Neural Decoding of EEG Signals with Machine Learning: A Systematic Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Research Questions (RQs)
- RQ1: What classification tasks have received the most attention with the introduction of ML and DL algorithms and the use of EEG brain data?
- RQ2: Which feature extraction methods were used for each task to extract appropriate inputs for ML and DL classifiers?
- RQ3: What are the most frequently used ML and DL algorithms for EEG data processing?
- RQ4: Which specific ML and DL models are suitable for classifying EEG data involving different types of tasks?
2.2. Search Strategy
2.3. Criteria for Identification of Studies
3. Theoretical Background
3.1. EEG Data Acquisition
3.2. Artifacts in EEG Signals and Preprocessing
3.2.1. Regression Methods
3.2.2. Blind Source Separation Methods
3.2.3. Wavelet Transform
3.2.4. Filtering Methods
Frequency Filtering
Adaptive Filtering
Wiener Filtering
3.3. Feature Extraction Methods
3.3.1. Principal Component Analysis
3.3.2. Autoregressive Model
3.3.3. Fast Fourier Transform
3.3.4. Wavelet Transform
3.3.5. Common Spatial Pattern
3.4. Classification Algorithms
3.4.1. Conventional Classification Algorithms
3.4.2. Deep Learning Algorithms
4. Results
4.1. Literature Search
4.2. Quality Assessment
4.3. Study Characteristics
4.4. Which Feature Extraction Methods Were Used for Each Task to Extract Appropriate Inputs for Machine Learning and Deep Learning Classifiers?
4.5. What Are the Most Frequently Used Machine Learning and Deep Learning Algorithms for EEG Data Processing?
5. Discussion
5.1. Emotion Recognition Task
5.2. Mental Workload Task
5.3. Motor Imagery Task
5.4. Seizure Detection Task
5.5. Sleep Stage Scoring Task
5.6. Neurodegenerative Disease Task
6. Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Article | Year | Task Information | Database | Feature Extraction Method | ML/DL Algorithm | Performance |
|---|---|---|---|---|---|---|
| [245] | 2010 | MWL 7 subj. 6 channels | Own database | Entropy | IFWSVM | Accuracy = 97.5 |
| [176] | 2020 | MI 30 channels | BCI Competition III and IV | CSP | SVM | Accuracy = 92.2 |
| [239] | 2018 | MWL 8 subj. 1 channel | Own database | FFT | Cubic SVM KNN LDA | Accuracy = 95 |
| [81] | 2014 | MWL 50 subj. | Own database | FFT | ANNs | Accuracy = 88.9 |
| [284] | 2019 | AD 100 subj. 24 channels | Own database | Entropy | SVM KNN | Accuracy = 96 |
| [244] | 2016 | MWL 6 subj. | Own database | HHT | SVM LDA QDA KNN | Accuracy = 89.07 |
| [253] | 2010 | MWL 13 subj. 20 channels | Own database | PCA | RBF-SVM | - |
| [195] | 2019 | MI 11 subj. 3 channels | PhysioNet database | N/A | LR SVM KNN | Accuracy = 90 |
| [118] | 2016 | SD 22 subj. 128 channels | BONN database | WT | H-MSVM | Accuracy = 94 |
| [192] | 2017 | SD 24 subj. 23 channels | CHB-MIT database | EMD | SVM | Sensitivity = 92.2 |
| [191] | 2011 | SD 22 subj. | CHB-MIT database | N/A | SVM | Sensitivity = 96 |
| [102] | 2018 | AD 109 subj. 19 channels | Own database | DWT/FFT | DT J48 | Accuracy = 91.7 |
| [243] | 2009 | MWL 4 subj. 6 channels | Own database | AR/DWT | DSTKNN | Accuracy = 93.04 |
| [279] | 2008 | SS 7 subj. | Sleep-EDF database | WT | MLP | Accuracy = 93 |
| [248] | 2012 | MWL 6 subj. 6 channels | Own database | EMD | SVM LDC QDC KNN | Accuracy = 94.3 |
| [104] | 2018 | MI 5 subj. | BCI Competition III | WPD | RF KNN SVM | Accuracy = 98.45 |
| [225] | 2020 | ER 40 subj. 14 channels | Own database | TQWT | PNN ELM KNN RF DT | Accuracy = 96.16 |
| [179] | 2018 | SD 25 subj. 100 channels | BONN database | DWT | RBF-SVM KNN NB | Accuracy = 100 |
| [275] | 2007 | SS 41 subj. 1 channel | Own database | FFT | MLP KNN | Accuracy = 71.56 |
| [32] | 2019 | MWL 8 subj. 14 channels | Own database | N/A | XGBoost MLP KNN SVM DT NB | Accuracy = 88 |
| [183] | 2020 | SD | BONN database | DWT | ANNs SVM KNN NB | Accuracy = 97.82 |
| [194] | 2013 | MI 6 subj. 8 channels | PhysioNet database | ICA | SVM ANNs | Accuracy = 97.1 |
| [54] | 2008 | MWL 15 subj. 60 channels | Own database | PCA | SVM | Accuracy = 71.7 |
| [251] | 2007 | MWL 7 subj. 6 channels | Own database | AR | RBF-SVM | Accuracy = 70 |
| [229] | 2020 | ER 28 subj. 64 channels | Own database | DWT/EMD | ANNs KNN SVM | Accuracy = 94.3 |
| [297] | 2016 | ERP 52 subj. 64 channels | Own database | WT | SVM | Accuracy = 87 |
| [151] | 2014 | ERP 7 subj. 49 channels | Own database | ICA | SVM | Accuracy = 87 |
| [228] | 2004 | ER 12 subj. 3 channels | Own database | Statistics | SVM | Accuracy = 66.7 |
| [249] | 2011 | MWL 6 subj. 32 channels | Own database | PCA | SVR RBF-NN LR | Accuracy = 86.92 |
| [252] | 2006 | MWL 7 subj. | Own database | AR | SVM ELM | Accuracy = 67.57 |
| [274] | 2021 | AD 32 subj. 19 channels | Own database | DWT | Bagging DT KNN SVM | Accuracy = 96.5 |
| [286] | 2014 | AD 100 subj. 19 channels | Own database | FFT | DT J48 SVM | Accuracy = 90 |
| [189] | 2015 | SD 22 subj. | CHB-MIT database | PCA | KNN | Sensitivity = 93 |
| [218] | 2007 | ER 17 subj. | Own database | N/A | KNN | Accuracy = 82.27 |
| [61] | 2017 | MI 5 subj. 30 channels | BCI Competition III | DWT/AR | NB LDA SVM | Accuracy = 95.47 |
| [174] | 2013 | MI 3 channels | BCI Competition II | WT | SVM LDA MLP | Accuracy = 90 |
| [84] | 2020 | MWL 20 subj. | Own database | HHT | SVM | Accuracy = 84.8 |
| [289] | 2019 | SZ 81 subj. 9 channels | Own database | N/A | RF | Accuracy = 81.1 |
| [298] | 2019 | MI 12 subj. 8 channels | Own database | N/A | ANNs | Accuracy = 90 |
| [212] | 2021 | SZ 28 subj. 19 channels | Own database | RMSFMS filter | KNN SVM Bagged trees Boosted trees | Accuracy = 99.21 |
| [199] | 2014 | SD 224 subj. 6 channels | European Epilepsy database | WT | ANNs SVM | Sensitivity = 73.08 |
| [149] | 2017 | ERP 108 subj. | Own database | TFHA | SVM-RFE | Accuracy = 99 |
| [207] | 2012 | SD 19 subj. 6 channels | Freiburg database | PCA | SVM | Sensitivity = 85.5 |
| [276] | 2015 | SS 15 subj. 21 channels | Own database | Entropy | Dendrogram-SVM | Accuracy = 92 |
| [55] | 2018 | MWL 8 subj. 14 channels | Own database | PCA | KNN SVM LR DT | Accuracy = 70.6 |
| [167] | 2018 | ER 32 subj. 32 channels | DEAP database | DWT | KNN | Accuracy = 87.1 |
| [261] | 2020 | MI 9 subj. 22 channels | BCI Competition IV | MRC | MLP | Accuracy = 76 |
| [277] | 2011 | SS 20 subj. 6 channels | Own database | Entropy/AR | LDA | Sensitivity = 89.1 |
| [202] | 2019 | SS 67 subj. 2 channels | Sleep-EDF database | WT | Dendrogram-SVM | Accuracy = 91.4 |
| [31] | 2012 | ER 32 subj. 32 channels | DEAP database | N/A | NB | Accuracy = 66.5 |
| [299] | 2016 | MWL 15 subj. 3 channels | Own database | N/A | SVM KNN ANNs | Accuracy = 95.21 |
| [33] | 2018 | Depression 23 subj. 19 channels | Own database | HFD/Entropy | MLP LR SVM DT RF NB | Accuracy = 97.56 |
| [58] | 2019 | AD 189 subj. 19 channels | Own database | CWT | MLP LR SVM | Accuracy = 95.76 |
| [164] | 2020 | ER 32 subj. 7 channels | DEAP database | PCA | KNN LR DT SVM LDA | Accuracy = 74.25 |
| [124] | 2021 | MI 23 channels | BCI Competition IV | CSP | LDA | Accuracy = 89.84 |
| [163] | 2018 | ER 32 subj. 10 channels | DEAP database | PCA | RBF-SVM KNN ANNs | Accuracy = 91.2 |
| [52] | 2018 | MI 9 subj. 2 channels | BCI Competition IV | DWT | NB KNN LDA | Accuracy = 73 |
| [300] | 2010 | MI 6 channels | N/A | Statistics | LDA BPNN SVM | Accuracy = 88.6 |
| [162] | 2019 | ER 32 subj. 32 channels | DEAP database | Entropy | RF RBF-SVM LDA KNN | Accuracy = 90 |
| [301] | 2016 | MWL 20 subj. 63 channels | Own database | Statistics | SVM | Accuracy = 70 |
| [168] | 2020 | ER 32 subj. 14 channels | DEAP database | Statistics | SVM KNN DT | Accuracy = 77.6–78.9 |
| [153] | 2019 | Anxiety 28 subj. 4 channels | Own database | N/A | RF LR MLP | Accuracy = 78.5 |
| [66] | 2016 | Depression 27 subj. 30 channels | Own database | WT/Statistics | RBF-SVM | Accuracy > 80 |
| [152] | 2012 | ERP 3 subj. 14 channels | Own database | Statistics | ANNs | Accuracy = 80 |
| [203] | 2016 | SS | Sleep-EDF database | EMD | AdaBoost NB LDA ANNs SVM KNN | Accuracy = 92.24 |
| [53] | 2019 | MI 6 subj. 14 channels | Own database | CSP | SVM | - |
| [182] | 2015 | SD 22 subj. 100 channels | BONN database | EMD | SVM | - |
| [34] | 2020 | MI 8 subj. 8 channels | Own database | CSP | LDA | Accuracy = 74.69 |
| [159] | 2019 | Sleep Apnea 16 subj. | MIT-BIH database | HHT | SVM KNN ANNs | Accuracy = 99 |
| [209] | 2018 | PD 20 subj. 14 channels | Own database | HOS | SVM DT KNN NB PNN | Accuracy = 99.6 |
| [219] | 2009 | ER 10 subj. 62 channels | Own database | CSP | SVM | Accuracy = 93.5 |
| [226] | 2008 | ER 10 subj. | Own database | Statistics | SVM ANNs NB | Accuracy = 80 |
| [59] | 2020 | SS 125 subj. | Sleep-EDF database | Entropy | RF SVM DT | Accuracy = 97.8 |
| [180] | 2018 | SD 25 subj. 100 channels | BONN database | PCA | RBF-SVM | Accuracy = 100 |
| [185] | 2021 | SD | BONN database | VMD | RF | Accuracy = 98.7–100 |
| [204] | 2020 | SS 2 channels | Sleep-EDF database | FFT | RBF-SVM | Accuracy = 87.8 |
| [196] | 2020 | MWL 36 subj. 19 channels | PhysioBank database | FFT | KNN SVM | Accuracy = 99.4 |
| [154] | 2020 | Stress 33 subj. 5 channels | Own database | FFT | SVM LR NB KNN DT | Accuracy = 85.2 |
| [165] | 2020 | ER 32 subj. | DEAP database | STFT | SVM KNN DT | - |
| [210] | 2016 | MI 10 subj. 64 channels | Own database | Kolmogorov complexity (Kc) | AdaBoost-ELM | Accuracy = 79.5 |
| [260] | 2005 | MI 4 subj. 62 channels | Own database | ICA/PCA | SVM ANNs | Accuracy = 77.3 |
| [171] | 2015 | MI | BCI Competition III and IV | CSP | Twin-SVM | Accuracy = 100 |
| [150] | 2017 | ERP 69 subj. 4 channels | Own database | FFT | GB RF RBF-SVM | Accuracy = 74 |
| [287] | 2019 | PD 18 subj. 128 channels | Own database | FFT | KNN SVM | Accuracy = 88 |
| [258] | 2020 | MI 5 subj. 64 channels | Own database | CSP | LDA SVM LR NB | Accuracy = 81 |
| [259] | 2020 | MI | BCI Competition III database (1) Autocalibration and Recurrent Adaptation database (2) | WPD | SVM LDA KNN | Accuracy = 93.46 (1) Accuracy = 86 (2) |
| [157] | 2016 | Tinnitus 22 subj. 129 channels | Own database | FFT | SVM | Accuracy = 90.9 |
| [278] | 2016 | SS 5 subj. | Own database | FFT/DWT | AdaBoost | - |
| [198] | 2017 | SD 216 subj. 6 channels | European Epilepsy database | DWT | SVM | - |
| [156] | 2020 | Alcoholism detection 2 subj. 64 channels | UCI database | EWD | SVM NB KNN | Accuracy = 98.75 |
| [155] | 2013 | Depression 90 subj. 19 channels | Own database | FFT | LR LDA KNN | Accuracy = 90 |
| [267] | 2020 | SD | Own database | DWT | RBF-SVM KNN DT LR | Accuracy = 100 |
| [119] | 2019 | ER 6 subj. 32 channels | DEAP database | Statistics | RF RBF-SVM KNN NB ANNs DT | Accuracy = 98.2 |
| [117] | 2011 | AD 32 subj. | Own database | FFT | SVM | Accuracy = 86.97 |
| [116] | 2020 | SD 24 subj. 22 channels | N/A | EMD | SVM | Accuracy > 90 |
| [191] | 2016 | SD 22 subj. | CHB-MIT database | PCA | RF | Accuracy = 98.3 |
| [166] | 2013 | ER 32 subj. 32 channels | DEAP database | PCA | ANNs RBF-SVM | Accuracy > 60 |
| [211] | 2018 | MWL 10 subj. 14 channels | Own database | FFT/WT | RF SVM MLP | Accuracy = 85–99 |
| [200] | 2015 | SD 24 subj. 6 channels | European Epilepsy database | MDADH | SVM | - |
| [230] | 2020 | ER 20 subj. 24 channels | Own database | TQWT | MC-LSVM | Accuracy = 95.7 |
| [175] | 2019 | MI 4 subj. 59 channels | BCI Competition 2008 | CSP | BPNN SVM | Accuracy = 91.6 |
| [186] | 2021 | SD | BONN database | Dissimilarity-based TFD | LDA ANNs SVM | Accuracy = 98 |
| [240] | 2018 | MWL | OneR database | FFT/Entropy | RF SVM | Accuracy = 87.2 |
| [65] | 2011 | ER 20 subj. 62 channels | Own database | DWT | KNN | Accuracy = 82.87 |
| [241] | 2017 | MWL | Own database | FFT | KNN SVM ANNs | Accuracy = 90.5 |
| [173] | 2014 | MI 7 subj. 22 channels | BCI Competition 2008 | EMD | RBF-SVM | Accuracy = 100 |
| [181] | 2017 | SD 10 subj. | BONN database | WPD | ELM | Accuracy = 97.7 |
| [268] | 2017 | SD 23 subj. | Own database | FFT | KNN | Sensitivity = 80.9 |
| [187] | 2018 | SD | BONN database | 1D-TP | RF SVM ANNs | Accuracy > 94 |
| [227] | 2019 | ER 28 subj. 2 channels | Own database | N/A | KNN MLP RF | Accuracy = 86.7 |
| [172] | 2017 | MI 5 subj. | BCI Competition III | CSP/DWT | LDA SVM ANNs | Accuracy = 84.8 |
| [269] | 2020 | SD 10 subj. | Own database | DFT | WBCKNN | Accuracy = 99 |
| [160] | 2020 | Creativity 20 subj. 32 channels | Own database | CSP | QDA SVM | Accuracy = 82 |
| [250] | 2020 | MWL 7 subj. 6 channels | Keirn and Aunon database | WT/EMD | SVM KNN | Accuracy = 80–100 |
| [184] | 2020 | SD 10 subj. | BONN database | FT/WT | SVM KNN | Accuracy = 100 |
| [158] | 2020 | ADHD 97 subj. 19 channels | Own database | PSR | NDC EPNN SVM | Accuracy = 100 |
| [177] | 2020 | MI 5 subj. | BCI Competition III | PCA | H-KELM | Accuracy = 96.5 |
| [224] | 2017 | ER 32 subj. 8 channels | DEAP database | STFT | CNN ReLU and Softmax (FC) as activation functions | Accuracy = 87.3 |
| [285] | 2016 | AD | Own database | WT | MC-DCNN Sigmoid and Softmax (FC) | Accuracy = 82 |
| [302] | 2020 | MI | BCI Competition IV | CSP | DNN | Accuracy = 83.98 |
| [262] | 2020 | MI | BCI Competition IV (1) TJU database (2) | SSD | CNN ReLU as activation functions | Accuracy = 79.3 (1) Accuracy = 85.7 (2) |
| [265] | 2019 | SD | BONN database (1) Bern-Barcelon database (2) | N/A | Residual-CNN ReLU and Softmax (FC) as activation functions | Accuracy = 99 (1) Accuracy = 92 (2) |
| [247] | 2020 | MWL 48 subj. | STEW database | N/A | LSTM CNN + LSTM | Accuracy = 61.08 |
| [263] | 2021 | MI | BCI Competition IV (1) HGD (2) | N/A | CNN | Accuracy = 81.6 (1) Accuracy = 95.5 (2) |
| [266] | 2018 | SD 5 subj. | BONN database | N/A | P-1D-CNN ReLU and Softmax (FC) as activation functions | Accuracy = 99.1 |
| [242] | 2018 | MWL 15 subj. | Own database | FFT | CNN ReLU and Softmax (FC) as activation functions | Accuracy = 90 |
| [264] | 2019 | MI | BCI Competition IV (1) Own database (2) | STFT | CNN-VAE | Kappa = 0.564 (1) Kappa = 0.603 (2) |
References
- Herculano-Houzel, S. The human brain in numbers: A linearly scaled-up primate brain. Front. Hum. Neurosci. 2009, 3, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pakkenberg, B.; Pelvig, D.; Marner, L.; Bundgaard, M.J.; Gundersen, H.J.G.; Nyengaard, J.R.; Regeur, L. Aging and the human neocortex. Exp. Gerontol. 2003, 38, 95–99. [Google Scholar] [CrossRef]
- Al-Kadi, M.I.; Reaz, M.B.I.; Ali, M.A.M. Evolution of electroencephalogram signal analysis techniques during anesthesia. Sensors 2013, 13, 6605–6635. [Google Scholar] [CrossRef] [Green Version]
- Molfese, D.L.; Molfese, V.J.; Kelly, S. The use of brain electrophysiology techniques to study language: A basic guide for the beginning consumer of electrophysiology information. Learn. Disabil. Q. 2001, 24, 177–188. [Google Scholar] [CrossRef]
- Kayser, J.; Tenke, C.E. In search of the Rosetta Stone for scalp EEG: Converging on reference-free techniques. Clin. Neurophysiol. Off. J. Int. Fed. Clin. Neurophysiol. 2010, 121, 1973. [Google Scholar] [CrossRef] [Green Version]
- Ismail, L.E.; Karwowski, W. A graph theory-based modeling of functional brain connectivity based on eeg: A systematic review in the context of neuroergonomics. IEEE Access 2020, 8, 155103–155135. [Google Scholar] [CrossRef]
- Henry, J.C. Electroencephalography: Basic principles, clinical applications, and related fields. Neurology 2006, 67, 2092. [Google Scholar] [CrossRef]
- Buzsaki, G. Rhythms of the Brain; Oxford University Press: Oxford, UK, 2006. [Google Scholar]
- Douglas, P.K.; Douglas, D.B. Reconsidering Spatial Priors In EEG Source Estimation: Does White Matter Contribute to EEG Rhythms? In Proceedings of the 2019 7th International Winter Conference on Brain-Computer Interface (BCI), Gangwon, Korea, 18–20 February 2019; pp. 1–12. [Google Scholar]
- Kumral, D.; Cesnaite, E.; Beyer, F.; Hofmann, S.M.; Hensch, T.; Sander, C.; Hegerl, U.; Haufe, S.; Villringer, A.; Witte, A.V.; et al. Relationship between Regional White Matter Hyperintensities and Alpha Oscillations in Older Adults. bioRxiv 2021. [Google Scholar] [CrossRef]
- Siuly, S.; Li, Y.; Zhang, Y. EEG signal analysis and classification. IEEE Trans. Neural Syst. Rehabil. Eng. 2016, 11, 141–144. [Google Scholar]
- Cohen, M.X. Analyzing Neural Time Series Data: Theory and Practice; MIT Press: Cambridge, MA, USA, 2014. [Google Scholar]
- Teplan, M. Fundamentals of EEG measurement. Meas. Sci. Rev. 2002, 2, 1–11. [Google Scholar]
- Kerr, W.T.; Anderson, A.; Lau, E.P.; Cho, A.Y.; Xia, H.; Bramen, J.; Douglas, P.K.; Braun, E.S.; Stern, J.M.; Cohen, M.S. Automated diagnosis of epilepsy using EEG power spectrum. Epilepsia 2012, 53, e189–e192. [Google Scholar] [CrossRef] [Green Version]
- Cooray, G.K.; Sengupta, B.; Douglas, P.; Englund, M.; Wickstrom, R.; Friston, K. Characterising seizures in anti-NMDA-receptor encephalitis with dynamic causal modelling. Neuroimage 2015, 118, 508–519. [Google Scholar] [CrossRef] [Green Version]
- Grosse-Wentrup, M.; Liefhold, C.; Gramann, K.; Buss, M. Beamforming in noninvasive brain–computer interfaces. IEEE Trans. Biomed. Eng. 2009, 56, 1209–1219. [Google Scholar] [CrossRef]
- Douglas, P.; Lau, E.; Anderson, A.; Kerr, W.; Head, A.; Wollner, M.A.; Moyer, D.; Durnhofer, M.; Li, W.; Bramen, J.; et al. Single trial decoding of belief decision making from EEG and fMRI data using independent components features. Front. Hum. Neurosci. 2013, 7, 392. [Google Scholar] [CrossRef] [Green Version]
- Kriegeskorte, N.; Douglas, P.K. Interpreting encoding and decoding models. Curr. Opin. Neurobiol. 2019, 55, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Hasenstab, K.; Scheffler, A.; Telesca, D.; Sugar, C.A.; Jeste, S.; DiStefano, C.; Şentürk, D. A multi-dimensional functional principal components analysis of EEG data. Biometrics 2017, 73, 999–1009. [Google Scholar] [CrossRef] [Green Version]
- Craik, A.; He, Y.; Contreras-Vidal, J.L. Deep learning for electroencephalogram (EEG) classification tasks: A review. J. Neural Eng. 2019, 16, 031001. [Google Scholar] [CrossRef] [PubMed]
- Gilles, J.; Meyer, T.; Douglas, P. Leveraging Sparsity: A Low-Rank+ Sparse Decomposition (LR+ SD) Method for Automatic EEG Artifact Removal. Proc. Int. Work. Sparsity Tech. Med. Imaging 2014, 2014, 80–88. [Google Scholar]
- Bigdely-Shamlo, N.; Mullen, T.; Kothe, C.; Su, K.M.; Robbins, K.A. The PREP pipeline: Standardized preprocessing for large-scale EEG analysis. Front. Neuroinform. 2015, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Jas, M.; Engemann, D.A.; Bekhti, Y.; Raimondo, F.; Gramfort, A. Autoreject: Automated artifact rejection for MEG and EEG data. NeuroImage 2017, 159, 417–429. [Google Scholar] [CrossRef] [Green Version]
- Cole, S.; Voytek, B. Cycle-by-cycle analysis of neural oscillations. J. Neurophysiol. 2019, 122, 849–861. [Google Scholar] [CrossRef]
- Gramfort, A.; Strohmeier, D.; Haueisen, J.; Hämäläinen, M.S.; Kowalski, M. Time-frequency mixed-norm estimates: Sparse M/EEG imaging with non-stationary source activations. NeuroImage 2013, 70, 410–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clerc, M.; Bougrain, L.; Lotte, F. Brain-Computer Interfaces 1: Methods and Perspectives; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Sen, P.C.; Hajra, M.; Ghosh, M. Supervised classification algorithms in machine learning: A survey and review. In Emerging Technology in Modelling and Graphics; Springer: Berlin/Heidelberg, Germany, 2020; pp. 99–111. [Google Scholar]
- Kumar, S.; Yger, F.; Lotte, F. Towards adaptive classification using Riemannian geometry approaches in brain-computer interfaces. In Proceedings of the 2019 7th International Winter Conference on Brain-Computer Interface (BCI), Gangwon, Korea, 18–20 February 2019; pp. 1–6. [Google Scholar]
- Lotte, F.; Bougrain, L.; Clerc, M. Electroencephalography (EEG)-Based Brain-Computer Interfaces; Wiley: Hoboken, NJ, USA, 2015. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Xiao, J.; Wu, C. Design of Deep Learning Model for Task-Evoked fMRI Data Classification. Comput. Intell. Neurosci. 2021, 2021, 6660866. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.Y.; Yoon, H.J. Affective classification using Bayesian classifier and supervised learning. In Proceedings of the 2012 12th International Conference on Control, Automation and Systems, Jeju, Korea, 17–21 October 2012; pp. 1768–1771. [Google Scholar]
- Anand, V.; Sreeja, S.; Samanta, D. An automated approach for task evaluation using EEG signals. arXiv 2019, arXiv:1911.02966. [Google Scholar]
- Cukic, M.; Pokrajac, D.; Stokic, M.; Radivojevic, V.; Ljubisavljevic, M. EEG machine learning with Higuchi fractal dimension and Sample Entropy as features for successful detection of depression. arXiv 2018, arXiv:1803.05985. [Google Scholar]
- Riquelme-Ros, J.V.; Rodríguez-Bermúdez, G.; Rodríguez-Rodríguez, I.; Rodríguez, J.V.; Molina-García-Pardo, J.M. On the better performance of pianists with motor imagery-based brain-computer interface systems. Sensors 2020, 20, 4452. [Google Scholar] [CrossRef]
- Garofalo, S.; Timmermann, C.; Battaglia, S.; Maier, M.E.; Di Pellegrino, G. Mediofrontal negativity signals unexpected timing of salient outcomes. J. Cogn. Neurosci. 2017, 29, 718–727. [Google Scholar] [CrossRef]
- Alexander, W.H.; Brown, J.W. Medial prefrontal cortex as an action-outcome predictor. Nat. Neurosci. 2011, 14, 1338–1344. [Google Scholar] [CrossRef]
- Battaglia, S.; Garofalo, S.; di Pellegrino, G.; Starita, F. Revaluing the role of vmPFC in the acquisition of Pavlovian threat conditioning in humans. J. Neurosci. 2020, 40, 8491–8500. [Google Scholar] [CrossRef] [PubMed]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [Green Version]
- Higgins, J.P.; Altman, D.G.; Gøtzsche, P.C.; Jüni, P.; Moher, D.; Oxman, A.D.; Savović, J.; Schulz, K.F.; Weeks, L.; Sterne, J.A. The Cochrane Collaboration’s tool for assessing risk of bias in randomised trials. BMJ 2011, 343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Wu, J.; Xia, Y.; He, Q.; Jin, H. Review of semi-dry electrodes for EEG recording. J. Neural Eng. 2020, 17, 051004. [Google Scholar] [CrossRef]
- Lopez-Gordo, M.A.; Sanchez-Morillo, D.; Valle, F.P. Dry EEG electrodes. Sensors 2014, 14, 12847–12870. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Wang, S.; Li, M.; Duan, Y.Y. Towards real-life EEG applications: Novel superporous hydrogel-based semi-dry EEG electrodes enabling automatically ‘charge–discharge’electrolyte. J. Neural Eng. 2021, 18, 046016. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Huang, S.; Xu, Z.; Wang, P.; Wu, X.; Zhang, D. Cognitive Workload Recognition Using EEG Signals and Machine Learning: A Review. IEEE Trans. Cogn. Dev. Syst. 2021. [Google Scholar] [CrossRef]
- Dehais, F.; Duprès, A.; Blum, S.; Drougard, N.; Scannella, S.; Roy, R.N.; Lotte, F. Monitoring pilot’s mental workload using ERPs and spectral power with a six-dry-electrode EEG system in real flight conditions. Sensors 2019, 19, 1324. [Google Scholar] [CrossRef] [Green Version]
- Hu, L.; Zhang, Z. EEG Signal Processing and Feature Extraction; Springer: Berlin/Heidelberg, Germany, 2019. [Google Scholar]
- Jiang, X.; Bian, G.B.; Tian, Z. Removal of artifacts from EEG signals: A review. Sensors 2019, 19, 987. [Google Scholar] [CrossRef] [Green Version]
- Johal, P.K.; Jain, N. Artifact removal from EEG: A comparison of techniques. In Proceedings of the 2016 International Conference on Electrical, Electronics, and Optimization Techniques (ICEEOT), Chennai, India, 3–5 March 2016; pp. 2088–2091. [Google Scholar]
- Urigüen, J.A.; Garcia-Zapirain, B. EEG artifact removal—state-of-the-art and guidelines. J. Neural Eng. 2015, 12, 031001. [Google Scholar] [CrossRef]
- Zhang, C.; Tong, L.; Zeng, Y.; Jiang, J.; Bu, H.; Yan, B.; Li, J. Automatic artifact removal from electroencephalogram data based on a priori artifact information. BioMed Res. Int. 2015, 2015, 720450. [Google Scholar] [CrossRef] [Green Version]
- Klados, M.A.; Papadelis, C.; Braun, C.; Bamidis, P.D. REG-ICA: A hybrid methodology combining blind source separation and regression techniques for the rejection of ocular artifacts. Biomed. Signal Process. Control 2011, 6, 291–300. [Google Scholar] [CrossRef]
- Cashero, Z. Comparison of EEG Preprocessing Methods to Improve the Classification of P300 Trials. Master’s Thesis, Colorado State University, Fort Collins, CO, USA, 2011. [Google Scholar]
- Maswanganyi, C.; Tu, C.; Owolawi, P.; Du, S. Discrimination of Motor Imagery Task using Wavelet Based EEG Signal Features. In Proceedings of the 2018 International Conference on Intelligent and Innovative Computing Applications (ICONIC), Mon Tresor, Mauritius, 6–7 December 2018; pp. 1–4. [Google Scholar]
- Zhang, Y.; Li, Q.; Yan, H.; Geng, X. The Research of the feature extraction and classification algorithm Based on EEG signal of motor imagery. In Proceedings of the 2019 International Conference on Virtual Reality and Intelligent Systems (ICVRIS), Jishou, China, 14–15 September 2019; pp. 187–190. [Google Scholar]
- Besserve, M.; Philippe, M.; Florence, G.; Laurent, F.; Garnero, L.; Martinerie, J. Prediction of performance level during a cognitive task from ongoing EEG oscillatory activities. Clin. Neurophysiol. 2008, 119, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Ghosh, R. Classification and Prediction of Human Cognitive Skills Using EEG Signals. In Proceedings of the 2018 Fourth International Conference on Biosignals, Images and Instrumentation (ICBSII), Chennai, India, 22–24 March 2018; pp. 206–212. [Google Scholar]
- Bin, G.; Gao, X.; Yan, Z.; Hong, B.; Gao, S. An online multi-channel SSVEP-based brain–computer interface using a canonical correlation analysis method. J. Neural Eng. 2009, 6, 046002. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, H.; Mandic, D. Blind source separation and artefact cancellation for single channel bioelectrical signal. In Proceedings of the 2016 IEEE 13th International Conference on Wearable and Implantable Body Sensor Networks (BSN), San Francisco, CA, USA, 14–17 June 2016; pp. 177–182. [Google Scholar]
- Ieracitano, C.; Mammone, N.; Bramanti, A.; Marino, S.; Hussain, A.; Morabito, F.C. A time-frequency based machine learning system for brain states classification via eeg signal processing. In Proceedings of the 2019 International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–8. [Google Scholar]
- Santaji, S.; Desai, V. Analysis of EEG signal to classify sleep stages using machine learning. Sleep Vigil. 2020, 4, 145–152. [Google Scholar] [CrossRef]
- Sifuzzaman, M.; Islam, M.R.; Ali, M. Application of Wavelet Transform and Its Advantages Compared to Fourier Transform; Vidyasagar University: Midnapore, India, 2009. [Google Scholar]
- Sreeja, S.; Rabha, J.; Nagarjuna, K.; Samanta, D.; Mitra, P.; Sarma, M. Motor imagery EEG signal processing and classification using machine learning approach. In Proceedings of the 2017 International Conference on New Trends in Computing Sciences (ICTCS), Amman, Jordan, 11–13 October 2017; pp. 61–66. [Google Scholar]
- Kotte, S.; Dabbakuti, J.K. Methods for removal of artifacts from EEG signal: A review. J. Phys. Conf. Ser. 2020, 1706, 012093. [Google Scholar] [CrossRef]
- Sweeney, K.T.; Ward, T.E.; McLoone, S.F. Artifact removal in physiological signals—Practices and possibilities. IEEE Trans. Inf. Technol. Biomed. 2012, 16, 488–500. [Google Scholar] [CrossRef]
- Kher, R.; Gandhi, R. Adaptive filtering based artifact removal from electroencephalogram (EEG) signals. In Proceedings of the 2016 International Conference on Communication and Signal Processing (ICCSP), Tamilnadu, India, 6–8 April 2016; pp. 561–564. [Google Scholar]
- Murugappan, M. Human emotion classification using wavelet transform and KNN. In Proceedings of the 2011 International Conference on Pattern Analysis and Intelligence Robotics, Kuala Lumpur, Malaysia, 28–29 June 2011; Volume 1, pp. 148–153. [Google Scholar]
- Puk, K.M.; Gandy, K.C.; Wang, S.; Park, H. Pattern classification and analysis of memory processing in depression using EEG signals. In Proceedings of the International Conference on Brain Informatics, Omaha, NE, USA, 13–16 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 124–137. [Google Scholar]
- Al-Fahoum, A.S.; Al-Fraihat, A.A. Methods of EEG signal features extraction using linear analysis in frequency and time-frequency domains. Int. Sch. Res. Not. 2014, 2014, 730218. [Google Scholar] [CrossRef] [Green Version]
- Aggarwal, S.; Chugh, N. Signal processing techniques for motor imagery brain computer interface: A review. Array 2019, 1, 100003. [Google Scholar] [CrossRef]
- Boostani, R.; Karimzadeh, F.; Nami, M. A comparative review on sleep stage classification methods in patients and healthy individuals. Comput. Methods Programs Biomed. 2017, 140, 77–91. [Google Scholar] [CrossRef]
- Torres, E.P.; Torres, E.A.; Hernández-Álvarez, M.; Yoo, S.G. EEG-based BCI emotion recognition: A survey. Sensors 2020, 20, 5083. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhang, X.; Wang, W.F.; Gu, Y. A parallel algorithm framework for feature extraction of EEG signals on MPI. Comput. Math. Methods Med. 2020, 2020, 9812019. [Google Scholar] [CrossRef] [PubMed]
- Aboalayon, K.A.I.; Faezipour, M.; Almuhammadi, W.S.; Moslehpour, S. Sleep stage classification using EEG signal analysis: A comprehensive survey and new investigation. Entropy 2016, 18, 272. [Google Scholar] [CrossRef]
- Lakshmi, M.R.; Prasad, T.; Prakash, D.V.C. Survey on EEG signal processing methods. Int. J. Adv. Res. Comput. Sci. Softw. Eng. 2014, 4, 84–91. [Google Scholar]
- Mane, A.R.; Biradar, S.; Shastri, R. Review paper on feature extraction methods for EEG signal analysis. Int. J. Emerg. Trend Eng. Basic Sci. 2015, 2, 545–552. [Google Scholar]
- Xie, Y.; Oniga, S. A Review of Processing Methods and Classification Algorithm for EEG Signal. Carpathian J. Electron. Comput. Eng. 2020, 12, 23–29. [Google Scholar] [CrossRef]
- Mirzaei, S.; Ghasemi, P. EEG motor imagery classification using dynamic connectivity patterns and convolutional autoencoder. Biomed. Signal Process. Control 2021, 68, 102584. [Google Scholar] [CrossRef]
- Rajaguru, H.; Prabhakar, S.K. Sparse PCA and soft decision tree classifiers for epilepsy classification from EEG signals. In Proceedings of the 2017 International Conference of Electronics, Communication and Aerospace Technology (ICECA), Coimbatore, India, 20–22 April 2017; Volume 1, pp. 581–584. [Google Scholar]
- Zhang, Y.; Liu, B.; Ji, X.; Huang, D. Classification of EEG signals based on autoregressive model and wavelet packet decomposition. Neural Process. Lett. 2017, 45, 365–378. [Google Scholar] [CrossRef]
- Hengstler, S.; Sand, S.; Costa, A.H. Adaptive autoregressive modeling for time-frequency analysis. In Proceedings of the Third International Conference on Information, Communications & Signal Processing (ICICS 2001), Chongqing, China, 19–21 November 2001; pp. 241–244. [Google Scholar]
- Shakeel, A.; Tanaka, T.; Kitajo, K. Time-series prediction of the oscillatory phase of EEG signals using the least mean square algorithm-based AR model. Appl. Sci. 2020, 10, 3616. [Google Scholar] [CrossRef]
- Jahidin, A.H.; Ali, M.M.; Taib, M.N.; Tahir, N.M.; Yassin, I.M.; Lias, S. Classification of intelligence quotient via brainwave sub-band power ratio features and artificial neural network. Comput. Methods Programs Biomed. 2014, 114, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Joshi, S.D.; Patney, R.K.; Saha, K. The Fourier decomposition method for nonlinear and non-stationary time series analysis. Proc. R. Soc. A Math. Phys. Eng. Sci. 2017, 473, 20160871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dragomiretskiy, K.; Zosso, D. Variational mode decomposition. IEEE Trans. Signal Process. 2013, 62, 531–544. [Google Scholar] [CrossRef]
- Peng, C.J.; Chen, Y.C.; Chen, C.C.; Chen, S.J.; Cagneau, B.; Chassagne, L. An EEG-based attentiveness recognition system using Hilbert–Huang transform and support vector machine. J. Med. Biol. Eng. 2020, 40, 230–238. [Google Scholar] [CrossRef] [Green Version]
- Jahankhani, P.; Kodogiannis, V.; Revett, K. EEG signal classification using wavelet feature extraction and neural networks. In Proceedings of the IEEE John Vincent Atanasoff 2006 International Symposium on Modern Computing (JVA’06), Sofia, Bulgaria, 3–6 October 2006; pp. 120–124. [Google Scholar]
- Wang, Y.; Veluvolu, K.C.; Lee, M. Time-frequency analysis of band-limited EEG with BMFLC and Kalman filter for BCI applications. J. Neuroeng. Rehabil. 2013, 10, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Rutkowski, G.; Patan, K.; Leśniak, P. Comparison of time-frequency feature extraction methods for EEG signals classification. In Proceedings of the International Conference on Artificial Intelligence and Soft Computing, Zakopane, Poland, 9–13 June 2013; Springer: Berlin/Heidelberg, Germany, 2013; pp. 320–329. [Google Scholar]
- Jin, J.; Xiao, R.; Daly, I.; Miao, Y.; Wang, X.; Cichocki, A. Internal feature selection method of CSP based on L1-norm and dempster-shafer theory. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 4814–4825. [Google Scholar] [CrossRef] [PubMed]
- Fu, R.; Han, M.; Tian, Y.; Shi, P. Improvement motor imagery EEG classification based on sparse common spatial pattern and regularized discriminant analysis. J. Neurosci. Methods 2020, 343, 108833. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, A.; Agrawal, R. Relevant feature extraction by combining independent components analysis and common spatial patterns for EEG based motor imagery classification. Int. J. Eng. Res. Technol. 2014, 3, 246–252. [Google Scholar]
- Blankertz, B.; Tomioka, R.; Lemm, S.; Kawanabe, M.; Muller, K.R. Optimizing spatial filters for robust EEG single-trial analysis. IEEE Signal Process. Mag. 2007, 25, 41–56. [Google Scholar] [CrossRef]
- Ang, K.K.; Chin, Z.Y.; Wang, C.; Guan, C.; Zhang, H. Filter bank common spatial pattern algorithm on BCI competition IV datasets 2a and 2b. Front. Neurosci. 2012, 6, 39. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Sharma, A.; Tsunoda, T. An improved discriminative filter bank selection approach for motor imagery EEG signal classification using mutual information. BMC Bioinform. 2017, 18, 545. [Google Scholar] [CrossRef] [Green Version]
- Lemm, S.; Blankertz, B.; Curio, G.; Muller, K.R. Spatio-spectral filters for improving the classification of single trial EEG. IEEE Trans. Biomed. Eng. 2005, 52, 1541–1548. [Google Scholar] [CrossRef]
- Cho, H.; Ahn, M.; Ahn, S.; Jun, S.C. Invariant common spatio-spectral patterns. In Proceedings of the 3rd TOBI Workshop, Würzburg, Germany, 20–22 March 2012; pp. 31–32. [Google Scholar]
- Dornhege, G.; Blankertz, B.; Krauledat, M.; Losch, F.; Curio, G.; Muller, K.R. Combined optimization of spatial and temporal filters for improving brain-computer interfacing. IEEE Trans. Biomed. Eng. 2006, 53, 2274–2281. [Google Scholar] [CrossRef]
- Novi, Q.; Guan, C.; Dat, T.H.; Xue, P. Sub-band common spatial pattern (SBCSP) for brain-computer interface. In Proceedings of the 2007 3rd International IEEE/EMBS Conference on Neural Engineering, Kohala Coast, HI, USA, 2–5 May 2007; pp. 204–207. [Google Scholar]
- Higashi, H.; Tanaka, T. Simultaneous design of FIR filter banks and spatial patterns for EEG signal classification. IEEE Trans. Biomed. Eng. 2012, 60, 1100–1110. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cang, S.; Yu, H. A survey on wearable sensor modality centred human activity recognition in health care. Expert Syst. Appl. 2019, 137, 167–190. [Google Scholar] [CrossRef]
- Bengio, Y.; Courville, A.C.; Vincent, P. Unsupervised feature learning and deep learning: A review and new perspectives. arXiv 2012, arXiv:1206.5538v1. [Google Scholar]
- Fiscon, G.; Weitschek, E.; Cialini, A.; Felici, G.; Bertolazzi, P.; De Salvo, S.; Bramanti, A.; Bramanti, P.; De Cola, M.C. Combining EEG signal processing with supervised methods for Alzheimer’s patients classification. BMC Med. Inform. Decis. Mak. 2018, 18, 35. [Google Scholar] [CrossRef] [PubMed]
- Haykin, S.; Network, N. A comprehensive foundation. Neural Netw. 2004, 2, 41. [Google Scholar]
- Behri, M.; Subasi, A.; Qaisar, S.M. Comparison of machine learning methods for two class motor imagery tasks using EEG in brain-computer interface. In Proceedings of the 2018 Advances in Science and Engineering Technology International Conferences (ASET). Abu Dhabi, United Arab Emirates, 6 February–5 April 2018; pp. 1–5. [Google Scholar]
- Tan, P.N.; Steinbach, M.; Kumar, V. Classification: Basic concepts, decision trees, and model evaluation. Introd. Data Min. 2006, 1, 145–205. [Google Scholar]
- Hosseini, M.P.; Hosseini, A.; Ahi, K. A Review on machine learning for EEG Signal processing in bioengineering. IEEE Rev. Biomed. Eng. 2020, 14, 204–218. [Google Scholar] [CrossRef]
- Oktavia, N.Y.; Wibawa, A.D.; Pane, E.S.; Purnomo, M.H. Human emotion classification based on EEG signals using Naïve bayes method. In Proceedings of the 2019 International Seminar on Application for Technology of Information and Communication (iSemantic), Semarang, Indonesia, 21–22 September 2019; pp. 319–324. [Google Scholar]
- Lestari, F.P.; Haekal, M.; Edison, R.E.; Fauzy, F.R.; Khotimah, S.N.; Haryanto, F. Epileptic Seizure Detection in EEGs by Using Random Tree Forest, Naïve Bayes and KNN Classification. J. Phys. Conf. Ser. 2020, 1505, 012055. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y. Detection of motor imagery EEG signals employing Naïve Bayes based learning process. Measurement 2016, 86, 148–158. [Google Scholar]
- Palaniappan, R.; Sundaraj, K.; Sundaraj, S. A comparative study of the svm and k-nn machine learning algorithms for the diagnosis of respiratory pathologies using pulmonary acoustic signals. BMC Bioinform. 2014, 15, 223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osowski, S.; Siwek, K.; Markiewicz, T. MLP and SVM networks-a comparative study. In Proceedings of the 6th Nordic Signal Processing Symposium, Espoo, Finland, 9–11 June 2004; pp. 37–40. [Google Scholar]
- Beyer, K.; Goldstein, J.; Ramakrishnan, R.; Shaft, U. When is “nearest neighbor” meaningful? In Proceedings of the International Conference on Database Theory, Jerusalem, Israel, 10–12 January 1999; Springer: Berlin/Heidelberg, Germany, 1999; pp. 217–235. [Google Scholar]
- Du, S.C.; Huang, D.L.; Wang, H. An adaptive support vector machine-based workpiece surface classification system using high-definition metrology. IEEE Trans. Instrum. Meas. 2015, 64, 2590–2604. [Google Scholar] [CrossRef]
- Huang, C.L.; Wang, C.J. A GA-based feature selection and parameters optimizationfor support vector machines. Expert Syst. Appl. 2006, 31, 231–240. [Google Scholar] [CrossRef]
- Wang, G.; Qiu, Y.F.; Li, H.X. Temperature forecast based on SVM optimized by PSO algorithm. In Proceedings of the 2010 International Conference on Intelligent Computing and Cognitive Informatics, Kuala Lumpur, Malaysia, 22–23 June 2010; pp. 259–262. [Google Scholar]
- Moctezuma, L.A.; Molinas, M. Classification of low-density EEG for epileptic seizures by energy and fractal features based on EMD. J. Biomed. Res. 2020, 34, 180. [Google Scholar] [CrossRef] [PubMed]
- Trambaiolli, L.R.; Lorena, A.C.; Fraga, F.J.; Kanda, P.A.; Anghinah, R.; Nitrini, R. Improving Alzheimer’s disease diagnosis with machine learning techniques. Clin. EEG Neurosci. 2011, 42, 160–165. [Google Scholar] [CrossRef]
- Murugavel, A.M.; Ramakrishnan, S. Hierarchical multi-class SVM with ELM kernel for epileptic EEG signal classification. Med. Biol. Eng. Comput. 2016, 54, 149–161. [Google Scholar] [CrossRef]
- Ramzan, M.; Dawn, S. Learning-based classification of valence emotion from electroencephalography. Int. J. Neurosci. 2019, 129, 1085–1093. [Google Scholar] [CrossRef]
- Hosmer, D.W., Jr.; Lemeshow, S.; Sturdivant, R.X. Applied Logistic Regression; John Wiley & Sons: Hoboken, NJ, USA, 2013; Volume 398. [Google Scholar]
- Bandos, T.V.; Bruzzone, L.; Camps-Valls, G. Classification of hyperspectral images with regularized linear discriminant analysis. IEEE Trans. Geosci. Remote Sens. 2009, 47, 862–873. [Google Scholar] [CrossRef]
- Tharwat, A.; Gaber, T.; Ibrahim, A.; Hassanien, A.E. Linear discriminant analysis: A detailed tutorial. AI Commun. 2017, 30, 169–190. [Google Scholar] [CrossRef] [Green Version]
- Lotte, F.; Congedo, M.; Lécuyer, A.; Lamarche, F.; Arnaldi, B. A review of classification algorithms for EEG-based brain–computer interfaces. J. Neural Eng. 2007, 4, R1. [Google Scholar] [CrossRef]
- Molla, M.K.I.; Saha, S.K.; Yasmin, S.; Islam, M.R.; Shin, J. Trial Regeneration With Subband Signals for Motor Imagery Classification in BCI Paradigm. IEEE Access 2021, 9, 7632–7642. [Google Scholar] [CrossRef]
- Bostanov, V. BCI competition 2003-data sets Ib and IIb: Feature extraction from event-related brain potentials with the continuous wavelet transform and the t-value scalogram. IEEE Trans. Biomed. Eng. 2004, 51, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Scherer, R.; Muller, G.; Neuper, C.; Graimann, B.; Pfurtscheller, G. An asynchronously controlled EEG-based virtual keyboard: Improvement of the spelling rate. IEEE Trans. Biomed. Eng. 2004, 51, 979–984. [Google Scholar] [CrossRef]
- Garcia, G.N.; Ebrahimi, T.; Vesin, J.M. Support vector EEG classification in the Fourier and time-frequency correlation domains. In Proceedings of the First International IEEE EMBS Conference on Neural Engineering, Capri, Italy, 20–22 March 2003; pp. 591–594. [Google Scholar]
- Balakrishnama, S.; Ganapathiraju, A.; Picone, J. Linear discriminant analysis for signal processing problems. In Proceedings of the IEEE Southeastcon’99. Technology on the Brink of 2000 (Cat. No. 99CH36300), Lexington, KY, USA, 25–28 March 1999; pp. 78–81. [Google Scholar]
- Sakhavi, S.; Guan, C.; Yan, S. Learning temporal information for brain-computer interface using convolutional neural networks. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 5619–5629. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, W.S.; Pitts, W. A logical calculus of the ideas immanent in nervous activity. Bull. Math. Biophys. 1943, 5, 115–133. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- van de Ven, G.M.; Siegelmann, H.T.; Tolias, A.S. Brain-inspired replay for continual learning with artificial neural networks. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef]
- Borgomaneri, S.; Battaglia, S.; Garofalo, S.; Tortora, F.; Avenanti, A.; di Pellegrino, G. State-dependent TMS over prefrontal cortex disrupts fear-memory reconsolidation and prevents the return of fear. Curr. Biol. 2020, 30, 3672–3679. [Google Scholar] [CrossRef]
- Parisi, G.I.; Kemker, R.; Part, J.L.; Kanan, C.; Wermter, S. Continual lifelong learning with neural networks: A review. Neural Netw. 2019, 113, 54–71. [Google Scholar] [CrossRef]
- Valliani, A.A.A.; Ranti, D.; Oermann, E.K. Deep learning and neurology: A systematic review. Neurol. Ther. 2019, 8, 351–365. [Google Scholar] [CrossRef] [Green Version]
- Abiodun, O.I.; Jantan, A.; Omolara, A.E.; Dada, K.V.; Mohamed, N.A.; Arshad, H. State-of-the-art in artificial neural network applications: A survey. Heliyon 2018, 4, e00938. [Google Scholar] [CrossRef] [Green Version]
- Al-Saegh, A.; Dawwd, S.A.; Abdul-Jabbar, J.M. Deep learning for motor imagery EEG-based classification: A review. Biomed. Signal Process. Control 2021, 63, 102172. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Maddikunta, P.K.R.; Pham, Q.V.; Gadekallu, T.R.; Chowdhary, C.L.; Alazab, M.; Piran, M.J. Deep learning and medical image processing for coronavirus (COVID-19) pandemic: A survey. Sustain. Cities Soc. 2021, 65, 102589. [Google Scholar] [CrossRef] [PubMed]
- Fiok, K.; Karwowski, W.; Gutierrez, E.; Saeidi, M.; Aljuaid, A.M.; Davahli, M.R.; Taiar, R.; Marek, T.; Sawyer, B.D. A Study of the Effects of the COVID-19 Pandemic on the Experience of Back Pain Reported on Twitter in the United States: A Natural Language Processing Approach. Int. J. Environ. Res. Public Health 2021, 18, 4543. [Google Scholar] [CrossRef] [PubMed]
- Fiok, K.; Karwowski, W.; Gutierrez-Franco, E.; Liciaga, T.; Belmonte, A.; Capobianco, R.; Saeidi, M. Automated Detection of Leadership Qualities Using Textual Data at the Message Level. IEEE Access 2021, 9, 57141–57148. [Google Scholar] [CrossRef]
- Faust, O.; Hagiwara, Y.; Hong, T.J.; Lih, O.S.; Acharya, U.R. Deep learning for healthcare applications based on physiological signals: A review. Comput. Methods Programs Biomed. 2018, 161, 1–13. [Google Scholar] [CrossRef]
- Miao, Y.; Gowayyed, M.; Metze, F. EESEN: End-to-end speech recognition using deep RNN models and WFST-based decoding. In Proceedings of the 2015 IEEE Workshop on Automatic Speech Recognition and Understanding (ASRU), Scottsdale, AZ, USA, 13–17 December 2015; pp. 167–174. [Google Scholar]
- Wiggins, W.F.; Kitamura, F.; Santos, I.; Prevedello, L.M. Natural Language Processing of Radiology Text Reports: Interactive Text Classification. Radiol. Artif. Intell. 2021, 2021, e210035. [Google Scholar] [CrossRef]
- Tan, J.H.; Hagiwara, Y.; Pang, W.; Lim, I.; Oh, S.L.; Adam, M.; San Tan, R.; Chen, M.; Acharya, U.R. Application of stacked convolutional and long short-term memory network for accurate identification of CAD ECG signals. Comput. Biol. Med. 2018, 94, 19–26. [Google Scholar] [CrossRef]
- Graves, A. Long short-term memory. In Supervised Sequence Labelling with Recurrent Neural Networks; Springer: Berlin/Heidelberg, Germany, 2012; pp. 37–45. [Google Scholar]
- Wang, P.; Jiang, A.; Liu, X.; Shang, J.; Zhang, L. LSTM-based EEG classification in motor imagery tasks. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 2086–2095. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Kiral-Kornek, I.; Harrer, S. ChronoNet: A deep recurrent neural network for abnormal EEG identification. In Proceedings of the Conference on Artificial Intelligence in Medicine in Europe, Poznan, Poland, 26–29 June 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 47–56. [Google Scholar]
- Krause, B.; Lu, L.; Murray, I.; Renals, S. Multiplicative LSTM for sequence modelling. arXiv 2016, arXiv:1609.07959. [Google Scholar]
- Öztoprak, H.; Toycan, M.; Alp, Y.K.; Arıkan, O.; Doğutepe, E.; Karakaş, S. Machine-based classification of ADHD and nonADHD participants using time/frequency features of event-related neuroelectric activity. Clin. Neurophysiol. 2017, 128, 2400–2410. [Google Scholar] [CrossRef]
- Papakostas, M.; Tsiakas, K.; Giannakopoulos, T.; Makedon, F. Towards predicting task performance from EEG signals. In Proceedings of the 2017 IEEE International Conference on Big Data (Big Data), Boston, MA, USA, 11–14 December 2017; pp. 4423–4425. [Google Scholar]
- Stewart, A.X.; Nuthmann, A.; Sanguinetti, G. Single-trial classification of EEG in a visual object task using ICA and machine learning. J. Neurosci. Methods 2014, 228, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vo, T.; Gedeon, T.; Tran, D. Modeling the mental differentiation task with EEG. In Proceedings of the International Conference on Neural Information Processing, Doha, Qatar, 12–15 November 2012; Springer: Berlin/Heidelberg, Germany, 2012; pp. 357–364. [Google Scholar]
- Arsalan, A.; Majid, M.; Anwar, S.M. Electroencephalography based machine learning framework for anxiety classification. In Proceedings of the International Conference on Intelligent Technologies and Applications, Bahawalpur, Pakistan, 6–8 November 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 187–197. [Google Scholar]
- Saeed, S.M.U.; Anwar, S.M.; Khalid, H.; Majid, M.; Bagci, U. EEG based classification of long-term stress using psychological labeling. Sensors 2020, 20, 1886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hosseinifard, B.; Moradi, M.H.; Rostami, R. Classifying depression patients and normal subjects using machine learning techniques and nonlinear features from EEG signal. Comput. Methods Programs Biomed. 2013, 109, 339–345. [Google Scholar] [CrossRef]
- Anuragi, A.; Sisodia, D.S. Empirical wavelet transform based automated alcoholism detecting using EEG signal features. Biomed. Signal Process. Control 2020, 57, 101777. [Google Scholar] [CrossRef]
- Li, P.Z.; Li, J.H.; Wang, C.D. A SVM-based EEG signal analysis: An auxiliary therapy for tinnitus. In Proceedings of the International Conference on Brain Inspired Cognitive Systems, Beijing, China, 28–30 November 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 207–219. [Google Scholar]
- Kaur, S.; Singh, S.; Arun, P.; Kaur, D.; Bajaj, M. Phase space reconstruction of EEG signals for classification of ADHD and control adults. Clin. EEG Neurosci. 2020, 51, 102–113. [Google Scholar] [CrossRef]
- Vimala, V.; Ramar, K.; Ettappan, M. An intelligent sleep apnea classification system based on EEG signals. J. Med. Syst. 2019, 43, 36. [Google Scholar] [CrossRef]
- Stevens Jr, C.E.; Zabelina, D.L. Classifying creativity: Applying machine learning techniques to divergent thinking EEG data. NeuroImage 2020, 219, 116990. [Google Scholar] [CrossRef]
- Koelstra, S.; Muhl, C.; Soleymani, M.; Lee, J.S.; Yazdani, A.; Ebrahimi, T.; Pun, T.; Nijholt, A.; Patras, I. Deap: A database for emotion analysis; using physiological signals. IEEE Trans. Affect. Comput. 2011, 3, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Bălan, O.; Moise, G.; Moldoveanu, A.; Leordeanu, M.; Moldoveanu, F. Fear level classification based on emotional dimensions and machine learning techniques. Sensors 2019, 19, 1738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bazgir, O.; Mohammadi, Z.; Habibi, S.A.H. Emotion recognition with machine learning using EEG signals. In Proceedings of the 2018 25th National and 3rd International Iranian Conference on Biomedical Engineering (ICBME), Qom, Iran, 29–30 November 2018; pp. 1–5. [Google Scholar]
- Doma, V.; Pirouz, M. A comparative analysis of machine learning methods for emotion recognition using EEG and peripheral physiological signals. J. Big Data 2020, 7, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Jaswanth, V.; Naren, J. A System for the Study of Emotions with EEG Signals Using Machine Learning and Deep Learning. In Cognitive Informatics and Soft Computing; Springer: Berlin/Heidelberg, Germany, 2020; pp. 59–65. [Google Scholar]
- Rozgić, V.; Vitaladevuni, S.N.; Prasad, R. Robust EEG emotion classification using segment level decision fusion. In Proceedings of the 2013 IEEE International Conference on Acoustics, Speech and Signal Processing, Vancouver, BC, Canada, 26–31 May 2013; pp. 1286–1290. [Google Scholar]
- Shukla, S.; Chaurasiya, R.K. Emotion Analysis Through EEG and Peripheral Physiological Signals Using KNN Classifier. In International Conference on ISMAC in Computational Vision and Bio-Engineering; Springer: Berlin/Heidelberg, Germany, 2018; pp. 97–106. [Google Scholar]
- Nawaz, R.; Cheah, K.H.; Nisar, H.; Yap, V.V. Comparison of different feature extraction methods for EEG-based emotion recognition. Biocybern. Biomed. Eng. 2020, 40, 910–926. [Google Scholar] [CrossRef]
- Blankertz, B.; Muller, K.R.; Krusienski, D.J.; Schalk, G.; Wolpaw, J.R.; Schlogl, A.; Pfurtscheller, G.; Millan, J.R.; Schroder, M.; Birbaumer, N. The BCI competition III: Validating alternative approaches to actual BCI problems. IEEE Trans. Neural Syst. Rehabil. Eng. 2006, 14, 153–159. [Google Scholar] [CrossRef]
- Leeb, R.; Brunner, C.; Müller-Putz, G.; Schlögl, A.; Pfurtscheller, G. BCI Competition 2008–Graz Data Set B; Graz University of Technology: Graz, Austria, 2008; pp. 1–6. [Google Scholar]
- Soman, S. High performance EEG signal classification using classifiability and the Twin SVM. Appl. Soft Comput. 2015, 30, 305–318. [Google Scholar] [CrossRef]
- Aljalal, M.; Djemal, R. A Comparative Study of Wavelet and CSP Features Classified Using LDA, SVM and ANN in EEG Based Motor Imagery. In Proceedings of the 2017 9th IEEE-GCC Conference and Exhibition (GCCCE), Manama, Bahrain, 8–11 May 2017; pp. 1–9. [Google Scholar]
- El-Kafrawy, N.M.; Hegazy, D.; Tolba, M.F. Features extraction and classification of EEG signals using empirical mode decomposition and support vector machine. In Proceedings of the International Conference on Advanced Machine Learning Technologies and Applications, Cairo, Egypt, 28–30 November 2014; Springer: Berlin/Heidelberg, Germany, 2014; pp. 189–198. [Google Scholar]
- Ines, H.; Slim, Y.; Noureddine, E. EEG classification using support vector machine. In Proceedings of the 10th International Multi-Conferences on Systems, Signals & Devices 2013 (SSD13), Hammamet, Tunisia, 18–21 March 2013; pp. 1–4. [Google Scholar]
- Jia, H.; Wang, S.; Zheng, D.; Qu, X.; Fan, S. Comparative study of motor imagery classification based on BP-NN and SVM. J. Eng. 2019, 2019, 8646–8649. [Google Scholar] [CrossRef]
- Molla, M.K.I.; Al Shiam, A.; Islam, M.R.; Tanaka, T. Discriminative feature selection-based motor imagery classification using EEG signal. IEEE Access 2020, 8, 98255–98265. [Google Scholar] [CrossRef]
- Venkatachalam, K.; Devipriya, A.; Maniraj, J.; Sivaram, M.; Ambikapathy, A.; Iraj, S.A. A Novel Method of motor imagery classification using eeg signal. Artif. Intell. Med. 2020, 103, 101787. [Google Scholar]
- Andrzejak, R.G.; Lehnertz, K.; Mormann, F.; Rieke, C.; David, P.; Elger, C.E. Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: Dependence on recording region and brain state. Phys. Rev. E 2001, 64, 061907. [Google Scholar] [CrossRef] [Green Version]
- Hamad, A.; Hassanien, A.E.; Fahmy, A.A.; Houssein, E.H. A hybrid automated detection of epileptic seizures in EEG based on wavelet and machine learning techniques. arXiv 2018, arXiv:1807.10723. [Google Scholar]
- Jaiswal, A.K.; Banka, H. Epileptic seizure detection in EEG signal using machine learning techniques. Australas. Phys. Eng. Sci. Med. 2018, 41, 81–94. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, X.; Hou, Z.; Liu, H. Epileptic seizure detection based on the kernel extreme learning machine. Technol. Health Care 2017, 25, 399–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riaz, F.; Hassan, A.; Rehman, S.; Niazi, I.K.; Dremstrup, K. EMD-based temporal and spectral features for the classification of EEG signals using supervised learning. IEEE Trans. Neural Syst. Rehabil. Eng. 2015, 24, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Mardini, W.; Yassein, M.M.B.; Al-Rawashdeh, R.; Aljawarneh, S.; Khamayseh, Y.; Meqdadi, O. Enhanced detection of epileptic seizure using EEG signals in combination with machine learning classifiers. IEEE Access 2020, 8, 24046–24055. [Google Scholar] [CrossRef]
- Savadkoohi, M.; Oladunni, T.; Thompson, L. A machine learning approach to epileptic seizure prediction using Electroencephalogram (EEG) Signal. Biocybern. Biomed. Eng. 2020, 40, 1328–1341. [Google Scholar] [CrossRef]
- Chakraborty, M.; Mitra, D. Epilepsy seizure detection using kurtosis based VMD’s parameters selection and bandwidth features. Biomed. Signal Process. Control 2021, 64, 102255. [Google Scholar]
- Ech-Choudany, Y.; Scida, D.; Assarar, M.; Landré, J.; Bellach, B.; Morain-Nicolier, F. Dissimilarity-based time–frequency distributions as features for epileptic EEG signal classification. Biomed. Signal Process. Control 2021, 64, 102268. [Google Scholar] [CrossRef]
- Kaya, Y.; Ertuğrul, Ö.F. A stable feature extraction method in classification epileptic EEG signals. Australas. Phys. Eng. Sci. Med. 2018, 41, 721–730. [Google Scholar] [CrossRef]
- Moody, G.B.; Mark, R.G.; Goldberger, A.L. PhysioNet: Physiologic signals, time series and related open source software for basic, clinical, and applied research. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 8327–8330. [Google Scholar]
- Fergus, P.; Hignett, D.; Hussain, A.; Al-Jumeily, D.; Abdel-Aziz, K. Automatic epileptic seizure detection using scalp EEG and advanced artificial intelligence techniques. BioMed Res. Int. 2015, 2015, 986736. [Google Scholar] [CrossRef]
- Pinto-Orellana, M.A.; Cerqueira, F.R. Patient-specific epilepsy seizure detection using random forest classification over one-dimension transformed EEG data. In Proceedings of the International Conference on Intelligent Systems Design and Applications, Porto, Portugal, 16–18 December 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 519–528. [Google Scholar]
- Shoeb, A.; Kharbouch, A.; Soegaard, J.; Schachter, S.; Guttag, J. A machine-learning algorithm for detecting seizure termination in scalp EEG. Epilepsy Behav. 2011, 22, S36–S43. [Google Scholar] [CrossRef]
- Usman, S.M.; Usman, M.; Fong, S. Epileptic seizures prediction using machine learning methods. Comput. Math. Methods Med. 2017, 2017, 9074759. [Google Scholar] [CrossRef] [Green Version]
- Goldberger, A.L.; Amaral, L.A.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef] [Green Version]
- Alomari, M.H.; Samaha, A.; AlKamha, K. Automated classification of L/R hand movement EEG signals using advanced feature extraction and machine learning. arXiv 2013, arXiv:1312.2877. [Google Scholar]
- Moreira, J.; Moreira, M.; Pombo, N.; Silva, B.M.; Garcia, N.M. Identification of real and imaginary movements in EEG using machine learning models. In Proceedings of the International Conference on Medical and Biological Engineering, Banja Luka, 16 ̶̶ 18 May 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 469–474. [Google Scholar]
- Priya, T.H.; Mahalakshmi, P.; Naidu, V.; Srinivas, M. Stress detection from EEG using power ratio. In Proceedings of the 2020 International Conference on Emerging Trends in Information Technology and Engineering (ic-ETITE), Vellore, India, 24–25 February 2020; pp. 1–6. [Google Scholar]
- Ihle, M.; Feldwisch-Drentrup, H.; Teixeira, C.A.; Witon, A.; Schelter, B.; Timmer, J.; Schulze-Bonhage, A. EPILEPSIAE—A European epilepsy database. Comput. Methods Programs Biomed. 2012, 106, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Direito, B.; Teixeira, C.A.; Sales, F.; Castelo-Branco, M.; Dourado, A. A realistic seizure prediction study based on multiclass SVM. Int. J. Neural Syst. 2017, 27, 1750006. [Google Scholar] [CrossRef]
- Teixeira, C.A.; Direito, B.; Bandarabadi, M.; Le Van Quyen, M.; Valderrama, M.; Schelter, B.; Schulze-Bonhage, A.; Navarro, V.; Sales, F.; Dourado, A. Epileptic seizure predictors based on computational intelligence techniques: A comparative study with 278 patients. Comput. Methods Programs Biomed. 2014, 114, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Bandarabadi, M.; Teixeira, C.A.; Rasekhi, J.; Dourado, A. Epileptic seizure prediction using relative spectral power features. Clin. Neurophysiol. 2015, 126, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Kemp, B.; Zwinderman, A.H.; Tuk, B.; Kamphuisen, H.A.; Oberye, J.J. Analysis of a sleep-dependent neuronal feedback loop: The slow-wave microcontinuity of the EEG. IEEE Trans. Biomed. Eng. 2000, 47, 1185–1194. [Google Scholar] [CrossRef]
- Ravan, M. Machine Learning Approach to Measure Sleep Quality using EEG Signals. In Proceedings of the 2019 IEEE Signal Processing in Medicine and Biology Symposium (SPMB), Philadelphia, PA, USA, 7 December 2019; pp. 1–6. [Google Scholar]
- Hassan, A.R.; Bhuiyan, M.I.H. Automatic sleep scoring using statistical features in the EMD domain and ensemble methods. Biocybern. Biomed. Eng. 2016, 36, 248–255. [Google Scholar] [CrossRef]
- Delimayanti, M.K.; Purnama, B.; Nguyen, N.G.; Faisal, M.R.; Mahmudah, K.R.; Indriani, F.; Kubo, M.; Satou, K. Classification of brainwaves for sleep stages by high-dimensional FFT features from EEG signals. Appl. Sci. 2020, 10, 1797. [Google Scholar] [CrossRef] [Green Version]
- Ichimaru, Y.; Moody, G. Development of the polysomnographic database on CD-ROM. Psychiatry Clin. Neurosci. 1999, 53, 175–177. [Google Scholar] [CrossRef] [Green Version]
- Winterhalder, M.; Schelter, B.; Maiwald, T.; Brandt, A.; Schad, A.; Schulze-Bonhage, A.; Timmer, J. Spatio-temporal patient–individual assessment of synchronization changes for epileptic seizure prediction. Clin. Neurophysiol. 2006, 117, 2399–2413. [Google Scholar] [CrossRef]
- Williamson, J.R.; Bliss, D.W.; Browne, D.W.; Narayanan, J.T. Seizure prediction using EEG spatiotemporal correlation structure. Epilepsy Behav. 2012, 25, 230–238. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Xu, M.; Wang, Y.; Zhang, S.; Chen, L.; Ming, D. Enhance decoding of pre-movement EEG patterns for brain–computer interfaces. J. Neural Eng. 2020, 17, 016033. [Google Scholar] [CrossRef] [PubMed]
- Yuvaraj, R.; Acharya, U.R.; Hagiwara, Y. A novel Parkinson’s Disease Diagnosis Index using higher-order spectra features in EEG signals. Neural Comput. Appl. 2018, 30, 1225–1235. [Google Scholar] [CrossRef]
- Gao, L.; Cheng, W.; Zhang, J.; Wang, J. EEG classification for motor imagery and resting state in BCI applications using multi-class Adaboost extreme learning machine. Rev. Sci. Instruments 2016, 87, 085110. [Google Scholar] [CrossRef] [PubMed]
- Cheema, B.S.; Samima, S.; Sarma, M.; Samanta, D. Mental workload estimation from EEG signals using machine learning algorithms. In Proceedings of the International Conference on Engineering Psychology and Cognitive Ergonomics, Las Vegas, NV, USA, 15–20 July 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 265–284. [Google Scholar]
- Sharma, M.; Acharya, U.R. Automated detection of schizophrenia using optimal wavelet-based l1 norm features extracted from single-channel EEG. Cogn. Neurodyn. 2021, 15, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wu, W.; Gu, Z.; Yu, Z.; Qi, F.; Li, Y. Deep learning based on batch normalization for P300 signal detection. Neurocomputing 2018, 275, 288–297. [Google Scholar] [CrossRef]
- Blankertz, B.; Krusienski, D.; Schalk, G. Documentation Second Wadsworth BCI Dataset (P300 Evoked Potentials) Data Acquired Using BCI2000 P300 Speller Paradigm. BCI Classification Contest November. Available online: http://www.bbci.de/competition/ii/albany_desc/albany_desc_ii.pdf (accessed on 14 November 2018).
- Pereira, A.; Padden, D.; Jantz, J.; Lin, K.; Alcaide-Aguirre, R. Cross-Subject EEG Event-Related Potential Classification for Brain-Computer Interfaces Using Residual Networks. Available online: https://hal.archives-ouvertes.fr/hal-01878227 (accessed on 10 September 2019).
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A compact convolutional neural network for EEG-based brain–computer interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef] [Green Version]
- Stober, S.; Cameron, D.J.; Grahn, J.A. Using Convolutional Neural Networks to Recognize Rhythm Stimuli from Electroencephalography Recordings. Adv. Neural Inf. Process. Syst. 2014, 27, 1449–1457. [Google Scholar]
- Heraz, A.; Razaki, R.; Frasson, C. Using machine learning to predict learner emotional state from brainwaves. In Proceedings of the Seventh IEEE International Conference on Advanced Learning Technologies (ICALT 2007), Niigata, Japan, 18–20 July 2007; pp. 853–857. [Google Scholar]
- Li, M.; Lu, B.L. Emotion classification based on gamma-band EEG. In Proceedings of the 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Minneapolis, MN, USA, 3–6 September 2009; pp. 1223–1226. [Google Scholar]
- Grabowski, K.; Rynkiewicz, A.; Lassalle, A.; Baron-Cohen, S.; Schuller, B.; Cummins, N.; Baird, A.; Podgórska-Bednarz, J.; Pieniążek, A.; Lucka, I. Emotional expression in psychiatric conditions: New technology for clinicians. Psychiatry Clin. Neurosci. 2019, 73, 50–62. [Google Scholar] [CrossRef]
- Borgomaneri, S.; Vitale, F.; Battaglia, S.; Avenanti, A. Early Right Motor Cortex Response to Happy and Fearful Facial Expressions: A TMS Motor-Evoked Potential Study. Brain Sci. 2021, 11, 1203. [Google Scholar] [CrossRef]
- Ellena, G.; Starita, F.; Haggard, P.; Làdavas, E. The spatial logic of fear. Cognition 2020, 203, 104336. [Google Scholar] [CrossRef] [PubMed]
- Candini, M.; Battaglia, S.; Benassi, M.; di Pellegrino, G.; Frassinetti, F. The physiological correlates of interpersonal space. Sci. Rep. 2021, 11, 1–8. [Google Scholar]
- Qiao, R.; Qing, C.; Zhang, T.; Xing, X.; Xu, X. A novel deep-learning based framework for multi-subject emotion recognition. In Proceedings of the 2017 4th International Conference on Information, Cybernetics and Computational Social Systems (ICCSS), Dalian, China, 24–26 July 2017; pp. 181–185. [Google Scholar]
- Murugappan, M.; Alshuaib, W.; Bourisly, A.K.; Khare, S.K.; Sruthi, S.; Bajaj, V. Tunable Q wavelet transform based emotion classification in Parkinson’s disease using Electroencephalography. PLoS ONE 2020, 15, e0242014. [Google Scholar] [CrossRef] [PubMed]
- Horlings, R.; Datcu, D.; Rothkrantz, L.J. Emotion recognition using brain activity. In Proceedings of the 9th International Conference on Computer Systems and Technologies and workshop for PhD Students in Computing, Gabrovo, Bulgaria, 12–13 June 2008. [Google Scholar]
- Seo, J.; Laine, T.H.; Sohn, K.A. Machine learning approaches for boredom classification using EEG. J. Ambient Intell. Humaniz. Comput. 2019, 10, 3831–3846. [Google Scholar] [CrossRef]
- Takahashi, K. Remarks on SVM-based emotion recognition from multi-modal bio-potential signals. In Proceedings of the RO-MAN 2004, 13th IEEE International Workshop on Robot and Human Interactive Communication (IEEE Catalog No. 04TH8759), Kurashiki, Japan, 22 September 2004; pp. 95–100. [Google Scholar]
- Alex, M.; Tariq, U.; Al-Shargie, F.; Mir, H.S.; Al Nashash, H. Discrimination of Genuine and Acted Emotional Expressions Using EEG Signal and Machine Learning. IEEE Access 2020, 8, 191080–191089. [Google Scholar] [CrossRef]
- Khare, S.K.; Bajaj, V.; Sinha, G. Adaptive tunable Q wavelet transform-based emotion identification. IEEE Trans. Instrum. Meas. 2020, 69, 9609–9617. [Google Scholar] [CrossRef]
- Xiao, L.; Qiu, J.; Lu, J. A Study of Human Behavior and Mental Workload Based on Neural Network. In Proceedings of the International Conference on Human Aspects of IT for the Aged Population, Toronto, ON, Canada, 17–22 July 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 389–397. [Google Scholar]
- Feyen, R.G. Bridging the gap: Exploring interactions between digital human models and cognitive models. In Proceedings of the International Conference on Digital Human Modeling, Beijing, China, 22–27 July 2007; Springer: Berlin/Heidelberg, Germany, 2007; pp. 382–391. [Google Scholar]
- Stone, R.T.; Wei, C.S. Exploring the linkage between facial expression and mental workload for arithmetic tasks. In Proceedings of the Human Factors and Ergonomics Society Annual Meeting; SAGE Publications: Los Angeles, CA, USA, 2011; Volume 55, pp. 616–619. [Google Scholar]
- Garofalo, S.; Battaglia, S.; di Pellegrino, G. Individual differences in working memory capacity and cue-guided behavior in humans. Sci. Rep. 2019, 9, 1–14. [Google Scholar]
- Borgomaneri, S.; Serio, G.; Battaglia, S. Please, don’t do it! Ten years of progress of non-invasive brain stimulation in action inhibition. Cortex 2020, 132, 404–422. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, S.; Serio, G.; Scarpazza, C.; D’Ausilio, A.; Borgomaneri, S. Frozen in (e) motion: How reactive motor inhibition is influenced by the emotional content of stimuli in healthy and psychiatric populations. Behav. Res. Ther. 2021, 146, 103963. [Google Scholar] [CrossRef]
- Heine, T.; Lenis, G.; Reichensperger, P.; Beran, T.; Doessel, O.; Deml, B. Electrocardiographic features for the measurement of drivers’ mental workload. Appl. Ergon. 2017, 61, 31–43. [Google Scholar] [CrossRef] [PubMed]
- So, W.K.; Wong, S.W.; Mak, J.N.; Chan, R.H. An evaluation of mental workload with frontal EEG. PLoS ONE 2017, 12, e0174949. [Google Scholar] [CrossRef]
- Rashid, M.; Sulaiman, N.; Mustafa, M.; Khatun, S.; Bari, B.S. The classification of EEG signal using different machine learning techniques for BCI application. In Proceedings of the International Conference on Robot Intelligence Technology and Applications, Kuala Lumpur, Malaysia, 16–18 December 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 207–221. [Google Scholar]
- Bird, J.J.; Manso, L.J.; Ribeiro, E.P.; Ekárt, A.; Faria, D.R. A study on mental state classification using eeg-based brain-machine interface. In Proceedings of the 2018 International Conference on Intelligent Systems (IS), Funchal, Portugal, 25–27 September 2018; pp. 795–800. [Google Scholar]
- Rus, I.D.; Marc, P.; Dinsoreanu, M.; Potolea, R.; Muresan, R.C. Classification of EEG signals in an object recognition task. In Proceedings of the 2017 13th IEEE International Conference on Intelligent Computer Communication and Processing (ICCP), Cluj-Napoca, Romania, 7–9 September 2017; pp. 391–395. [Google Scholar]
- Jiao, Z.; Gao, X.; Wang, Y.; Li, J.; Xu, H. Deep convolutional neural networks for mental load classification based on EEG data. Pattern Recognit. 2018, 76, 582–595. [Google Scholar] [CrossRef]
- Yazdani, A.; Ebrahimi, T.; Hoffmann, U. Classification of EEG signals using Dempster Shafer theory and a k-nearest neighbor classifier. In Proceedings of the 2009 4th International IEEE/EMBS Conference on Neural Engineering, Antalya, Turkey, 29 April–2 May 2009; pp. 327–330. [Google Scholar]
- Vanitha, V.; Krishnan, P. Real Time Stress Detection System Based on EEG Signals. 2017, pp. S271–S275. Available online: http://www.biomedres.info/biomedical-research/real-time-stress-detectionsystem-based-on-eeg-signals.html (accessed on 18 April 2019).
- Guo, L.; Wu, Y.; Zhao, L.; Cao, T.; Yan, W.; Shen, X. Classification of mental task from EEG signals using immune feature weighted support vector machines. IEEE Trans. Magn. 2010, 47, 866–869. [Google Scholar] [CrossRef]
- Kumar, Y.; Dewal, M.; Anand, R. Features extraction of EEG signals using approximate and sample entropy. In Proceedings of the 2012 IEEE Students’ Conference on Electrical, Electronics and Computer Science, Bhopal, India, 1–2 March 2012; pp. 1–5. [Google Scholar]
- Pandey, V.; Choudhary, D.K.; Verma, V.; Sharma, G.; Singh, R.; Chandra, S. Mental Workload Estimation Using EEG. In Proceedings of the 2020 Fifth International Conference on Research in Computational Intelligence and Communication Networks (ICRCICN), Bangalore, India, 26–27 November 2020; pp. 83–86. [Google Scholar]
- Gupta, A.; Agrawal, R. Relevant feature selection from EEG signal for mental task classification. In Proceedings of the Pacific-Asia Conference on Knowledge Discovery and Data Mining, Kuala Lumpur, Malaysia, 29 May–1 June 2012; Springer: Berlin/Heidelberg, Germany, 2012; pp. 431–442. [Google Scholar]
- Wei, C.S.; Ko, L.W.; Chuang, S.W.; Jung, T.P.; Lin, C.T. EEG-based evaluation system for motion sickness estimation. In Proceedings of the 2011 5th International IEEE/EMBS Conference on Neural Engineering, Cancun, Mexico, 27 April–1 May 2011; pp. 100–103. [Google Scholar]
- Gupta, A.; Khan, R.U.; Singh, V.K.; Tanveer, M.; Kumar, D.; Chakraborti, A.; Pachori, R.B. A novel approach for classification of mental tasks using multiview ensemble learning (MEL). Neurocomputing 2020, 417, 558–584. [Google Scholar] [CrossRef]
- Hosni, S.M.; Gadallah, M.E.; Bahgat, S.F.; AbdelWahab, M.S. Classification of EEG signals using different feature extraction techniques for mental-task BCI. In Proceedings of the 2007 International Conference on Computer Engineering & Systems, Cairo, Egypt, 27–29 November 2007; pp. 220–226. [Google Scholar]
- Liang, N.Y.; Saratchandran, P.; Huang, G.B.; Sundararajan, N. Classification of mental tasks from EEG signals using extreme learning machine. Int. J. Neural Syst. 2006, 16, 29–38. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, A.; Almeida, C.; Georgieva, P.; Tomé, A.; Silva, F. Advances in EEG-based biometry. In Proceedings of the International Conference Image Analysis and Recognition, Póvoa de Varzin, Portugal, 21–23 June 2010; Springer: Berlin/Heidelberg, Germany, 2010; pp. 287–295. [Google Scholar]
- Tang, J.; Xu, M.; Han, J.; Liu, M.; Dai, T.; Chen, S.; Ming, D. Optimizing SSVEP-based BCI system towards practical high-speed spelling. Sensors 2020, 20, 4186. [Google Scholar] [CrossRef]
- Xiao, X.; Xu, M.; Jin, J.; Wang, Y.; Jung, T.P.; Ming, D. Discriminative canonical pattern matching for single-trial classification of ERP components. IEEE Trans. Biomed. Eng. 2019, 67, 2266–2275. [Google Scholar] [CrossRef]
- Ke, Y.; Liu, P.; An, X.; Song, X.; Ming, D. An online SSVEP-BCI system in an optical see-through augmented reality environment. J. Neural Eng. 2020, 17, 016066. [Google Scholar] [CrossRef]
- Mulder, T. Motor imagery and action observation: Cognitive tools for rehabilitation. J. Neural Transm. 2007, 114, 1265–1278. [Google Scholar] [CrossRef] [Green Version]
- Gannouni, S.; Belwafi, K.; Aboalsamh, H.; AlSamhan, Z.; Alebdi, B.; Almassad, Y.; Alobaedallah, H. EEG-Based BCI System to Detect Fingers Movements. Brain Sci. 2020, 10, 965. [Google Scholar] [CrossRef]
- Attallah, O.; Abougharbia, J.; Tamazin, M.; Nasser, A.A. A BCI system based on motor imagery for assisting people with motor deficiencies in the limbs. Brain Sci. 2020, 10, 864. [Google Scholar] [CrossRef]
- Hung, C.I.; Lee, P.L.; Wu, Y.T.; Chen, L.F.; Yeh, T.C.; Hsieh, J.C. Recognition of motor imagery electroencephalography using independent component analysis and machine classifiers. Ann. Biomed. Eng. 2005, 33, 1053–1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, P.; Wang, J.; Zhao, H.; Li, R. Mlp with riemannian covariance for motor imagery based eeg analysis. IEEE Access 2020, 8, 139974–139982. [Google Scholar] [CrossRef]
- Sun, B.; Zhao, X.; Zhang, H.; Bai, R.; Li, T. EEG motor imagery classification with sparse spectrotemporal decomposition and deep learning. IEEE Trans. Autom. Sci. Eng. 2020, 18, 541–551. [Google Scholar] [CrossRef]
- Liu, X.; Lv, L.; Shen, Y.; Xiong, P.; Yang, J.; Liu, J. Multiscale space-time-frequency feature-guided multitask learning CNN for motor imagery EEG classification. J. Neural Eng. 2021, 18, 026003. [Google Scholar] [CrossRef]
- Dai, M.; Zheng, D.; Na, R.; Wang, S.; Zhang, S. EEG classification of motor imagery using a novel deep learning framework. Sensors 2019, 19, 551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, D.; Triesch, J. Residual deep convolutional neural network for eeg signal classification in epilepsy. arXiv 2019, arXiv:1903.08100. [Google Scholar]
- Ullah, I.; Hussain, M.; Aboalsamh, H. An automated system for epilepsy detection using EEG brain signals based on deep learning approach. Expert Syst. Appl. 2018, 107, 61–71. [Google Scholar] [CrossRef] [Green Version]
- Janghel, R.R.; Verma, A.; Rathore, Y.K. Performance comparison of machine learning techniques for epilepsy classification and detection in EEG signal. In Data Management, Analytics and Innovation; Springer: Berlin/Heidelberg, Germany, 2020; pp. 425–438. [Google Scholar]
- Birjandtalab, J.; Pouyan, M.B.; Cogan, D.; Nourani, M.; Harvey, J. Automated seizure detection using limited-channel EEG and non-linear dimension reduction. Comput. Biol. Med. 2017, 82, 49–58. [Google Scholar] [CrossRef]
- Wang, Z.; Na, J.; Zheng, B. An improved knn classifier for epilepsy diagnosis. IEEE Access 2020, 8, 100022–100030. [Google Scholar] [CrossRef]
- Kaya, Y.; Uyar, M.; Tekin, R.; Yıldırım, S. 1D-local binary pattern based feature extraction for classification of epileptic EEG signals. Appl. Math. Comput. 2014, 243, 209–219. [Google Scholar] [CrossRef]
- Mormann, F.; Andrzejak, R.G.; Elger, C.E.; Lehnertz, K. Seizure prediction: The long and winding road. Brain 2007, 130, 314–333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, C.; Direito, B.; Feldwisch-Drentrup, H.; Valderrama, M.; Costa, R.; Alvarado-Rojas, C.; Nikolopoulos, S.; Le Van Quyen, M.; Timmer, J.; Schelter, B.; et al. EPILAB: A software package for studies on the prediction of epileptic seizures. J. Neurosci. Methods 2011, 200, 257–271. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Sotelo, J.L.; Osorio-Forero, A.; Jiménez-Rodríguez, A.; Cuesta-Frau, D.; Cirugeda-Roldán, E.; Peluffo, D. Automatic sleep stages classification using EEG entropy features and unsupervised pattern analysis techniques. Entropy 2014, 16, 6573–6589. [Google Scholar] [CrossRef] [Green Version]
- Oltu, B.; Akşahin, M.F.; Kibaroğlu, S. A novel electroencephalography based approach for Alzheimer’s disease and mild cognitive impairment detection. Biomed. Signal Process. Control 2021, 63, 102223. [Google Scholar] [CrossRef]
- Zoubek, L.; Charbonnier, S.; Lesecq, S.; Buguet, A.; Chapotot, F. Feature selection for sleep/wake stages classification using data driven methods. Biomed. Signal Process. Control 2007, 2, 171–179. [Google Scholar] [CrossRef]
- Lajnef, T.; Chaibi, S.; Ruby, P.; Aguera, P.E.; Eichenlaub, J.B.; Samet, M.; Kachouri, A.; Jerbi, K. Learning machines and sleeping brains: Automatic sleep stage classification using decision-tree multi-class support vector machines. J. Neurosci. Methods 2015, 250, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.E.; Liang, S.F. Automatic stage scoring of single-channel sleep EEG based on multiscale permutation entropy. In Proceedings of the 2011 IEEE Biomedical Circuits and Systems Conference (BioCAS), San Diego, CA, USA, 10–12 November 2011; pp. 448–451. [Google Scholar]
- Yetton, B.D.; Niknazar, M.; Duggan, K.A.; McDevitt, E.A.; Whitehurst, L.N.; Sattari, N.; Mednick, S.C. Automatic detection of rapid eye movements (REMs): A machine learning approach. J. Neurosci. Methods 2016, 259, 72–82. [Google Scholar] [CrossRef] [Green Version]
- Ebrahimi, F.; Mikaeili, M.; Estrada, E.; Nazeran, H. Automatic sleep stage classification based on EEG signals by using neural networks and wavelet packet coefficients. In Proceedings of the 2008 30th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Vancouver, BC, Canada, 21–24 August 2008; pp. 1151–1154. [Google Scholar]
- Phelps, C.E.; Navratilova, E.; Porreca, F. Cognition in the chronic pain experience: Preclinical insights. Trends Cogn. Sci. 2021, 25, 365–376. [Google Scholar] [CrossRef]
- Ramachandran, V.S. Encyclopedia of Human Behavior; Academic Press: Cambridge, MA, USA, 2012. [Google Scholar]
- Battaglia, S.; Garofalo, S.; di Pellegrino, G. Context-dependent extinction of threat memories: Influences of healthy aging. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, N.; Bairagi, V. Extracting salient features for EEG-based diagnosis of Alzheimer’s disease using support vector machine classifier. IETE J. Res. 2017, 63, 11–22. [Google Scholar] [CrossRef]
- Kulkarni, N. EEG Signal Analysis for Mild Alzheimer’s Disease Diagnosis by Means of Spectral-and Complexity-Based Features and Machine Learning Techniques. In Proceedings of the 2nd International Conference on Data Engineering and Communication Technology; Springer: Berlin/Heidelberg, Germany, 2019; pp. 395–403. [Google Scholar]
- Morabito, F.C.; Campolo, M.; Ieracitano, C.; Ebadi, J.M.; Bonanno, L.; Bramanti, A.; Desalvo, S.; Mammone, N.; Bramanti, P. Deep convolutional neural networks for classification of mild cognitive impaired and Alzheimer’s disease patients from scalp EEG recordings. In Proceedings of the 2016 IEEE 2nd International Forum on Research and Technologies for Society and Industry Leveraging a Better Tomorrow (RTSI), Bologna, Italy, 7–9 September 2016; pp. 1–6. [Google Scholar]
- Fiscon, G.; Weitschek, E.; Felici, G.; Bertolazzi, P.; De Salvo, S.; Bramanti, P.; De Cola, M.C. Alzheimer’s disease patients classification through EEG signals processing. In Proceedings of the 2014 IEEE Symposium on Computational Intelligence and Data Mining (CIDM), Orlando, FL, USA, 9–12 December 2014; pp. 105–112. [Google Scholar]
- Betrouni, N.; Delval, A.; Chaton, L.; Defebvre, L.; Duits, A.; Moonen, A.; Leentjens, A.F.; Dujardin, K. Electroencephalography-based machine learning for cognitive profiling in Parkinson’s disease: Preliminary results. Mov. Disord. 2019, 34, 210–217. [Google Scholar] [CrossRef]
- Chua, K.C.; Chandran, V.; Acharya, U.R.; Lim, C.M. Application of higher order statistics/spectra in biomedical signals—A review. Med. Eng. Phys. 2010, 32, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L. EEG signals classification using machine learning for the identification and diagnosis of schizophrenia. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 4521–4524. [Google Scholar]
- Jiang, X.; Gao, T. An EEG Emotion Classification System Based on One-Dimension Convolutional Neural Networks and Virtual Reality. In Proceedings of the International Conference on Innovative Mobile and Internet Services in Ubiquitous Computing, Lodz, Poland, 1–3 July 2020; Springer: Berlin/Heidelberg, Germany, 2020; pp. 194–202. [Google Scholar]
- Shin, H.C.; Roth, H.R.; Gao, M.; Lu, L.; Xu, Z.; Nogues, I.; Yao, J.; Mollura, D.; Summers, R.M. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans. Med. Imaging 2016, 35, 1285–1298. [Google Scholar] [CrossRef] [Green Version]
- Demir, A.; Koike-Akino, T.; Wang, Y.; Haruna, M.; Erdogmus, D. EEG-GNN: Graph Neural Networks for Classification of Electroencephalogram (EEG) Signals. arXiv 2021, arXiv:2106.09135. [Google Scholar]
- Wu, D.; Li, X.; Feng, J. Connectome-based individual prediction of cognitive behaviors via the graph propagation network reveals directed brain network topology. bioRxiv 2021. [Google Scholar] [CrossRef]
- Zhu, S.; Pan, S.; Zhou, C.; Wu, J.; Cao, Y.; Wang, B. Graph geometry interaction learning. arXiv 2020, arXiv:2010.12135. [Google Scholar]
- Zhou, J.; Cui, G.; Hu, S.; Zhang, Z.; Yang, C.; Liu, Z.; Wang, L.; Li, C.; Sun, M. Graph neural networks: A review of methods and applications. AI Open 2020, 1, 57–81. [Google Scholar] [CrossRef]
- Casas, S.; Gulino, C.; Liao, R.; Urtasun, R. Spagnn: Spatially-aware graph neural networks for relational behavior forecasting from sensor data. In Proceedings of the 2020 IEEE International Conference on Robotics and Automation (ICRA), Paris, France, 31 May–31 August 2020; pp. 9491–9497. [Google Scholar]
- Johannesen, J.K.; Bi, J.; Jiang, R.; Kenney, J.G.; Chen, C.M.A. Machine learning identification of EEG features predicting working memory performance in schizophrenia and healthy adults. Neuropsychiatr. Electrophysiol. 2016, 2, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Kurkin, S.; Pitsik, E.; Frolov, N. Artificial intelligence systems for classifying eeg responses to imaginary and real movements of operators. In Saratov Fall Meeting 2018: Computations and Data Analysis: From Nanoscale Tools to Brain Functions; International Society for Optics and Photonics: Bellingham, WA, USA, 2019; Volume 11067, p. 1106709. [Google Scholar]
- Islam, S.M.R.; Sajol, A.; Huang, X.; Ou, K.L. Feature extraction and classification of EEG signal for different brain control machine. In Proceedings of the 2016 3rd International Conference on Electrical Engineering and Information Communication Technology (ICEEICT), Dhaka, Bangladesh, 22–24 September 2016; pp. 1–6. [Google Scholar]
- Jian-Feng, H. Comparison of different classifiers for biometric system based on EEG signals. In Proceedings of the 2010 Second International Conference on Information Technology and Computer Science, Indianapolis, IN, USA, 30 November–3 December 2010; pp. 288–291. [Google Scholar]
- Plewan, T.; Wascher, E.; Falkenstein, M.; Hoffmann, S. Classifying response correctness across different task sets: A machine learning approach. PLoS ONE 2016, 11, e0152864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, H.; Zheng, Q.; Ma, K.; Li, H.; Zheng, Y. Deep representation-based domain adaptation for nonstationary EEG classification. IEEE Trans. Neural Netw. Learn. Syst. 2020, 32, 535–545. [Google Scholar] [CrossRef] [PubMed]
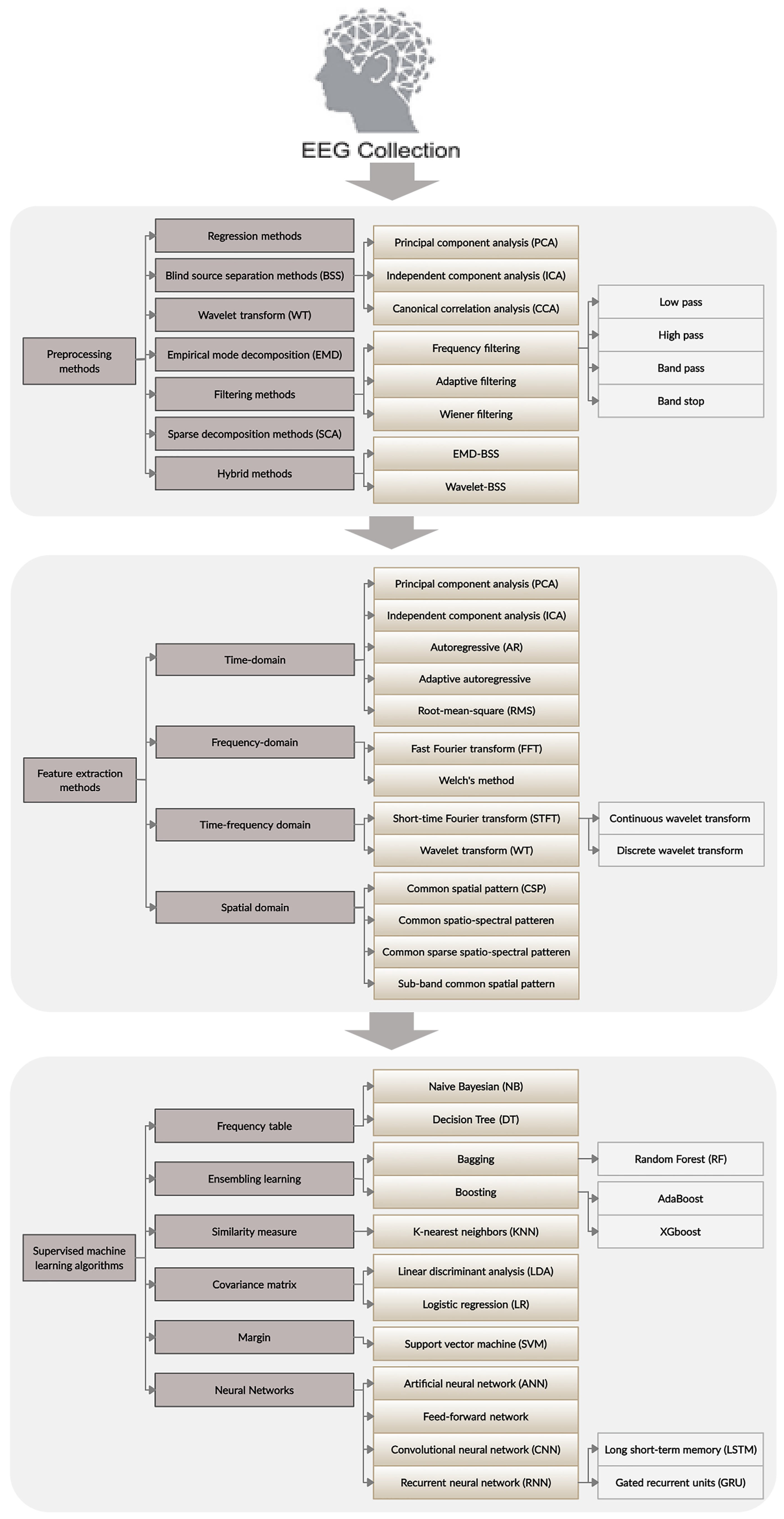
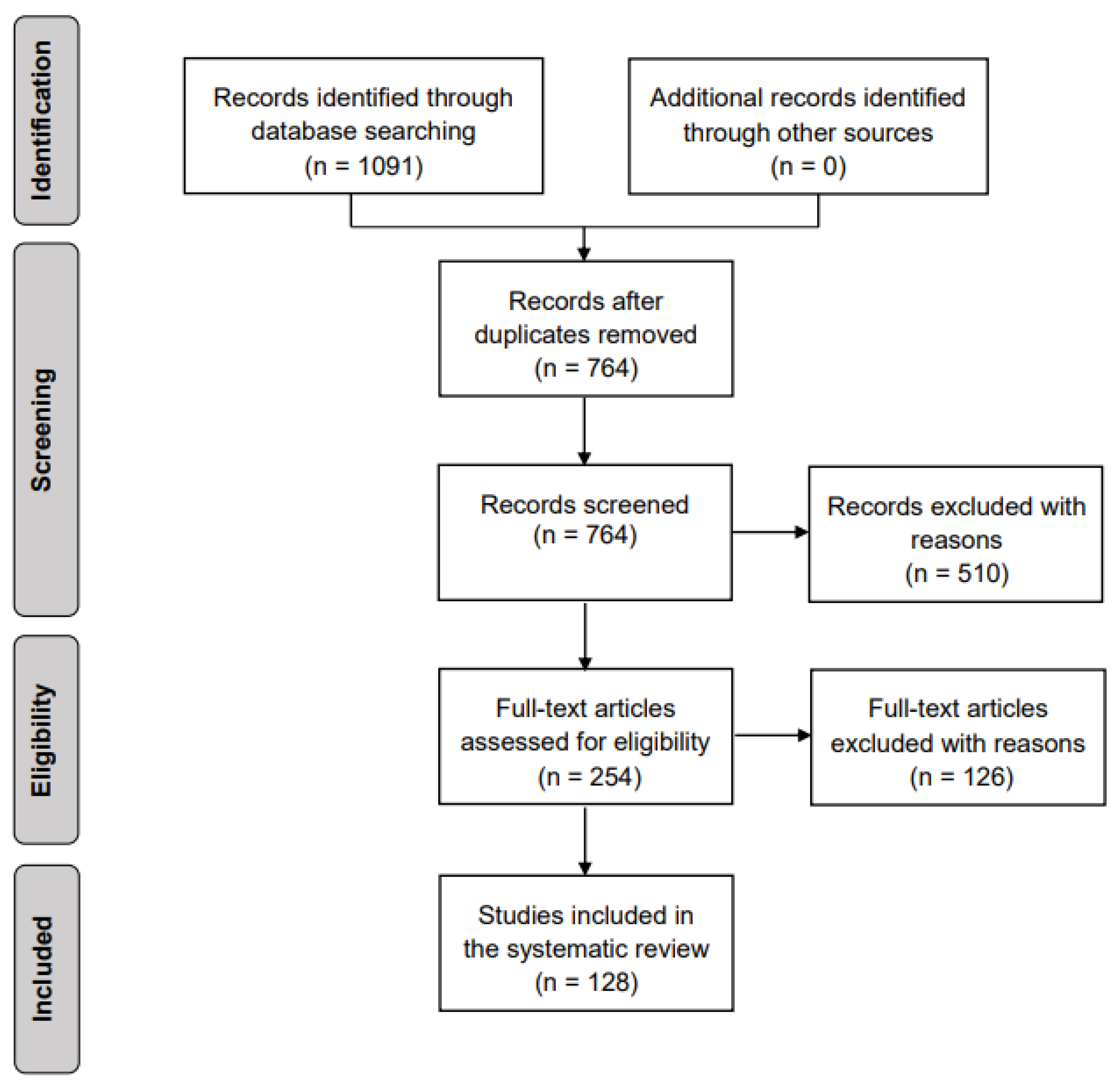
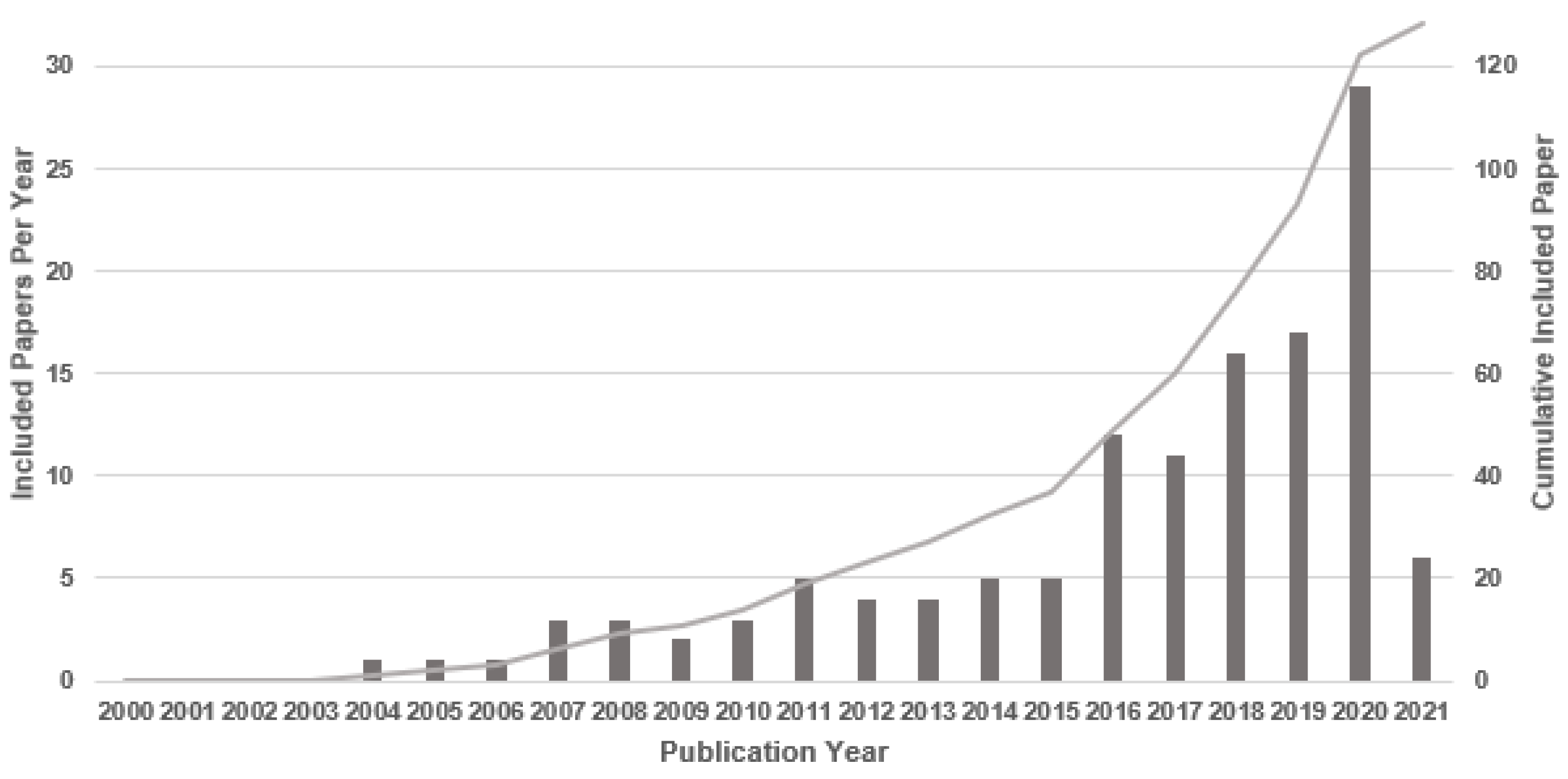
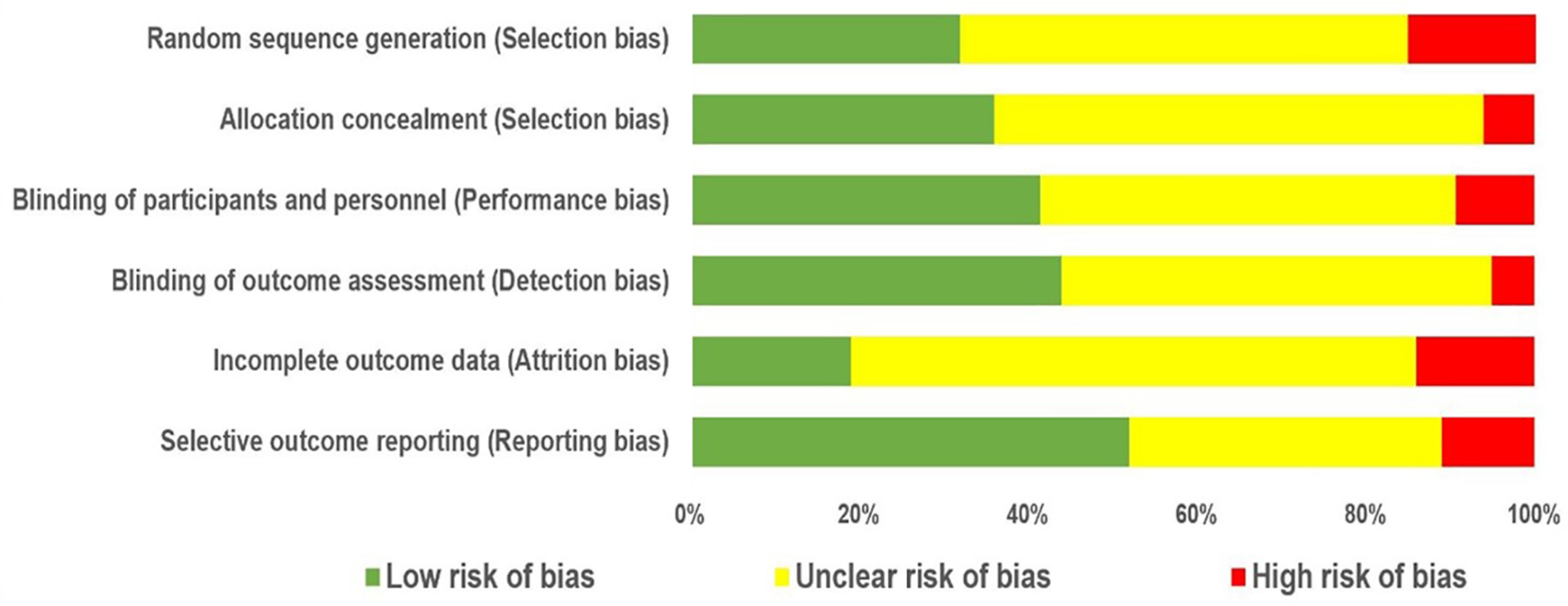
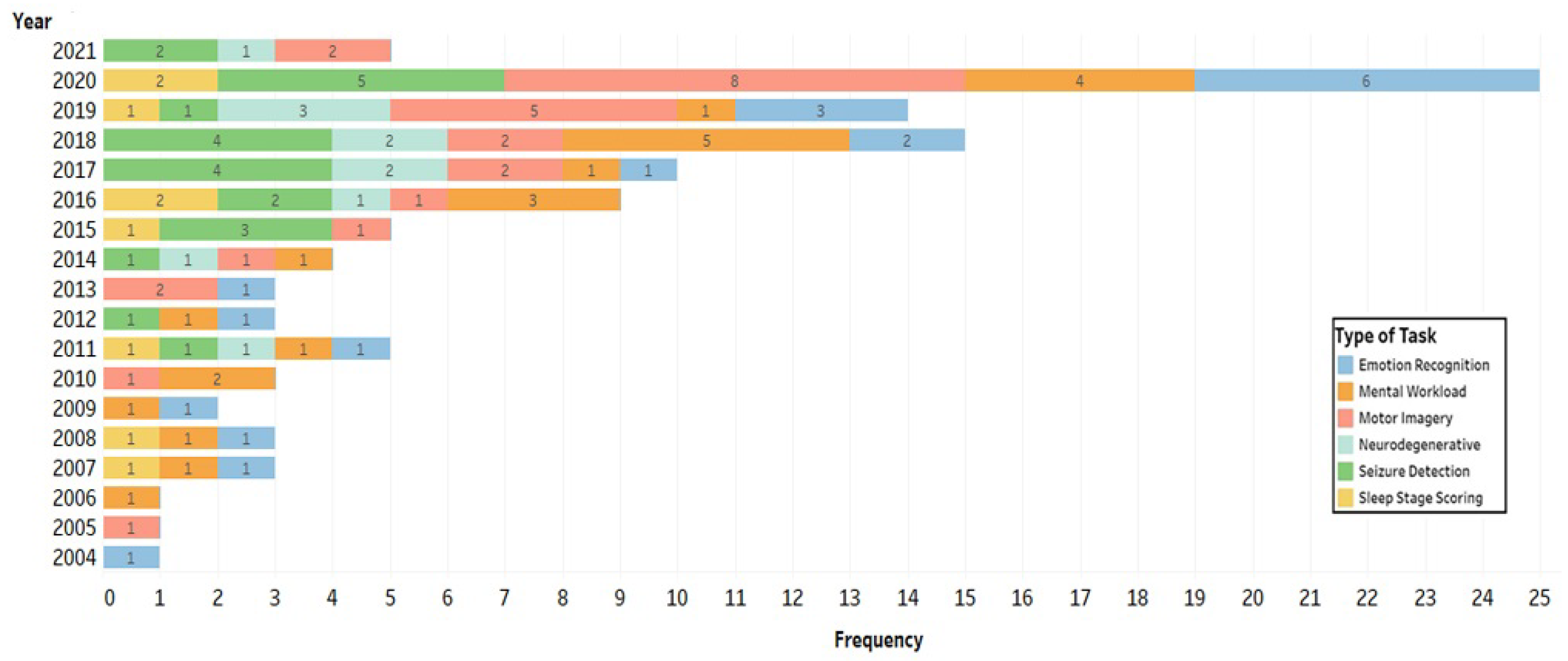
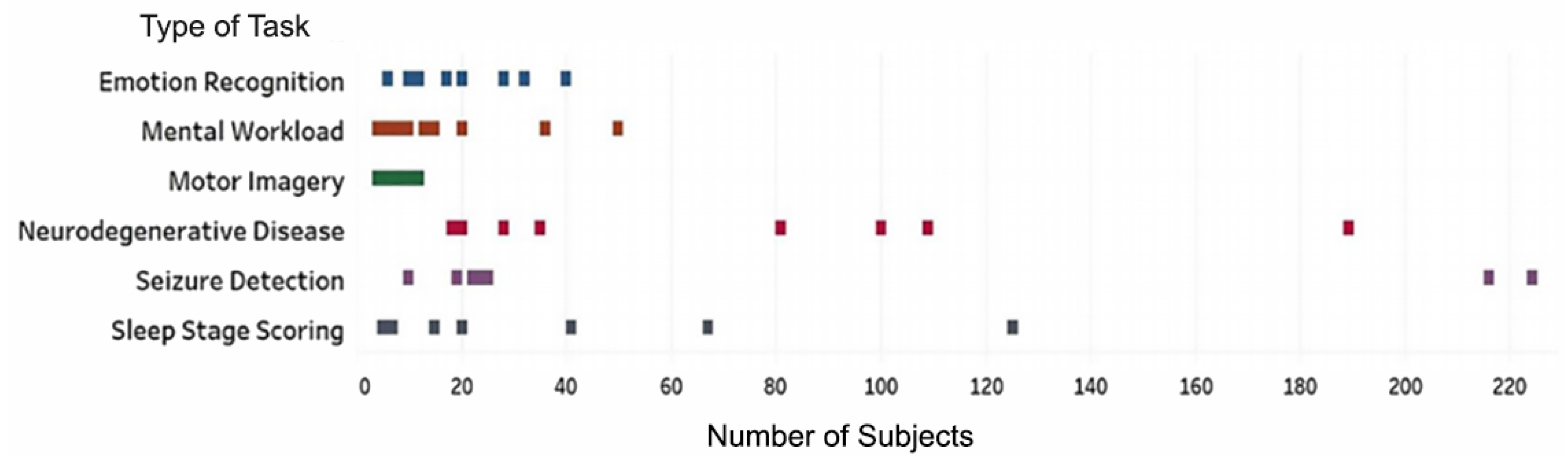
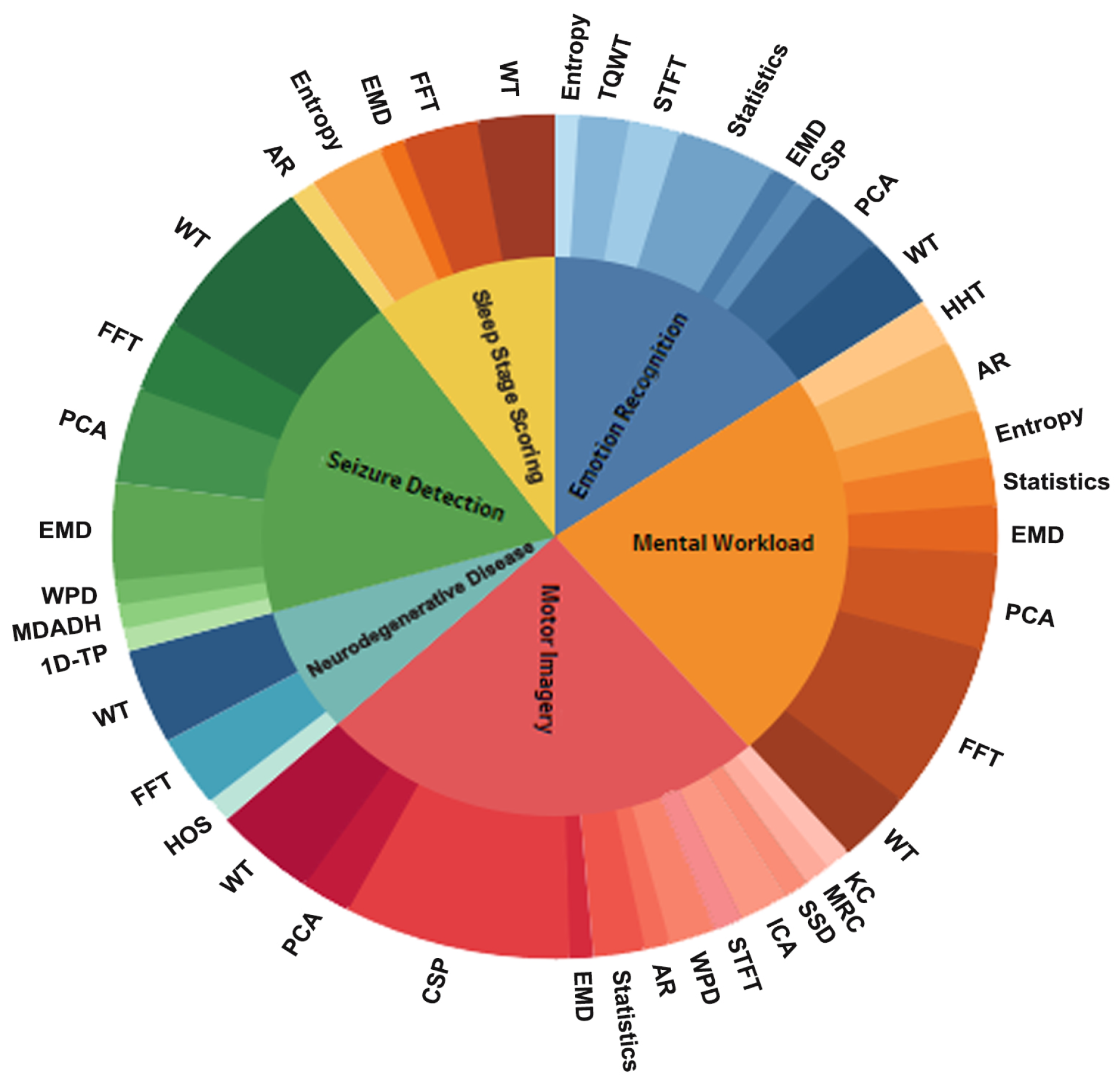
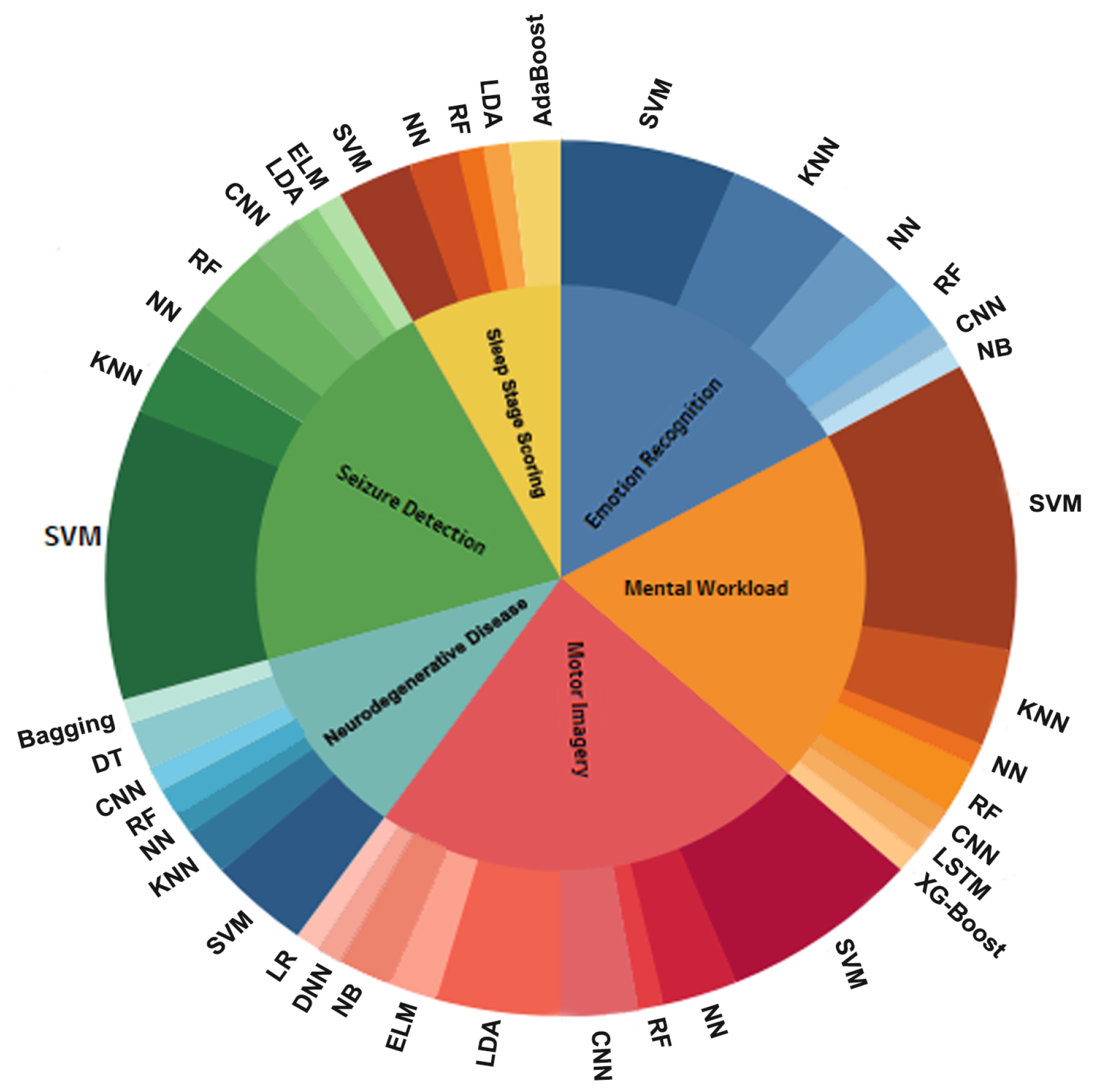

| Conventional Classification Algorithms | Deep Learning Algorithms | |
|---|---|---|
| Input features | Hand-crafted | Automatically based on representation learning |
| Feature selection process | Required | Not required |
| Model architecture | Based on statistical concepts | Consists of a diverse set of architecture based on sample data |
| Computational cost | Computational cost is based on the conventional classification models but is lower than that of deep learning algorithms | Computational cost is very high, because hyper parameters must be tuned |
| Source | Database | Studies Using This Data Set | Number of Subjects | Target Tasks |
|---|---|---|---|---|
| Koelstra et al. [161] | DEAP | [31,119,162,163,164,165,166,167,168] | 32 | Emotion recognition |
| Blankertz et al., Leeb et al. [169,170] | BCI Competition | [49,61,104,171,172,173,174,175,176,177] | 30 subjects in 4 different data sets | Motor imagery |
| Andrzejak et al. [178] | BONN | [118,179,180,181,182,183,184,185,186,187] | 5 | Seizure detection |
| Moody et al. [188] | CHB-MIT | [189,190,191,192] | 22 | Seizure detection |
| Goldberger et al. [193] | PhysioNet | [116,194,195,196] | 109 | Motor imagery/mental workload |
| Ihle et al. [197] | European Epilepsy | [198,199,200] | 300 | Seizure detection |
| Kemp et al. [201] | Sleep-EDF | [59,202,203,204] | 197 | Sleep stage scoring |
| Ichimaru et al. [205] | MIT-BIH | [159] | 16 | Sleep apnea detection |
| Winterhalder et al. [206] | Freiburg | [207] | 21 | Seizure detection |
| Authors | Year | Feature Extraction Method | Classification | Performance (%) |
|---|---|---|---|---|
| Ramzan and Dawn [119] | 2019 | Statistics | RF | Accuracy = 98.2 |
| Bazgir et al. [163] | 2018 | PCA | RBF-SVM | Accuracy = 91.1 (valence) Accuracy = 91.3 (arousal) |
| Balan et al. [162] | 2019 | Entropy | RF | Accuracy = 90.07 |
| Qiao et al. [224] | 2017 | STFT | CNN | Accuracy = 87.27 |
| Shukla and Chaurasiya [167] | 2018 | DWT | KNN | Accuracy = 87.1 |
| Nawaz et al. [168] | 2020 | Statistics | SVM | Accuracy = 77.62 (valence) Accuracy = 78.96 (arousal) Accuracy = 77.6 (dominance) |
| Doma and Pirouz [164] | 2020 | PCA | KNN | Accuracy = 74.25 |
| Chung and Yoon [31] | 2012 | N/A | NB | Accuracy = 66.6 (valence) Accuracy = 66.4 (arousal) |
| Rozgic et al. [166] | 2013 | PCA | ANNs | Accuracy > 60 |
| Authors | Year | Feature Extraction Method | Classification | Performance (%) |
|---|---|---|---|---|
| Guo et al. [245] | 2010 | Entropy | IFWSVM | Accuracy = 97.5 |
| Rashid et al. [239] | 2018 | FFT | Cubic SVM | Accuracy = 95 |
| Gupta and Agrawal [248] | 2012 | EMD | SVM | Accuracy = 94.3 |
| Vanitha and Krishnan [244] | 2016 | HHT | SVM | Accuracy = 89.07 |
| Wei et al. [249] | 2011 | PCA | SVR | Accuracy = 85.92 |
| Peng et al. [84] | 2020 | HHT | SVM | Accuracy = 84.8 |
| Gupta et al. [250] | 2020 | WT/EMD | Non-linear SVM | Accuracy = 80–100 |
| Hosni et al. [251] | 2017 | AR | RBF-SVM | Accuracy = 70 |
| Liang et al. [252] | 2006 | AR | SVM | Accuracy = 67.57 |
| Database | Authors | Year | Feature Extraction Method | Classification | Performance (%) |
|---|---|---|---|---|---|
| BONN | Hamed et al. [179] | 2018 | DWT | RBF-SVM | Accuracy = 100 |
| Savadkoohi et al. [184] | 2020 | FFT / WT | SVM | Accuracy = 100 | |
| Jaiswal and Banka [180] | 2018 | PCA | RBF-SVM | Accuracy = 100 | |
| Riaz et al. [182] | 2015 | EMD | SVM | High performance in the detection of seizures in case 1 and 2 | |
| Ullah et al. [266] | 2018 | - | 1D-CNN | Accuracy = 99.1 | |
| Lu and Triesch [265] | 2019 | - | Residual CNN | Accuracy = 99 | |
| Chakraborty and Mitra [185] | 2021 | VMD | RF | Accuracy = 98.7–100 | |
| Ech-Choudany et al. [186] | 2021 | Dissimilarity-based TFD | LDA | Accuracy = 98 | |
| Mardini et al. [183] | 2020 | DWT | ANNs | Accuracy = 97.8 | |
| Liu et al. [181] | 2017 | WPD | ELM | Accuracy = 97.7 | |
| Murugappan and Ramakrishnan [118] | 2016 | WT | H-MSVM | Accuracy = 94 | |
| Kaya and Ertugrul [187] | 2018 | 1D-TP | RF | Accuracy > 94 | |
| CHB-MIT | Pinto-Orellana and Cerqueira [190] | 2016 | PCA | RF | Sensitivity = 97.1 Specificity = 99.2 |
| Shoeb et al. [191] | 2011 | - | SVM | Sensitivity = 96 | |
| Fergus et al. [189] | 2015 | PCA | KNN | Sensitivity = 93 Specificity = 94 | |
| Usman et al. [192] | 2017 | EMD | SVM | Sensitivity = 92.2 Specificity = 93.4 | |
| European Epilepsy | Teixeira et al. [199] | 2014 | WT | ANNs | Sensitivity = 73.1 |
| Direito et al. [198] | 2017 | DWT | Linear-SVM | High performance in a small subset of participants | |
| Bandarabadi et al. [200] | 2015 | MDADH | SVM | Sensitivity = 73.98 |
| Authors | Year | Sleep Stages | Feature Extraction Method | Classification | Performance (%) |
|---|---|---|---|---|---|
| Santaji and Desai [59] | 2020 | S1, S2, REM | Entropy | RF | Accuracy = 97.8 |
| Ebrahimi et al. [279] | 2008 | Awake, S1 and REM, S2, SWS | WT | MLP | Accuracy = 93 |
| Hassan and Bhuiyan [203] | 2016 | Awake, S1, S2, S3, S4, REM | EMD | AdaBoost | Accuracy = 92.2 |
| Lajnef et al. [276] | 2015 | Awake, S1, S2, SWS, REM | Entropy | Dendrogram-SVM | Accuracy = 92 |
| Ravan [202] | 2019 | Awake, LS and REM, DS | WT | Dendrogram-SVM | Accuracy = 91.4 |
| Kuo and Liang [277] | 2011 | Awake, S1, S2, SWS, REM | Entropy/AR | LDA | Sensitivity = 89.1 |
| Delimayanti et al. [204] | 2020 | Awake, S1, S2, S3, S4, REM | FFT | RBF-SVM | Accuracy = 87.8 |
| Zoubek et al. [275] | 2007 | Awake, NREM1, NREM2, SWS, PS | FFT | MLP | Accuracy = 71.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saeidi, M.; Karwowski, W.; Farahani, F.V.; Fiok, K.; Taiar, R.; Hancock, P.A.; Al-Juaid, A. Neural Decoding of EEG Signals with Machine Learning: A Systematic Review. Brain Sci. 2021, 11, 1525. https://doi.org/10.3390/brainsci11111525
Saeidi M, Karwowski W, Farahani FV, Fiok K, Taiar R, Hancock PA, Al-Juaid A. Neural Decoding of EEG Signals with Machine Learning: A Systematic Review. Brain Sciences. 2021; 11(11):1525. https://doi.org/10.3390/brainsci11111525
Chicago/Turabian StyleSaeidi, Maham, Waldemar Karwowski, Farzad V. Farahani, Krzysztof Fiok, Redha Taiar, P. A. Hancock, and Awad Al-Juaid. 2021. "Neural Decoding of EEG Signals with Machine Learning: A Systematic Review" Brain Sciences 11, no. 11: 1525. https://doi.org/10.3390/brainsci11111525
APA StyleSaeidi, M., Karwowski, W., Farahani, F. V., Fiok, K., Taiar, R., Hancock, P. A., & Al-Juaid, A. (2021). Neural Decoding of EEG Signals with Machine Learning: A Systematic Review. Brain Sciences, 11(11), 1525. https://doi.org/10.3390/brainsci11111525









