Online Prediction of Lead Seizures from iEEG Data
Abstract
:1. Introduction
- •
- •
- Second, naturally occurring seizures are usually clustered in time. That is, multiple seizures occur within short time period (~cluster), and the time between successive clusters is much longer than duration of a cluster. It is clinically important to predict the first seizure in a cluster, also known as the lead seizure [21].
- •
- An SVM classifier is trained on windows data (yielding larger number of labeled training windows);
- •
- The number of input features (for encoding 20 s window) is much smaller than the number of features for 4 h segments (resulting in reduced input dimensionality).
- •
- Periodic retraining, in order to address non-stationarity of iEEG data.
- •
- Improving the quality of training data by removing training samples with noisy labels.
- •
- A new post-processing scheme during prediction (test) stage.
2. Materials and Methods
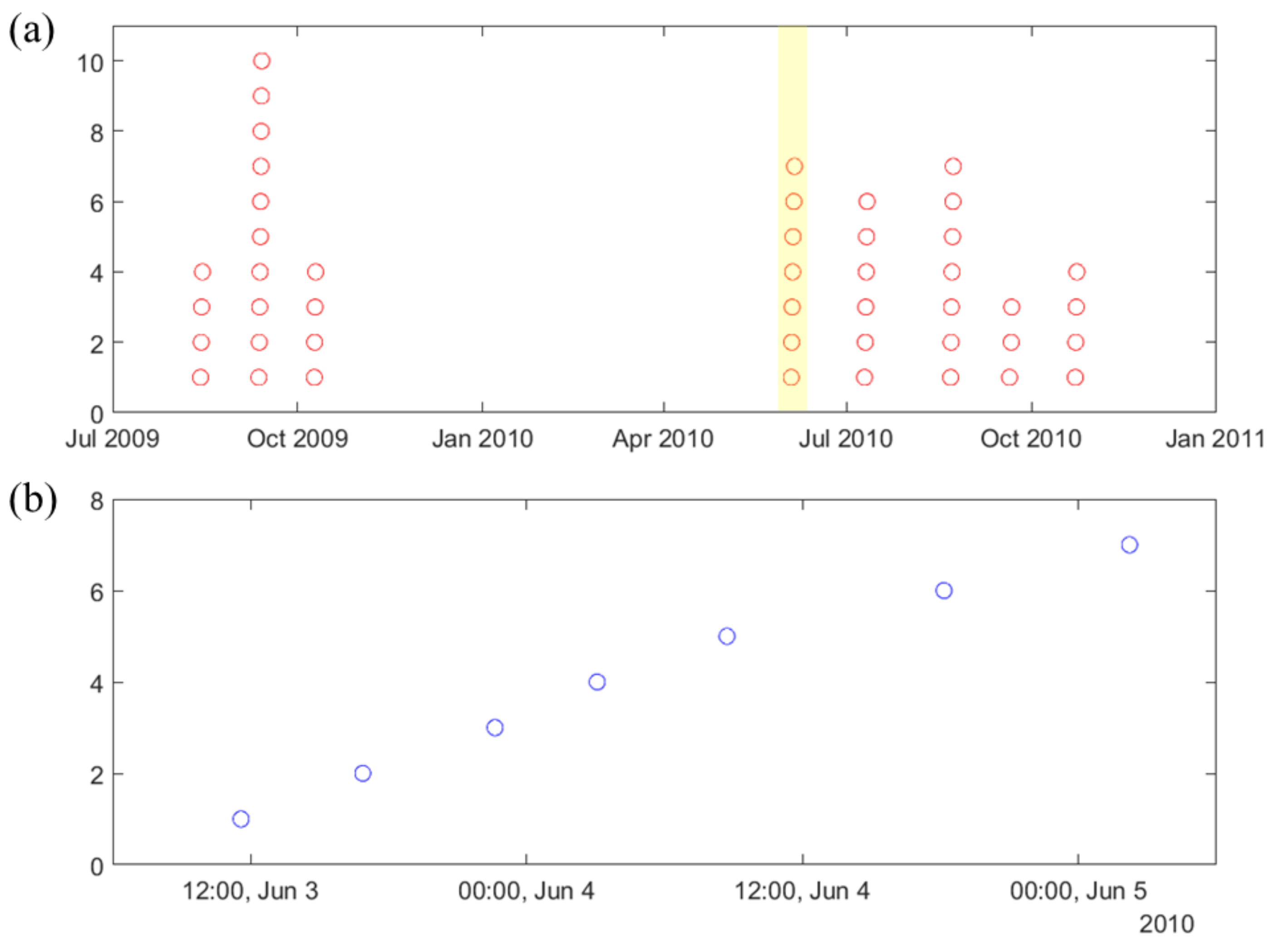
2.1. Lead Seizures
- •
- •
- 80 min seizure-free period in Assi et al. [25], even though it was not explicitly stated in their paper.
- •
- 8 h seizure free period was initially used in Cook et al. [34].
- •
- Later, the same research team adopted a 4 h seizure-free period in Kuhlmann et al. [35].
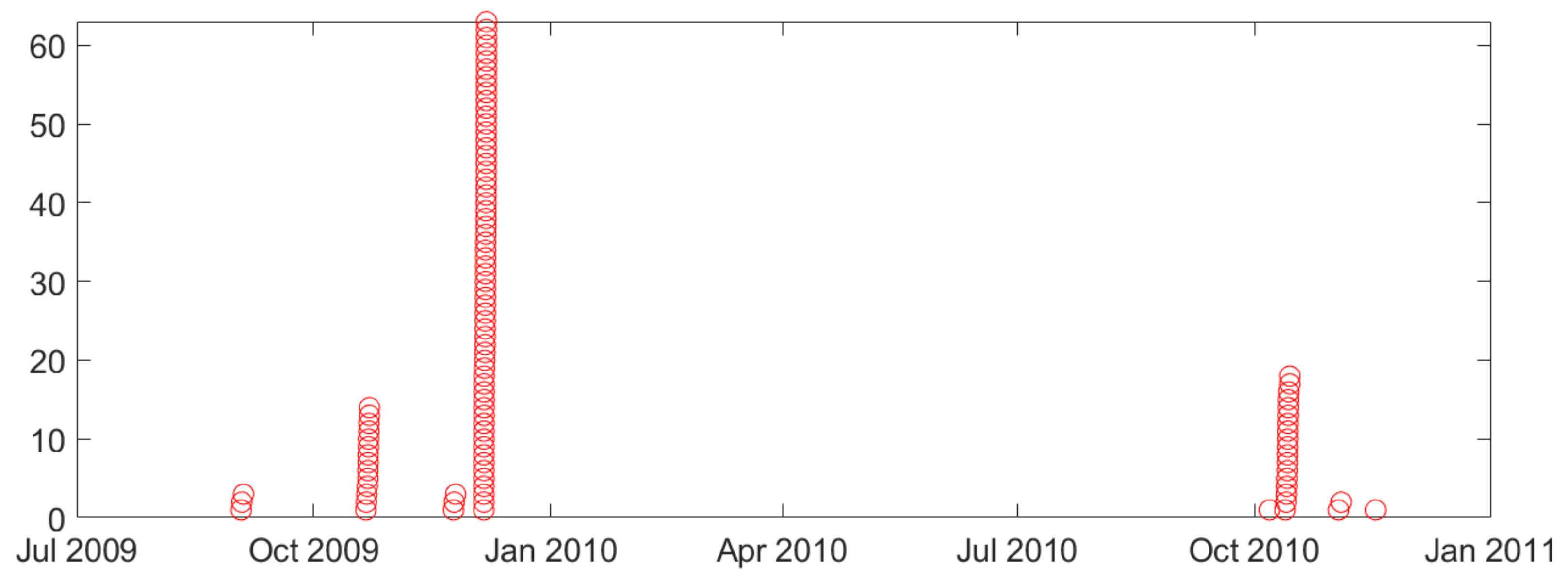
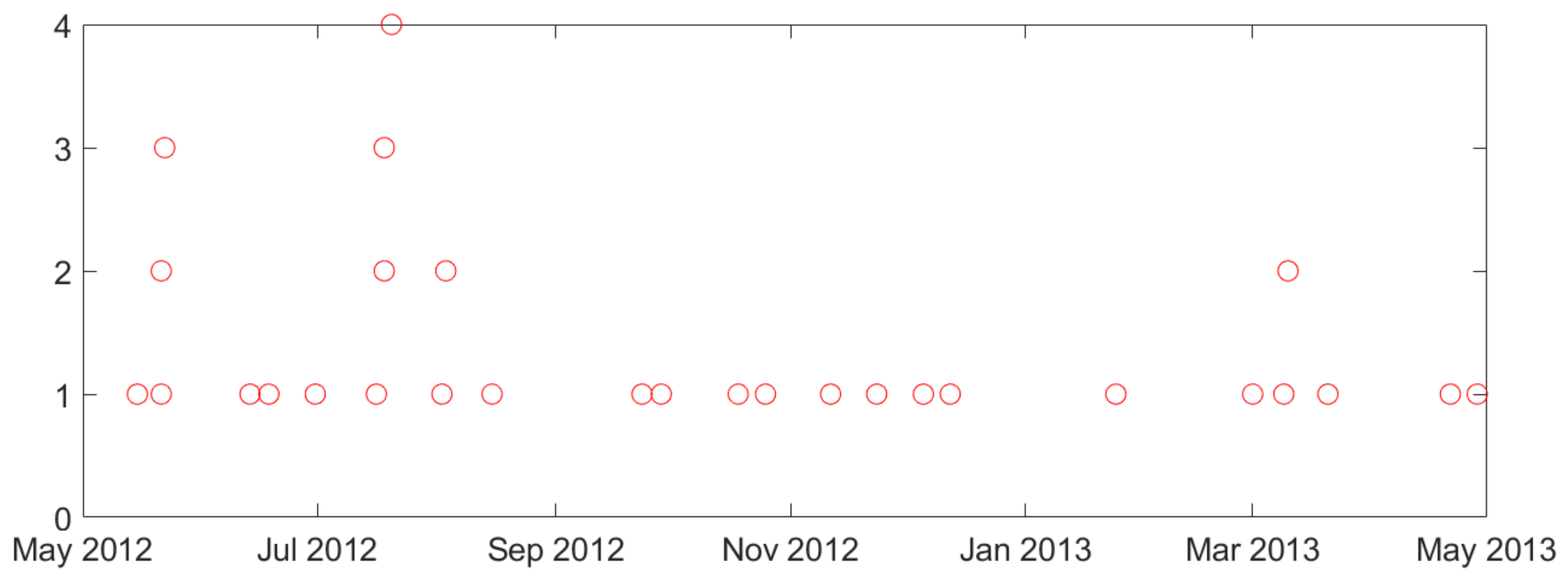
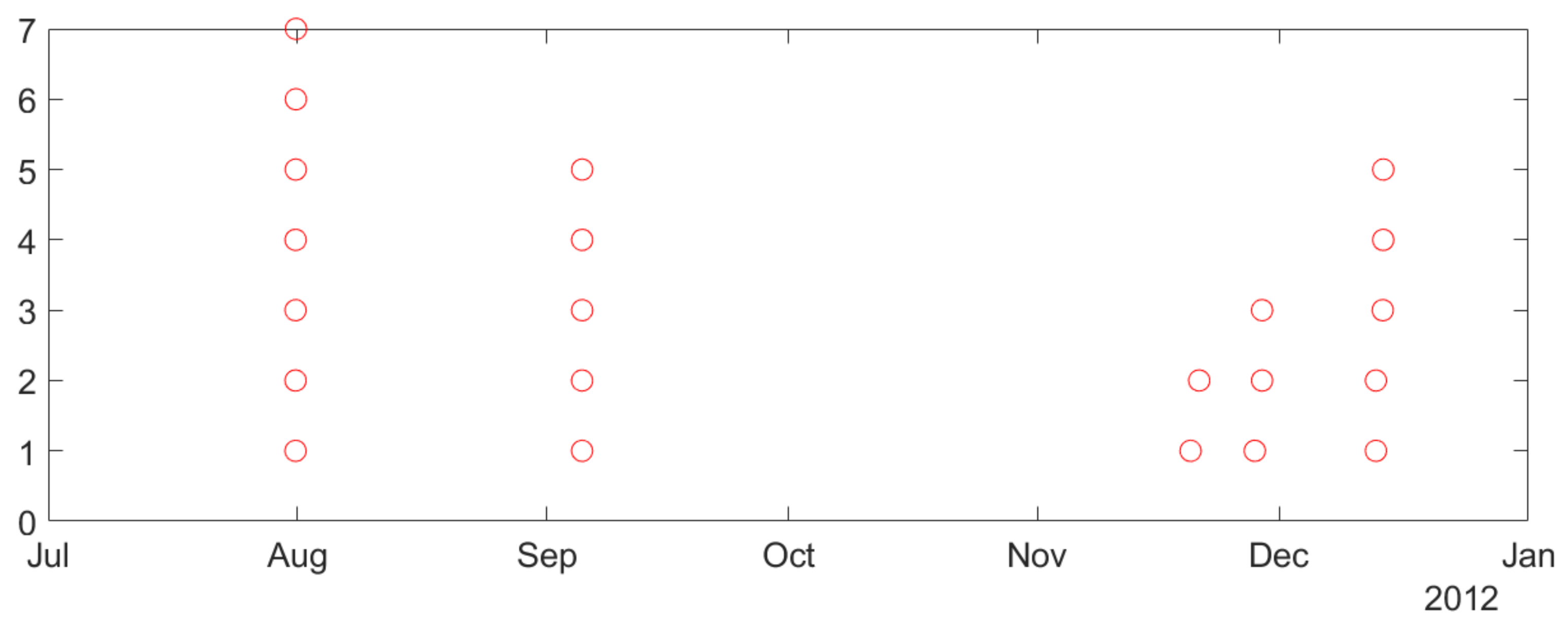
2.2. Description of Available Intracranial Electroencephalogram (iEEG) Data and Analysis of Seizure Clustering
- •
- Similarity of biological mechanisms of epilepsy in humans and dogs.
- •
- Difficulty of obtaining long-term seizure recordings for humans (e.g., typical human iEEG data recording is 1–3 days long, corresponding to patients’ stay in a hospital).
- •
- Smaller number of labeled (preictal) training samples.
- •
- Lower sensitivity, since lead seizures are harder to predict.
2.3. Online Modeling
- •
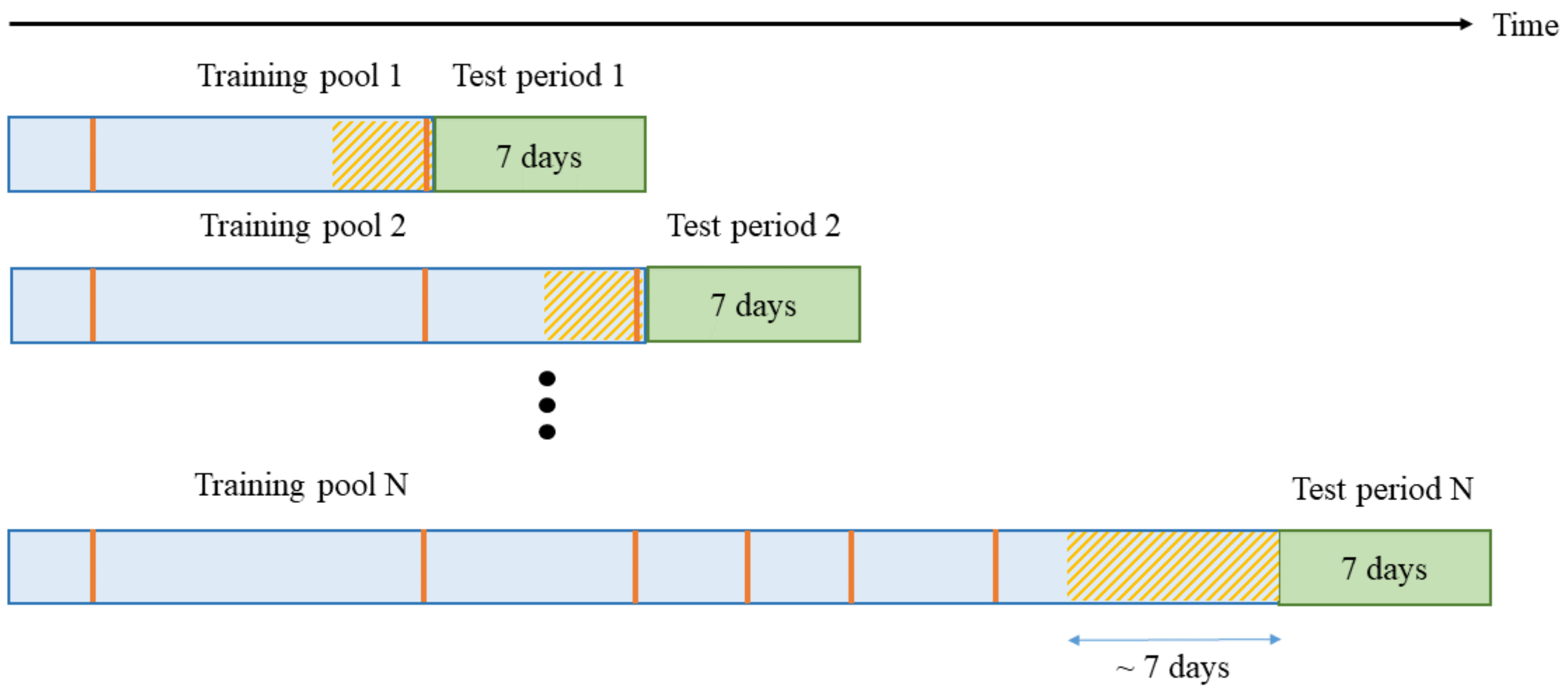
- Training data should be selected from the most recent iEEG segments.
- •
- A classifier needs to be periodically retrained (using most recent data).
- •
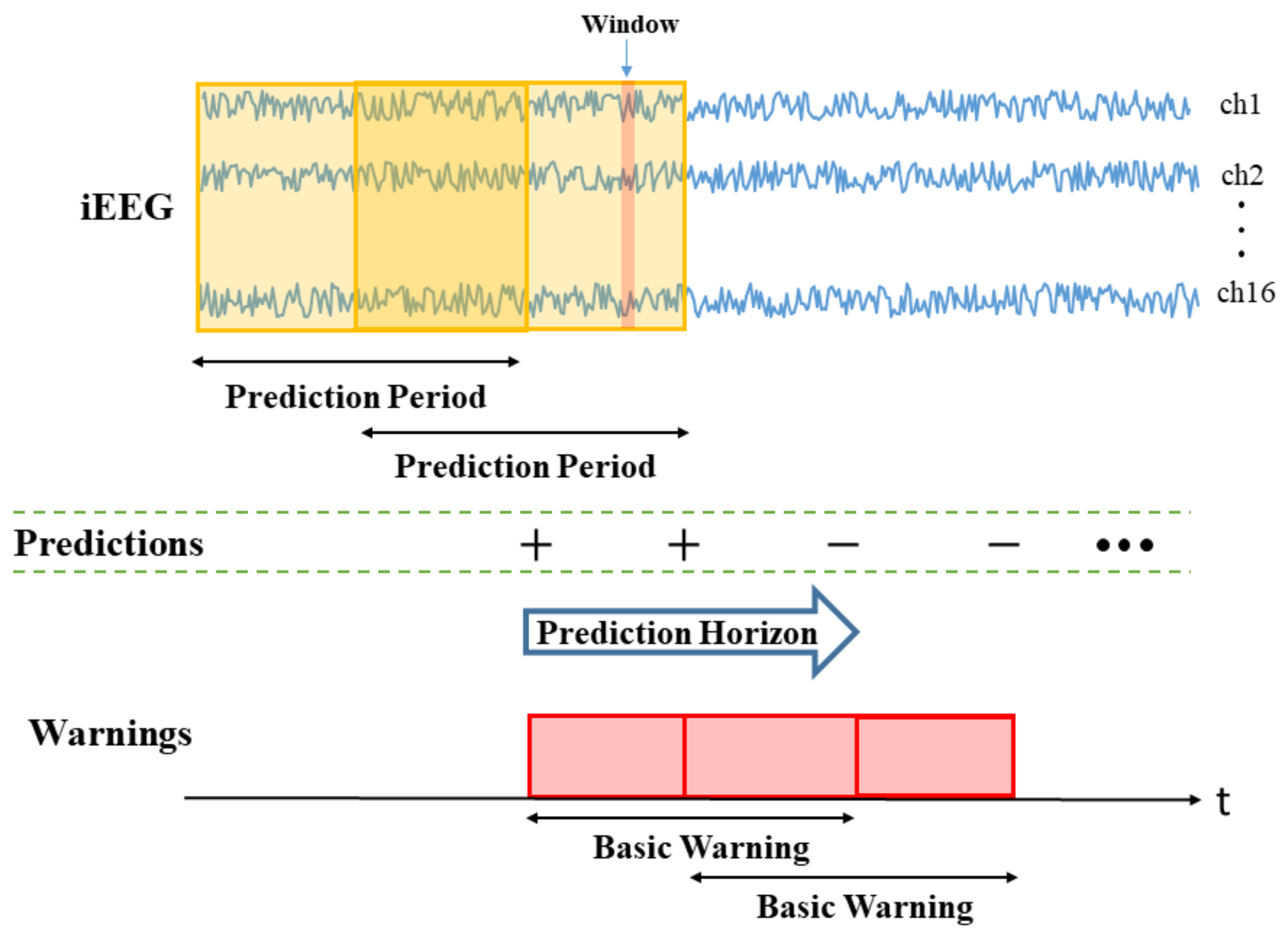
- Window size (W) is the duration of iEEG signal used to classify preictal vs. interictal states. It reflects the change of brain state preceding seizures, and its typical range is 10–30 s (the value of W = 20 s is used in this paper). During the test stage, the trained SVM classifier makes predictions for all consecutive windows within prediction period (PP).
- •
- Prediction Period (PP) is the length of the iEEG signal used for generating seizure prediction or warning during test stage. Its length obviously affects prediction performance; and different values of PPs were adopted in earlier studies (ranging from minutes to hours). Some studies do not clearly specify PP but effectively combine predictions from consecutive windows to generate single seizure prediction. The PP used in this study is 4 h, which equals the length of the iEEG segment used for training. The trained SVM classifier makes predictions for all windows within PP. Then predictions for all windows are combined via an adaptive post-processing scheme (described in Section 2.5.4) to make single prediction at the end of PP. These predictions are marked as positive (+) or negative (−) in Figure 3. A positive prediction indicates seizure warning for the duration of the next time period called prediction horizon (PH), whereas negative prediction indicates that there is no such warning. The system makes predictions every 2 h, so that each PP partially overlaps (50%) the previous one (see Figure 3).
- •
- Prediction Horizon (PH) is the time period during which positive/negative prediction holds. That is, each positive prediction triggers a basic seizure warning for the fixed duration of PH. The length of PH is determined based on clinical considerations. Typical PH values are in the range 0.5–4 h. In this study, PH = 4 h is used.
2.4. Performance Metrics for Online Prediction
- •
- TIW is defined as the duration of total warning time relative to total test period (i.e., the fraction of test period when the system generates warning).
- •
- FPR is defined as the number of false warnings per day. In a re-triggerable warning system, this definition assumes that each warning may be of variable duration (as explained earlier).
2.5. Proposed System for Online Seizure Prediction
- •
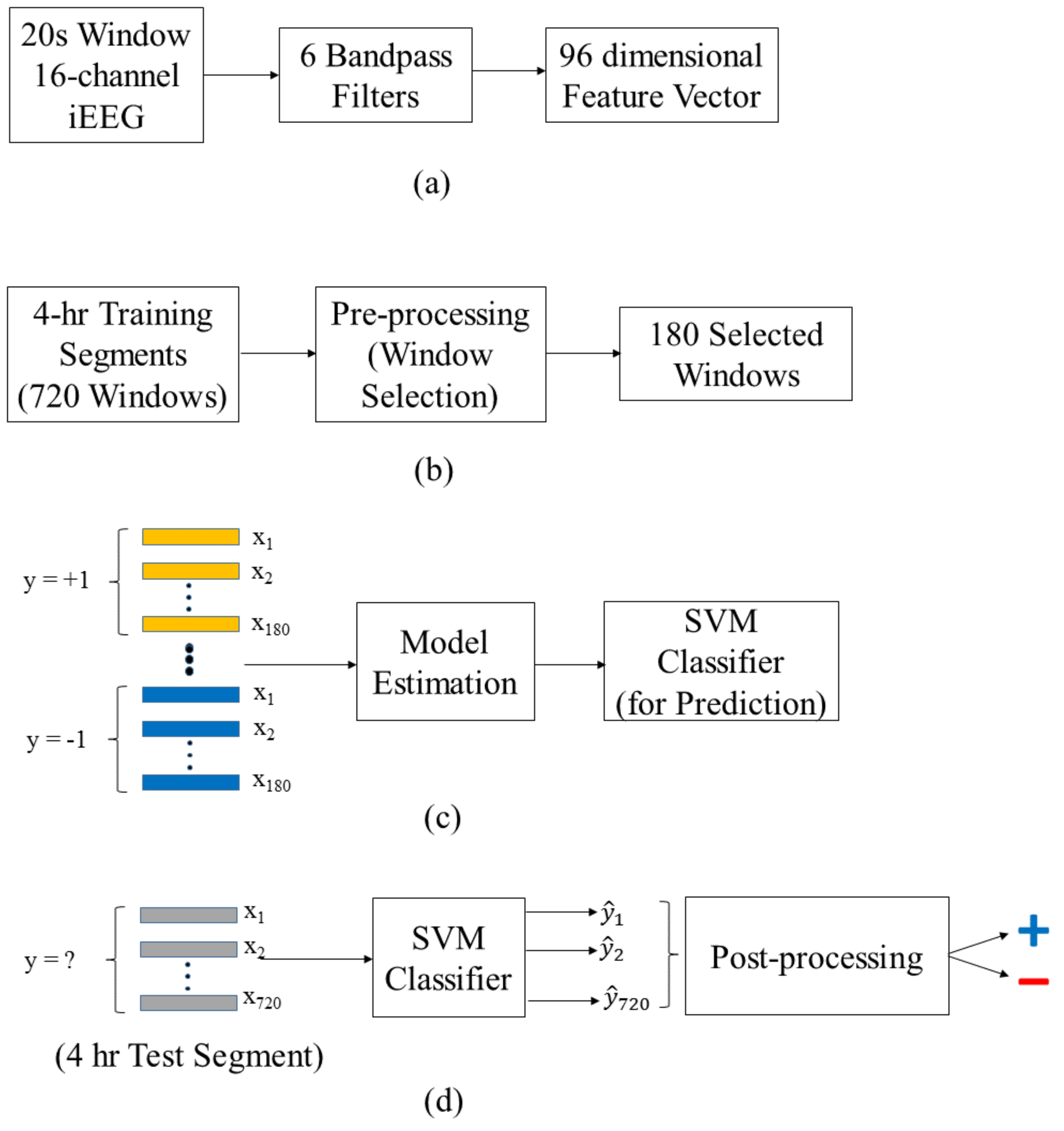
- First, annotation of iEEG recordings is performed for segments, rather than individual windows. That is, original labeling of data (by medical experts) applies to segments.
- •
- Second, classification (or model estimation) is performed for labeled windows, because (a) there are too few labeled segments, and (b) these segments are represented as high-dimensional feature vectors.
2.5.1. Data Preprocessing
2.5.2. Training Stage
2.5.3. Test Stage
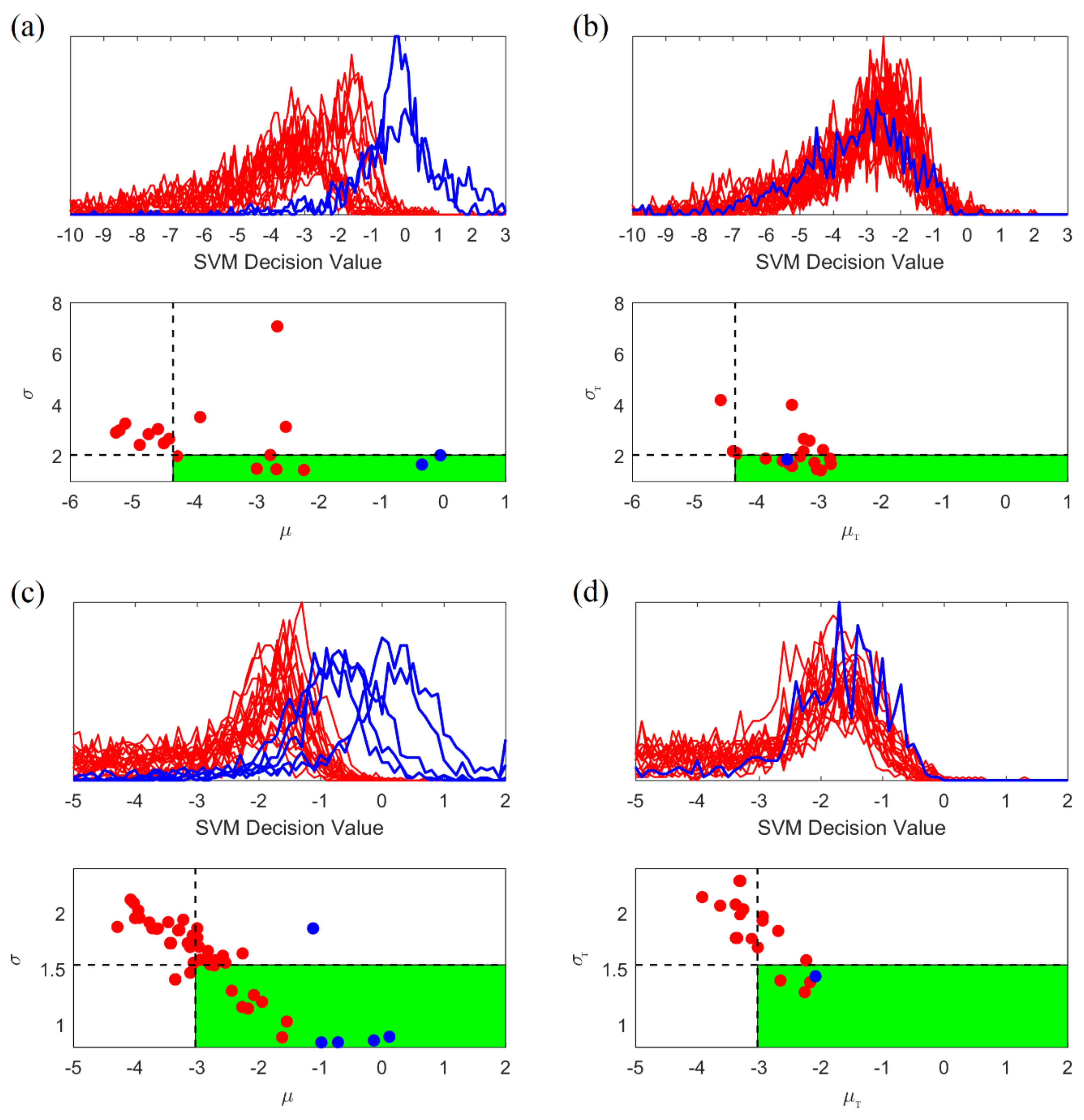
2.5.4. Adaptive Post-Processing for Group Learning
- •
- High variability of histograms, especially for preictal segments.
- •
- Large overlap between distributions (histograms) for preictal and interictal segments.
- 1.
- Generate the histogram of projections corresponding to SVM outputs (predictions) for all consecutive windows within 4-h test segment. Then calculate its mean and standard deviation .
- 2.
- Classify this unlabeled test segment as preictal (positive) if and only if:
- •
- Its mean is larger than 50% quantile of the values (for interictal training segments); and
- •
- Its standard deviation is smaller than 30% quantile of the values (for interictal training segments).
3. Results
3.1. Prediction Performance of Online Prediction System
3.2. Other Online Seizure Prediction Studies
- •
- Our study includes four canines with the same number of iEEG channels (16 channels). One canine (with severe signal loss in an entire channel) is not used for modeling.
- •
- •
- Nejedly et al. [33] use four canines’ data, including one dog with a missing channel. On the other hand, dog P2 with the smallest number of seizures is excluded from their study. This results in the largest number of seizures (available for modeling), and improves overall prediction performance.
- •
- •
- Our study uses 3-day seizure free period to define lead seizures. This is based on the previous study [8] and the statistical analysis of seizure clusters presented in Section 2.2.
- •
- •
- Assi et al. [25] apply only an 80 min seizure free period to define lead seizures. So, they have a large number of lead seizures, even though they use just a portion of iEEG recordings.
- •
- Howbert et al. [27] apply both 4 h and 80 min seizure free periods to define lead seizures for comparing the effect of using different of lead seizures on prediction performance.
- •
- As a result of these differences, each study uses a different average number of lead seizures (as shown in Table 5).
4. Discussion
4.1. Quality of iEEG Data
4.2. Non-Stationarity and Retraining
- (a)
- some studies used short-term recording period. For short-term recording period (~several days), the problem of non-stationarity of EEG signal is not as severe as for long-term recording (~several months).
- (b)
- these studies used batch modeling, where the problem of non-stationarity is alleviated because future data may be used for training.
4.3. Improving the Quality of Training Data
4.4. Adaptive Post-Processing
4.5. Methodological Issues
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
References
- Bhattacharyya, A.; Pachori, R.B. A Multivariate Approach for Patient-Specific EEG Seizure Detection Using Empirical Wavelet Transform. IEEE Trans. Biomed. Eng. 2017, 64, 2003–2015. [Google Scholar] [CrossRef]
- Deng, Z.; Xu, P.; Xie, L.; Choi, K.S.; Wang, S. Transductive Joint-Knowledge-Transfer TSK FS for Recognition of Epileptic EEG Signals. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 1481–1494. [Google Scholar] [CrossRef] [PubMed]
- Thodoroff, P.; Pineau, J.; Lim, A. Learning Robust Features using Deep Learning for Automatic Seizure Detection. In Proceedings of the 1st Machine Learning for Healthcare Conference, Los Angeles, CA, USA, 19–20 August 2016; Volume 56. [Google Scholar]
- Tian, X.; Deng, Z.; Ying, W.; Choi, K.S.; Wu, D.; Qin, B.; Wang, J.; Shen, H.; Wang, S. Deep Multi-View Feature Learning for EEG-Based Epileptic Seizure Detection. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 1962–1972. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Deng, Z.; Xu, P.; Choi, K.S.; Wang, S. Generalized Hidden-Mapping Transductive Transfer Learning for Recognition of Epileptic Electroencephalogram Signals. IEEE Trans. Cybern. 2018, 49, 2200–2214. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Deng, Z.; Choi, K.-S.; Jiang, Y.; Wang, S. Transductive domain adaptive learning for epileptic electroencephalogram recognition. Artif. Intell. Med. 2014, 62, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Deng, Z.; Choi, K.S.; Wang, S. Takagi-Sugeno-Kang transfer learning fuzzy logic system for the adaptive recognition of epileptic electroencephalogram signals. IEEE Trans. Fuzzy Syst. 2015, 24, 1079–1094. [Google Scholar] [CrossRef]
- Chen, H.H.; Cherkassky, V. Performance metrics for online seizure prediction. Neural Netw. 2020, 128, 22–32. [Google Scholar] [CrossRef]
- Lange, H.; Lieb, J.; Engel, J.J.; Crandall, P. Temporo-spatial patterns of pre-ictal spike activity in human temporal lobe epilepsy. Electroencephalogr. Clin. Neurophysiol. 1983, 56, 543–555. [Google Scholar] [CrossRef]
- Litt, B.; Echauz, J. Prediction of epileptic seizures. Lancet Neurol. 2002, 1, 22–30. [Google Scholar] [CrossRef]
- Litt, B.; Esteller, R.; Echauz, J.; D’Alessandro, M.; Shor, R.; Henry, T.; Pennell, P.; Epstein, C.; Bakay, R.; Dichter, M.; et al. Epileptic seizures may begin hours in advance of clinical onset: A report of five patients. Neuron 2001, 30, 51–64. [Google Scholar] [CrossRef] [Green Version]
- Petrosian, A. Kolmogorov complexity of finite sequences and recognition of different preictal EEG patterns. In Proceedings of the Proceedings Eighth IEEE Symposium on Computer-Based Medical Systems, Lubbock, TX, USA, 9–10 June 1995; pp. 212–217. [Google Scholar]
- Myers, M.H.; Padmanabha, A.; Bidelman, G.M.; Wheless, J.W. Seizure localization using EEG analytical signals. Clin. Neurophysiol. 2020, 131, 2131–2139. [Google Scholar] [CrossRef] [PubMed]
- Salam, M.T.; Sawan, M.; Nguyen, D.K. A Novel Low-Power-Implantable Epileptic Seizure-Onset Detector. IEEE Trans. Biomed. Circuits Syst. 2011, 5, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Cherkassky, V.; Veber, B.; Lee, J.; Shiao, H.T.; Patterson, N.; Worrell, G.A.; Brinkmann, B.H. Reliable seizure prediction from EEG data. In Proceedings of the 2015 International Joint Conference on Neural Networks (IJCNN), Killarney, Ireland, 12–17 July 2015. [Google Scholar] [CrossRef]
- Cherkassky, V.; Chen, H.H.; Shiao, H.T. Group Learning for High-Dimensional Sparse Data. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–10. [Google Scholar] [CrossRef]
- Gadhoumi, K.; Lina, J.M.; Mormann, F.; Gotman, J. Seizure prediction for therapeutic devices: A review. J. Neurosci. Methods 2016, 260, 270–282. [Google Scholar] [CrossRef]
- Mormann, F.; Andrzejak, R.G.; Elger, C.E.; Lehnertz, K. Seizure prediction: The long and winding road. Brain 2007, 130, 314–333. [Google Scholar] [CrossRef] [Green Version]
- Shiao, H.T.; Cherkassky, V.; Lee, J.; Veber, B.; Patterson, E.E.; Brinkmann, B.H.; Worrell, G.A. SVM-based system for prediction of epileptic seizures from iEEG signal. IEEE Trans. Biomed. Eng. 2016, 64, 1011–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuhlmann, L.; Lehnertz, K.; Richardson, M.P.; Schelter, B.; Zaveri, H.P. Seizure prediction—Ready for a new era. Nat. Rev. Neurol. 2018, 14, 618–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyder, D.E.; Echauz, J.; Grimes, D.B.; Litt, B. The statistics of a practical seizure warning system. J. Neural Eng. 2008, 5, 392–401. [Google Scholar] [CrossRef] [Green Version]
- Park, Y.; Luo, L.; Parhi, K.K.; Netoff, T. Seizure prediction with spectral power of EEG using cost-sensitive support vector machines. Epilepsia 2011, 52, 1761–1770. [Google Scholar] [CrossRef]
- Babloyantz, A.; Destexhe, A. Low-dimensional chaos in an instance of epilepsy. Proc. Natl. Acad. Sci. USA 1986, 83, 3513–3517. [Google Scholar] [CrossRef] [Green Version]
- Rosso, O.A.; Romanelli, L.; Blanco, S.; Romanelli, L.; Quiroga, R.Q.; Blanco, S.; Quiroga, R.Q.; Garcia, H.; Rosso, O.A. Stationarity of the EEG Series. IEEE Eng. Med. Biol. Mag. 1995, 14, 395–399. [Google Scholar] [CrossRef]
- Assi, E.B.; Nguyen, D.K.; Rihana, S.; Sawan, M. A Functional-genetic Scheme for Seizure Forecasting in Canine Epilepsy. IEEE Trans. Biomed. Eng. 2017, 65, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, B.H.; Patterson, E.E.; Vite, C.; Vasoli, V.M.; Crepeau, D.; Stead, M.; Jeffry Howbert, J.; Cherkassky, V.; Wagenaar, J.B.; Litt, B.; et al. Forecasting seizures using intracranial EEG measures and SVM in naturally occurring canine epilepsy. PLoS ONE 2015, 10, e0133900. [Google Scholar] [CrossRef]
- Howbert, J.J.; Patterson, E.E.; Stead, S.M.; Brinkmann, B.; Vasoli, V.; Crepeau, D.; Vite, C.H.; Sturges, B.; Ruedebusch, V.; Mavoori, J.; et al. Forecasting seizures in dogs with naturally occurring epilepsy. PLoS ONE 2014, 9, e81920. [Google Scholar] [CrossRef]
- Korshunova, I.; Kindermans, P.-J.; Degrave, J.; Verhoeven, T.; Brinkmann, B.H.; Dambre, J. Towards improved design and evaluation of epileptic seizure predictors. IEEE Trans. Biomed. Eng. 2018, 65, 502–510. [Google Scholar] [CrossRef] [Green Version]
- Truong, N.D.; Nguyen, A.D.; Kuhlmann, L.; Bonyadi, M.R.; Yang, J.; Ippolito, S.; Kavehei, O. Convolutional neural networks for seizure prediction using intracranial and scalp electroencephalogram. Neural Netw. 2018, 105, 104–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varatharajah, Y.; Iyer, R.K.; Berry, B.M.; Worrell, G.A.; Brinkmann, B.H. Seizure Forecasting and the Preictal State in Canine Epilepsy. Int. J. Neural Syst. 2017, 27, 1650046. [Google Scholar] [CrossRef]
- Chaovalitwongse, W.; Iasemidis, L.D.; Pardalos, P.M.; Carney, P.R.; Shiau, D.S.; Sackellares, J.C. Performance of a seizure warning algorithm based on the dynamics of intracranial EEG. Epilepsy Res. 2005, 64, 93–113. [Google Scholar] [CrossRef]
- Iasemidis, L.D.; Pardalos, P.; Sackellares, J.C.; Shiau, D.S. Quadratic Binary Programming and Dynamical System Approach to Determine the Predictability of Epileptic Seizures. J. Comb. Optim. 2001, 5, 9–26. [Google Scholar] [CrossRef]
- Nejedly, P.; Kremen, V.; Sladky, V.; Nasseri, M.; Guragain, H.; Klimes, P.; Cimbalnik, J.; Varatharajah, Y.; Brinkmann, B.H.; Worrell, G.A. Deep-learning for seizure forecasting in canines with epilepsy. J. Neural Eng. 2019, 16, 036031. [Google Scholar] [CrossRef]
- Cook, M.J.; O’Brien, T.J.; Berkovic, S.F.; Murphy, M.; Morokoff, A.; Fabinyi, G.; D’Souza, W.; Yerra, R.; Archer, J.; Litewka, L.; et al. Prediction of seizure likelihood with a long-term, implanted seizure advisory system in patients with drug-resistant epilepsy: A first-in-man study. Lancet Neurol. 2013, 12, 563–571. [Google Scholar] [CrossRef]
- Kuhlmann, L.; Karoly, P.; Freestone, D.R.; Brinkmann, B.H.; Temko, A.; Barachant, A.; Li, F.; Titericz, G.; Lang, B.W.; Lavery, D.; et al. Epilepsyecosystem.org: Crowd-sourcing reproducible seizure prediction with long-term human intracranial EEG. Brain 2018, 141, 2619–2630. [Google Scholar] [CrossRef] [Green Version]
- Gardner, A.B.; Krieger, A.M.; Vachtsevanos, G.; Litt, B. One-class novelty detection for seizure analysis from intracranial EEG. J. Mach. Learn. Res. 2006, 7, 1025–1044. [Google Scholar] [CrossRef]
- Fan, R.-E.; Chang, K.-W.; Hsieh, C.-J.; Wang, X.-R.; Lin, C.-J. LIBLINEAR: A Library for Large Linear Classification. J. Mach. Learn. Res. 2008, 9, 1871–1874. [Google Scholar] [CrossRef] [Green Version]
- Netoff, T.; Park, Y.; Parhi, K. Seizure prediction using cost-sensitive support vector machine. In Proceedings of the 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Minneapolis, MN, USA, 2–6 September 2009; pp. 3322–3325. [Google Scholar]
- Cherkassky, V.; Mulier, F. Learning from Data: Concepts, Theory and Methods, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2007; ISBN 9780470140529. [Google Scholar]
- Lin, Y.; Lee, Y.; Wahba, G. Support vector machines for classification in nonstandard situations. Mach. Learn. 2002, 46, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Domingos, P. MetaCost: A General Method for Making Classifiers Cost-Sensitive. In Proceedings of the Fifth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Diego, CA, USA, 15–18 August 1999; pp. 155–164. [Google Scholar]
- Elkan, C. The Foundations of Cost-Sensitive Learning The Foundations of Cost-Sensitive Learning. In Proceedings of the International Joint Conference on Artificial Intelligence, Seattle, WA, USA, 4–10 August 2001; Morgan Kaufmann Publishers Inc.: San Francisco, CA, USA, 2001; Volume 17, pp. 973–978. [Google Scholar]
- He, H.; Bai, Y.; Garcia, E.A.; Li, S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. In Proceedings of the 2008 IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–8 June 2008; pp. 1322–1328. [Google Scholar]
- Provost, F. Machine learning from imbalanced data sets 101. In Proceedings of the AAAI’2000 Workshop on Imbalanced Data Sets, Austin, TX, USA, 31 July 2000; AAAI Press: Menlo Park, CA, USA, 2000; Volume 68, pp. 1–3. [Google Scholar]
- Cherkassky, V. Predictive Learning. 2013. Available online: VCtextbook.com (accessed on 2 February 2017).
- Lotte, F.; Congedo, M.; Lécuyer, A.; Lamarche, F.; Arnaldi, B. A review of classification algorithms for EEG-based brain-computer interfaces. J. Neural Eng. 2007, 4, R1. [Google Scholar] [CrossRef]

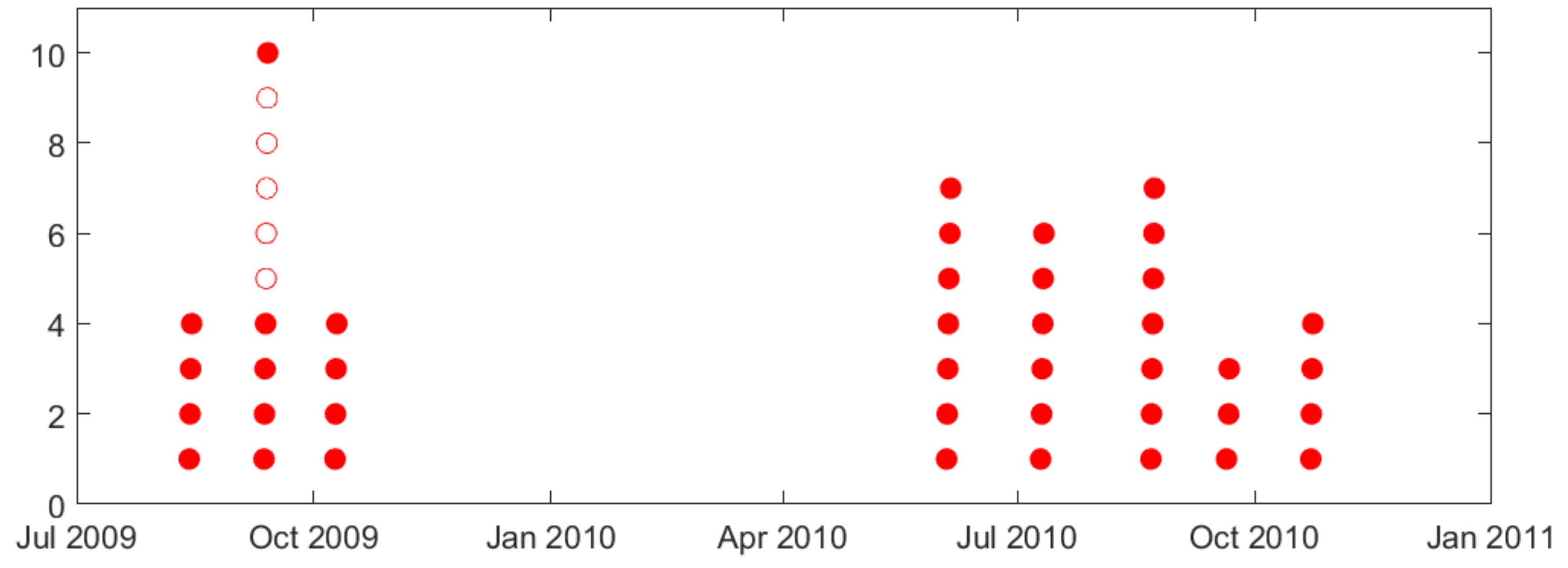
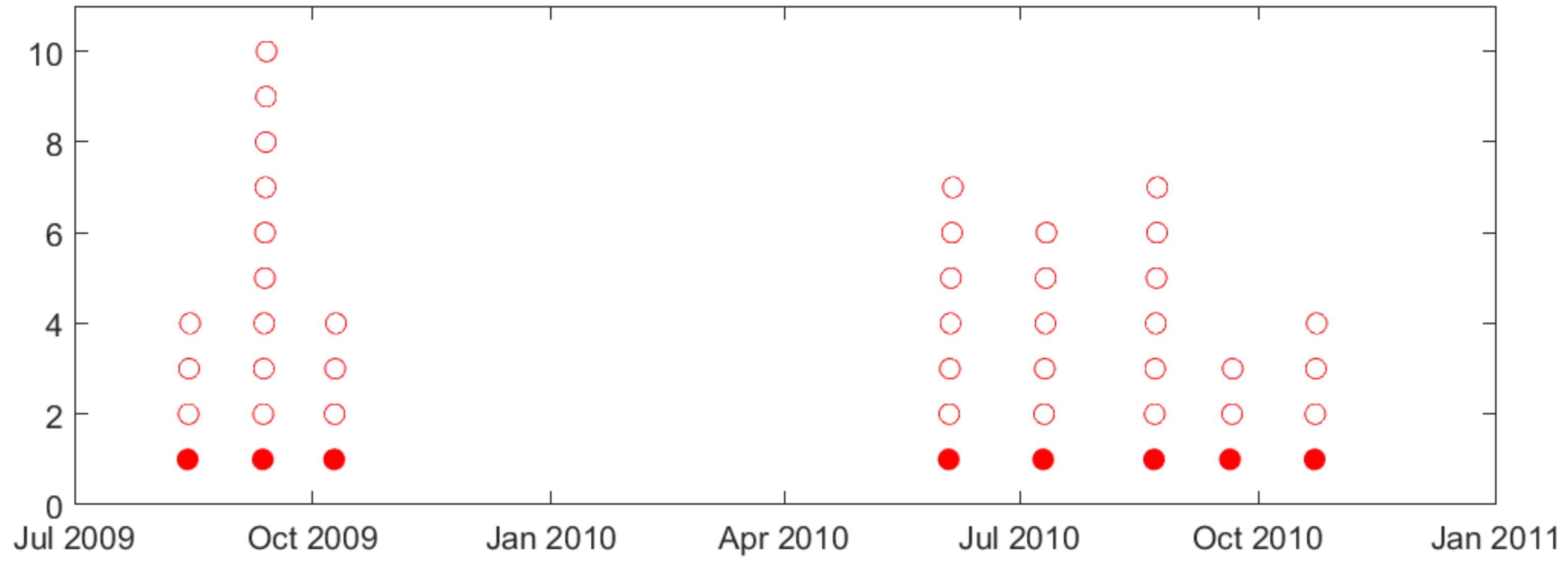
| (a) | ||||
|---|---|---|---|---|
| Dog ID | Recording Duration (Days) | Number of Annotated Seizures | Number of Gaps (>7 Days) | Number of Gaps (>1 h) |
| L2 | 475 | 45 | 4 | 87 |
| L7 | 451 | 105 | 4 | 26 |
| M3 | 394 | 29 | 4 | 155 |
| P2 | 294 | 22 | 7 | 93 |
| (b) | ||||
| Dog ID | Cluster Time Duration (Days) | Number of Seizures in Each Cluster (Ave.) | Number of Lead Seizures | Time between Lead Seizures (Ave. Days) |
| L2 | 1–2 | 5.6 | 8 | 62.2 |
| L7 | 1–2 | 17.2 | 8 | 62.9 |
| M3 | 1–4 | 2.8 | 22 | 16.6 |
| P2 | 0–1 | 4.4 | 5 | 33.6 |
| Dog ID | Test Period (Days) | Net Test Period (Days) | Number of Lead Seizures |
|---|---|---|---|
| L2 | 169 | 149 | 4 |
| L7 | 364 | 123 | 5 |
| M3 | 320 | 245 | 16 |
| P2 | 183 | 42 | 3 |
| Average | 259 ± 98 | 140 ± 84 | 7.0 ± 6.1 |
| (a) | |||
|---|---|---|---|
| Dog ID | Sensitivity | FPR/Day | TIW |
| L2 | 0.75 (3/4) | 0.85 | 0.27 |
| L7 | 0.80 (4/5) | 0.54 | 0.25 |
| M3 | 0.81 (13/16) | 0.70 | 0.29 |
| P2 | 1.00 (3/3) | 0.73 | 0.27 |
| Average | 0.84 ± 0.11 | 0.71 ± 0.13 | 0.27 ± 0.02 |
| (b) | |||
| Dog ID | Sensitivity | FPR/Day | TIW |
| L2 | 0.75 (3/4) | 0.96 | 0.40 |
| L7 | 0.80 (4/5) | 0.57 | 0.29 |
| M3 | 0.81 (13/16) | 0.88 | 0.37 |
| P2 | 1.00 (3/3) | 0.71 | 0.30 |
| Average | 0.84 ± 0.11 | 0.78 ± 0.17 | 0.34 ± 0.05 |
| Dog ID | Sensitivity | FPR/Day | TIW |
|---|---|---|---|
| L2 | 0.35 ± 0.49 | 0.61 ± 0.27 | 0.26 ± 0.21 |
| L7 | 0.64 ± 0.30 | 0.50 ± 0.11 | 0.35 ± 0.16 |
| M3 | 0.46 ± 0.14 | 0.63 ± 0.18 | 0.26 ± 0.14 |
| P2 | 0.67 ± 0.00 | 0.59 ± 0.19 | 0.19 ± 0.08 |
| Average | 0.53 ± 0.15 | 0.58 ± 0.06 | 0.26 ± 0.07 |
| Study | Number of Canines | Average Number of Lead Seizures | Sensitivity | TIW | FPR (Per Day) |
|---|---|---|---|---|---|
| Parameter T for lead seizures = 3 days | |||||
| This paper | 4 | 10.8 | 0.84 | 0.27 | 0.78 |
| Parameter T for lead seizures = 4 h | |||||
| Howbert et al. [27] | 3 | 17.7 | 0.60 | 0.30 | 2.06 |
| Brinkmann et al. [26] | 5 | 35.8 | 0.69 | 0.30 | 1.10 |
| Varatharajah et al. [30] | 5 | 35.8 | ~0.70 † | 0.25 | — |
| Nejedly et al. [33] | 4 | 42.8 | 0.79 | 0.18 | — |
| Parameter T for lead seizures = 80 min | |||||
| Howbert et al. [27] | 3 | 41.7 | 0.79 | 0.30 | 2.06 |
| Assi et al. [25] | 3 | 41.7 | 0.85 | 0.10 | — |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, H.-H.; Shiao, H.-T.; Cherkassky, V. Online Prediction of Lead Seizures from iEEG Data. Brain Sci. 2021, 11, 1554. https://doi.org/10.3390/brainsci11121554
Chen H-H, Shiao H-T, Cherkassky V. Online Prediction of Lead Seizures from iEEG Data. Brain Sciences. 2021; 11(12):1554. https://doi.org/10.3390/brainsci11121554
Chicago/Turabian StyleChen, Hsiang-Han, Han-Tai Shiao, and Vladimir Cherkassky. 2021. "Online Prediction of Lead Seizures from iEEG Data" Brain Sciences 11, no. 12: 1554. https://doi.org/10.3390/brainsci11121554
APA StyleChen, H.-H., Shiao, H.-T., & Cherkassky, V. (2021). Online Prediction of Lead Seizures from iEEG Data. Brain Sciences, 11(12), 1554. https://doi.org/10.3390/brainsci11121554






