K.; Holenya, P.; Shaked-Rabi, M.; Tucker, K.L.; Wölfl, S. Distinct and overlapping gene regulatory networks in BMP- and HDAC-controlled cell fate determination in the embryonic forebrain. BMC Genom. 2012, 13, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tarui, T.; Kim, A.; Flake, A.; Mcclain, L.; Stratigis, J.; Fried, I.; Newman, R.; Slonim, D.K.; Bianchi, D.W. Amniotic fluid transcriptomics reflects novel disease mechanisms in fetuses with myelomeningocele. Am. J. Obstet. Gynecol. 2017, 217, 587.e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasso, E.B.; Thornton, M.E.; Chmait, R.H.; Ouzounian, J.G.; Grubbs, B.H. Amylase concentration and activity in the amniotic fluid of fetal rats with retinoic acid induced myelomeningocele. J. Matern.-Fetal Neonatal Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Manohar, M.; Verma, A.K.; Venkateshaiah, S.U.; Sanders, N.L.; Mishra, A. Pathogenic mechanisms of pancreatitis. World J. Gastrointest. Pharmacol. Ther. 2017, 8, 10. [Google Scholar] [CrossRef] [PubMed]


| mRNA | Name | * Assay Code Number |
|---|---|---|
| Gsx1 | GS Homeobox1 | Rn01412792_g1 |
| Gsx2 | GS Homeobox2 | Rn03810223_m1 |
| Ngfr | Nerve Growth Factor Receptor | Rn00561634_m1 |
| Sparc | Secreted Protein, Acidic, Cysteine-Rich | Rn01470624_m1 |
| Pdpn | Podoplanin | Rn00571195_m1 |
| Hprt1 | Hypoxanthine Phosphoribosyl Transferase 1 | Rn01527840_m1 |
| Tnc | Tenascin C | Rn01454948_m1 |
| Pdgfa | Platelet-Derived Growth Factor Polypeptide | Rn00709363_m1 |
| E15 | E17 | E20 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Upregulated | Downregulated | Total | Upregulated | Downregulated | Total | Upregulated | Downregulated | |
| MMC vs. Vehicle | 3022 | 1324 (44%) | 1698 (56%) | 1167 | 312 (27%) | 855 (73%) | 302 | 133 (44%) | 169 (56%) |
| MMC vs. Control | 3777 | 2119 (56%) | 1658 (44%) | 1847 | 264 (14%) | 1583 (86%) | 469 | 154 (33%) | 315 (67%) |
| Control vs. Vehicle | 505 | 147 (29%) | 358 (71%) | 327 | 117 (56%) | 210 (44%) | 355 | 269 (76%) | 86 (24%) |
| Gene Symbol | Full Gene Name | Fold Change | p Value |
|---|---|---|---|
| Actc1 | Actin, alpha cardiac muscle 1 | 68.26 | |
| Tnni1 | Troponin I, slow skeletal muscle | 20.94 | |
| Rps20 | 40S ribosomal protein S20 | 20.21 | |
| Myh3 | Myosin-3 | 17.56 | |
| Tnnc2 | Troponin C, skeletal muscle | 15.43 | |
| Krt5 | Keratin, type II cytoskeletal 5 | 14.95 | |
| Myl1 | Myosin light chain 1/3, skeletal muscle isoform | 14.81 | |
| Myog | Myogenin | 13.50 | |
| Cxcl14 | C-X-C motif chemokine 14 precursor | 12.76 | |
| Twist2 | Twist-related protein 2 | 12.62 | |
| Tnnt1 | Troponin T, slow skeletal muscle | 12.24 | |
| Smoc2 | SPARC-related modular calcium-binding protein 2 precursor | 11.22 | |
| Fam167b | Family with sequence similarity 167, member B | 10.45 | |
| Agtr2 | Type-2 angiotensin II receptor | 10.28 | |
| Krt14 | Keratin, type I cytoskeletal 14 | 10.22 | |
| Myod1 | Myoblast determination protein 1 | 10.01 | |
| Kremen2 | Kremen protein 2 precursor | 9.82 | |
| Myl4 | Myosin light chain 4 | 9.67 | |
| Pitx1 | Pituitary homeobox 1 | 9.57 | |
| Hand2 | Heart- and neural crest derivatives-expressed protein 2 | 9.45 | |
| Krt15 | Keratin, type I cytoskeletal 15 | 9.23 | |
| Mylpf | Myosin regulatory light chain 2, skeletal muscle isoform | 8.91 | |
| Actn2 | Actinin alpha 2 | 7.50 | |
| Acta1 | Actin, alpha skeletal muscle | 6.50 | |
| Tbx15 | T-box transcription factor TBX15 | 6.44 | |
| EF1A | Elongation factor 1-alpha | −51.18 | |
| Lynx1 | Ly-6/neurotoxin-like protein 1 precursor | −13.24 | |
| Necab1 | N-terminal EF-hand calcium-binding protein 1 | −8.35 | |
| Slc6a13 | Sodium- and chloride-dependent GABA transporter 2 | −7.65 | |
| Bcan | Brevican core protein isoform 1 precursor | −7.18 | |
| Slc13a4 | Solute carrier family 13 member 4 precursor | −7.01 | |
| Clec3b | C-type lectin domain family 3, member b | -6.78 | |
| Hoxc8 | Homeobox protein Hox-C8 | −6.15 | |
| Slc6a11 | Sodium- and chloride-dependent GABA transporter 3 | −5.85 | |
| S100b | Protein S100-B | −5.63 | |
| Grm5 | Metabotropic glutamate receptor 5 | −5.40 | |
| Slc22a6 | Solute carrier family 22 member 6 | −4.86 | |
| Ptgds | Prostaglandin-H2 D-isomerase | −4.63 | |
| Vsnl1 | Visinin-like protein 1 | −4.61 | |
| Slc16a11 | Solute carrier family 16, member 11 | −4.26 | |
| Ccl19 | C-C motif chemokine 19 precursor | −4.20 | |
| Park2 | E3 ubiquitin-protein ligase parkin | −4.19 | |
| Cdh8 | Cadherin-8 precursor | −3.90 | |
| Sncb | Beta-synuclein | −3.80 | |
| Fmod | Fibromodulin precursor | −3.73 | |
| Hs6st3 | Heparan-sulfate 6-O-sulfotransferase 3 precursor | −3.72 | |
| Cyp26b1 | Cytochrome P450 26B1 | −3.69 | |
| Tac1 | Protachykinin-1 Substance P Neurokinin A Neuropeptide K Neuropeptide gamma C-terminal-flanking peptide | −3.69 | |
| Olig1 | Oligodendrocyte transcription factor 1 | −3.64 | |
| Chd5 | Chromodomain-helicase-DNA-binding protein 5 | −3.63 |
| Gene Symbol | Full Gene Name | Fold Change | p Value |
|---|---|---|---|
| Fam167b | Family with sequence similarity 167, member B | 8.18 | |
| Hoxc11 | Homeobox C11 | 7.72 | |
| Fam50a | Protein Fam50a | 6.96 | |
| Dbx1 | Homeobox protein DBX1 | 5.82 | |
| Capg | Macrophage-capping protein | 5.59 | |
| Hoxa11 | Protein Hoxa10 | 5.40 | |
| Tmem200b | Transmembrane protein 200B | 4.16 | |
| Cox6a2 | Cytochrome c oxidase subunit 6A2, mitochondrial | 4.06 | |
| Hbb-b2 | Hemoglobin subunit beta-2 | 3.97 | |
| Plvap | Plasmalemma vesicle-associated protein | 3.94 | |
| Hoxd12 | Homeo box D12 | 3.92 | |
| Rd3 | Retinal degeneration 3 | 3.67 | |
| Neurog2 | Protein Neurog2 | 3.67 | |
| Retsat | All-trans-retinol 13,14-reductase precursor | 3.41 | |
| Tnfrsf22 | Tumor necrosis factor superfamily member 22 | 3.37 | |
| Rps20 | 40S ribosomal protein S20 | 3.18 | |
| Hist1h2bc | Histone H2B | 3.14 | |
| Hoxc10 | Homeo box C10 | 3.13 | |
| Arx | Homeobox protein ARX | 2.97 | |
| ATP5F1E | ATP synthase subunit epsilon, mitochondrial | 2.96 | |
| Col15a1 | Collagen alpha-1(XV) chain precursor | 2.96 | |
| Hsd11b2 | Corticosteroid 11-beta-dehydrogenase isozyme 2 | 2.95 | |
| Wt1 | Wilms tumor protein homolog | 2.90 | |
| Hoxd10 | Homeo box D10 | 2.82 | |
| Cebpa | CCAAT/enhancer-binding protein alpha | 2.81 | |
| Ptgds | Prostaglandin-H2 D-isomerase | −49.27 | |
| Gm10068 | Chromobox 3, pseudogene 7 | −11.56 | 5.10 × 10−5 |
| Eno2 | Enolase 2 | −9.46 | |
| Calca | Calcitonin | −6.91 | |
| SDHAF1 | Succinate dehydrogenase assembly factor 1 | −6.07 | |
| Defb10 | Beta-defensin 10 | -5.15 | |
| Lynx1 | Ly-6/neurotoxin-like protein 1 precursor | −4.83 | |
| Reg3b | Regenerating islet-derived protein 3-beta | −4.55 | |
| Slc22a6 | Solute carrier family 22 member 6 | −4.41 | |
| Hoxc8 | Homeobox protein Hox-C8 | −4.35 | |
| Hoxc5 | Homeo box C5 | −4.16 | |
| Uts2d | Urotensin-2B | −4.06 | |
| Slc13a4 | Solute carrier family 13 member 4 precursor | −4.06 | |
| Gng4 | Guanine nucleotide-binding protein subunit gamma 4 | −4.01 | |
| NUPR1 | Nuclear protein 1 | −3.83 | |
| Slc6a13 | Sodium- and chloride-dependent GABA transporter 2 | −3.82 | |
| Nefh | Neurofilament heavy polypeptide | −3.73 | |
| Calcb | Calcitonin gene-related peptide 2 precursor | −3.62 | |
| Asgr1 | Asialoglycoprotein receptor 1 | −3.60 | |
| Gng13 | Guanine nucleotide binding protein, gamma 13 | −3.48 | |
| Ccl19 | C-C motif chemokine 19 precursor | −3.44 | |
| Hoxc6 | Homeo box C6 | −3.33 | |
| Snhg11 | Small nucleolar RNA host gene 11 (non-protein coding) | −3.32 | |
| Gjb6 | Gap junction beta-6 protein | −3.30 | |
| Synpr | Synaptoporin | −3.29 |
| Gene Symbol | Full Gene Name | Fold Change | p Value |
|---|---|---|---|
| EF1A | Elongation factor 1-alpha | 129.91 | |
| Krt5 | Keratin, type II cytoskeletal 5 | 51.61 | |
| Hoxc11 | Homeobox C11 | 29.97 | |
| HBB | Hemoglobin subunit beta-2 | 29.46 | |
| Krt14 | Keratin, type I cytoskeletal 14 | 25.30 | |
| Hoxa11 | Homeobox A11 | 9.35 | |
| RPS18 | 40S ribosomal protein S18 | 8.87 | |
| Hoxa11 | Protein Hoxa10; RCG52455 | 8.13 | |
| RT1-Da | RT1 class II, locus Da precursor | 6.32 | |
| MRPL36 | Mitochondrial ribosomal protein L36 | 5.85 | |
| Ntrk1 | High affinity nerve growth factor receptor | 5.70 | |
| Lgals3 | Galectin-3 | 5.36 | |
| Cd74 | H-2 class II histocompatibility antigen gamma chain | 5.23 | |
| FZD8 | Frizzled 8 | 4.84 | |
| Prdm12 | PR domain zinc finger protein 12 | 4.82 | |
| Tlx2 | T-cell leukemia, homeobox 2 | 4.82 | |
| Stfa2l1 | Stefin-3 | 4.70 | |
| Ccl12 | Chemokine (C-C motif) ligand 12 precursor | 4.52 | |
| Hcst | Hematopoietic cell signal transducer | 4.50 | |
| Tusc5 | Tumor suppressor candidate 5 homolog | 4.48 | |
| Scn10a | Sodium channel protein type 10 subunit alpha | 4.34 | |
| Slc2a3 | Solute carrier family 2, facilitated glucose transporter member 3 | 4.33 | |
| RT1-Bb | Rano class II histocompatibility antigen, B-1 beta chain precursor | 4.18 | |
| Fcnb | Ficolin-2 | 4.04 | |
| Sost | Sclerostin | 4.03 | |
| Pla2g3 | Group 3 secretory phospholipase A2 precursor | −9.01 | |
| Bcas1 | Breast carcinoma-amplified sequence 1 homolog | −5.78 | |
| Mbp | Myelin basic protein S | −5.72 | |
| Tnks2 | Tankyrase 2 | −5.59 | |
| Gpr17 | Uracil nucleotide/cysteinyl leukotriene receptor | −5.58 | |
| TSGA14 | Testis-Specific Gene A14 Protein | −5.56 | |
| Slc22a6 | Solute carrier family 22 member 6 | −5.19 | |
| Pnlip | Pancreatic triacylglycerol lipase precursor | −4.83 | |
| Slc6a13 | Sodium- and chloride-dependent GABA transporter 2 | −4.48 | |
| Ogn | Osteoglycin | −4.46 | |
| Paqr8 | Membrane progestin receptor beta | −4.44 | |
| Nkx6-2 | NK6 homeobox 2 | −4.35 | |
| Crygn | Gamma-crystallin N | −4.23 | |
| Scrg1 | Scrapie-responsive protein 1 | −4.09 | |
| Plp1 | Myelin proteolipid protein | −3.92 | |
| Zfp488 | Zinc Finger Protein 488 | −3.89 | |
| Fmod | Fibromodulin precursor | −3.80 | |
| Pmaip1 | Phorbol-12-myristate-13-acetate-induced protein 1 | −3.79 | |
| AKR1B10 | Aldo-keto reductase family 1, member B10 | −3.56 | |
| Fgf15 | Fibroblast growth factor 15 precursor | −3.50 | |
| Slc5a7 | High affinity choline transporter 1 | −3.47 | |
| Bmp6 | Bone morphogenetic protein 6 | −3.46 | |
| Lims2 | LIM and senescent cell antigen-like-containing domain protein 2 | −3.30 | |
| Pnpla1 | Patatin-like phospholipase domain-containing protein 1 | −3.29 | |
| Kcne1l | Potassium voltage-gated channel subfamily E member 1-like protein | −3.24 |
| GO Biological Process | GO Cellular Component | GO Molecular Function |
|---|---|---|
| Regulation of retroviral genome replication | Cleavage furrow | Extracellular matrix binding |
| DNA ligation | Ribonucleoside-diphosphate reductase complex | Single-stranded DNA-dependent ATPase activity |
| Tight junction assembly | DNA polymerase complex | Peptidase activator activity |
| Wound healing, spreading of cells | T-tubule | Histone kinase activity |
| DNA biosynthetic process | Contractile fiber | Caspase regulator activity |
| Cytokinesis | Contractile fiber part | Ribonucleoside-diphosphate reductase activity |
| Regulation of mesoderm development | Perinuclear region of cytoplasm | Flap endonuclease activity |
| Glial cell migration | Extracellular vesicular exosome | Cyclin-dependent protein kinase activity |
| Memory | Cyclin-dependent protein kinase holoenzyme complex | DNA N-glycosylase activity |
| Negative regulation of cytokine production | Nuclear envelope | Receptor signaling protein tyrosine kinase activity |
| GO Biological Process | GO Cellular Component | GO Molecular Function |
|---|---|---|
| Regulation of gamma-aminobutyric acid secretion | Sodium | Neurotransmitter binding |
| Positive regulation of epidermal growth factor receptor activity | Neuronal cell body membrane | Protein kinase C activity |
| Negative regulation of leukocyte migration | cAMP-dependent protein kinase complex | Adenylate cyclase activity |
| Protein localization to synapse | Heterotrimeric G-protein complex | Calcium-dependent protein kinase regulator activity |
| Regulation of ARF protein signal transduction | Periplasmic space | GABA receptor binding |
| Synaptic vesicle maturation | Protein phosphatase type 2A complex | Long-chain fatty acid-CoA ligase activity |
| Positive regulation of amine transport | Ionotropic glutamate receptor complex | Neurexin binding |
| Negative regulation of insulin receptor signaling pathway | Synapse part | Serotonin receptor activity |
| Cell junction maintenance | Recycling endosome membrane | Calcium-dependent protein binding |
| Innervation | Cell junction | Protein phosphatase regulator activity |
| GO Biological Process | GO Cellular Component | GO Molecular Function |
|---|---|---|
| Female pregnancy | Cortical cytoskeleton | mRNA 3′-UTR binding |
| Proximal/distal pattern formation | Myosin complex | Extracellular matrix structural constituent |
| Specification of loop of Henle identity | Stress fiber | Growth factor binding |
| Regulation of endocrine process | Nucleosome | Manganese ion binding |
| Platelet activation | Collagen | Protein dimerization activity |
| Somatic stem cell maintenance | Basement membrane | Hormone activity |
| Multicellular organism adhesion | Extracellular matrix | C2H2 zinc finger domain binding |
| Retinol metabolic process | Hemoglobin complex | Oxygen transporter activity |
| Endochondral ossification | Extracellular space | Oxygen binding |
| Negative regulation of cell fate specification | Nucleus | Actin filament binding |
| GO Biological Process | GO Cellular Component | GO Molecular Function |
|---|---|---|
| Regulation of response to food | Heterotrimeric G protein complex | Neuropeptide hormone activity |
| Gastric acid secretion | Perikaryon | Channel inhibitor activity |
| Negative regulation of tissue remodeling | Extrinsic to organelle membrane | GABA receptor binding |
| Neuronal action potential propagation | Endomembrane system | Retinoid binding |
| Peptide transport | Dendritic shaft | Neuropeptide receptor binding |
| Diol metabolic process | Axon part | Cyclic nucleotide binding |
| Regulation of cellular amine metabolic process | Myelin sheath | Tau protein binding |
| Regulation of glomerular filtration | Cell body | L amino acid transmembrane transporter activity |
| Negative regulation of muscle contraction | Extracellular region | Caspase regulator activity |
| Cellular glucose homeostasis | Ionotropic glutamate receptor complex | Oxidoreductase activity |
| GO Biological Process | GO Cellular Component | GO Molecular Function |
|---|---|---|
| Mechanical stimulus involved in sensory perception of pain | Lytic vacuole | Sodium channel activity |
| Antigen processing and presentation of antigen via MHC class II | External side of plasma membrane | Cytokine receptor binding |
| Cellular response to interferon-gamma | MHC class II protein complex | GTPase activity |
| Sodium ion transport | Integral to plasma membrane | Cytokine activity |
| Positive regulation of antigen processing and presentation | Vacuolar membrane | Sequence specific DNA binding |
| Regulation of cytokine production involved in immune response | Nucleosome | Sequence specific DNA binding transcription factor activity |
| Negative regulation of leukocyte activation | Cell surface | Ligand-gated ion channel activity |
| Regulation of T cell differentiation | Intracellular | Heme binding |
| Negative regulation of protein complex assembly | Extracellular region part | Structure specific DNA binding |
| Regulation of alpha-beta T cell activation | Keratin filament | Oxygen transporter activity |
| GO Biological Process | GO Cellular Component | GO Molecular Function |
|---|---|---|
| Response to toxin | Integral to plasma membrane | Carboxylic ester hydrolase activity |
| Myelination | Extracellular region | Carboxylic acid transmembrane transporter activity |
| Sterol transport | Caveola | Cytokine receptor binding |
| Regulation of symbiosis | Extracellular space | Amine transmembrane transporter activity |
| Negative regulation of neurogenesis | Extracellular matrix part | Voltage-gated ion channel activity |
| Regulation of cellular ketone metabolic process | Neuronal cell body | Secondary active transmembrane transporter activity |
| Regulation of steroid metabolic process | Extracellular matrix | Inorganic cation transmembrane transporter activity |
| Membrane organization | Nuclear membrane | Anion transmembrane transporter activity |
| Positive regulation of signal transduction | Cytokine activity | |
| Regulation of lipid transport |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murphy, K.P.; Pathak, B.; Peiro, J.L.; Oria, M. Time Course Transcriptome Analysis of Spina Bifida Progression in Fetal Rats. Brain Sci. 2021, 11, 1593. https://doi.org/10.3390/brainsci11121593
Murphy KP, Pathak B, Peiro JL, Oria M. Time Course Transcriptome Analysis of Spina Bifida Progression in Fetal Rats. Brain Sciences. 2021; 11(12):1593. https://doi.org/10.3390/brainsci11121593
Chicago/Turabian StyleMurphy, Kendall P., Bedika Pathak, Jose L. Peiro, and Marc Oria. 2021. "Time Course Transcriptome Analysis of Spina Bifida Progression in Fetal Rats" Brain Sciences 11, no. 12: 1593. https://doi.org/10.3390/brainsci11121593
APA StyleMurphy, K. P., Pathak, B., Peiro, J. L., & Oria, M. (2021). Time Course Transcriptome Analysis of Spina Bifida Progression in Fetal Rats. Brain Sciences, 11(12), 1593. https://doi.org/10.3390/brainsci11121593








