African Gene Flow Reduces Beta-Ionone Anosmia/Hyposmia Prevalence in Admixed Malagasy Populations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Material
2.3. Analysis
- Step 1: Perceptual evaluations and detection threshold
- Step 2: Genotyping
2.4. Olfactory Phenotype and Perception Specificity
2.4.1. Olfactory Phenotype
2.4.2. Testing the Perception Specificity
2.5. Genome Data Analysis
2.5.1. GWAS Analysis (Genome-Wide Association Study)
2.5.2. Estimating Local Ancestry: Inference of Ancestry for Each Allele
3. Results
3.1. Genotyping
3.2. Olfactory Phenotype and Perception Specificity
3.3. β-Ionone Perception according to Genotype
3.3.1. Perceptual Evaluations
3.3.2. Detection Scores
4. Genome Data Analysis
4.1. GWAS Analyses
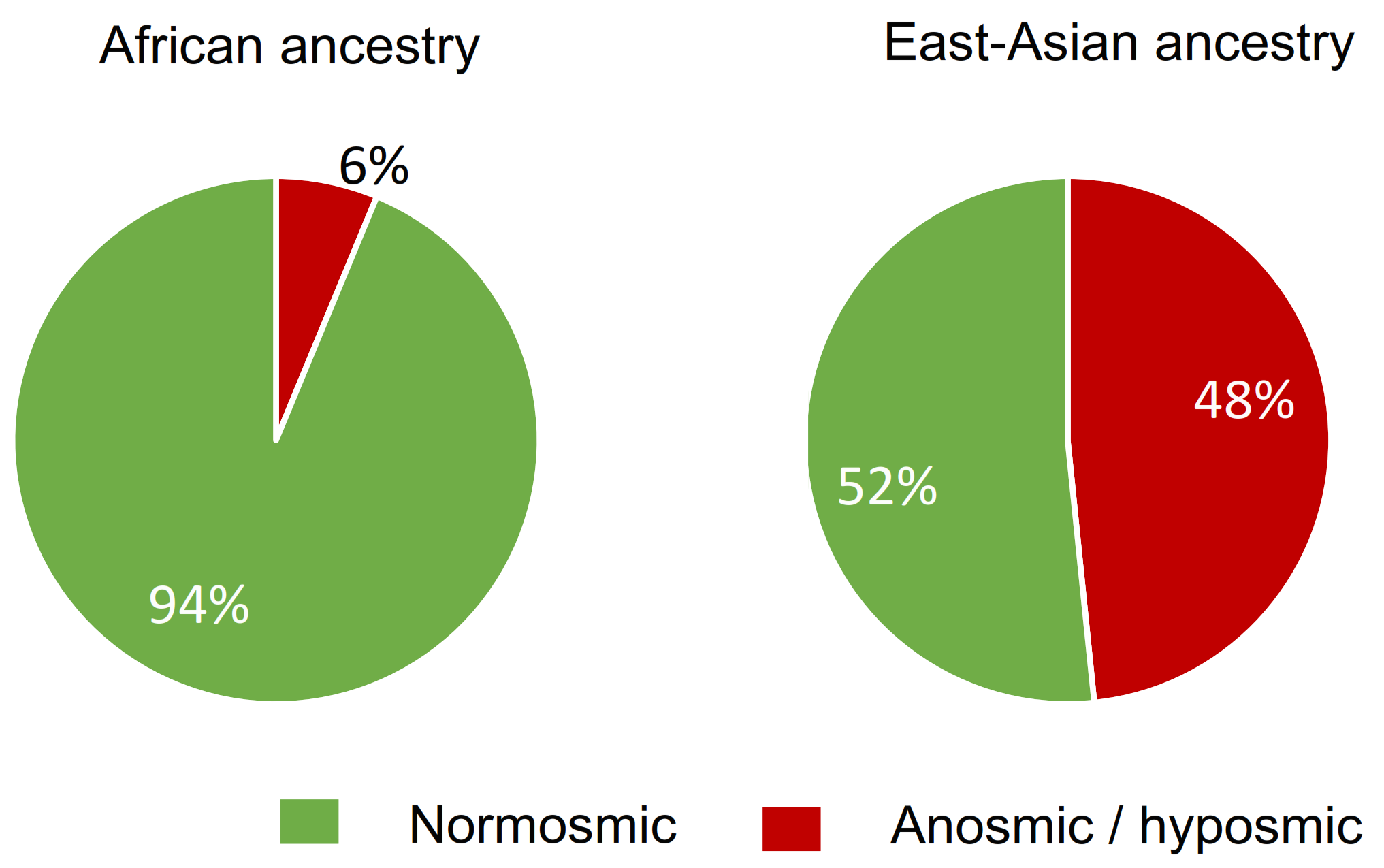
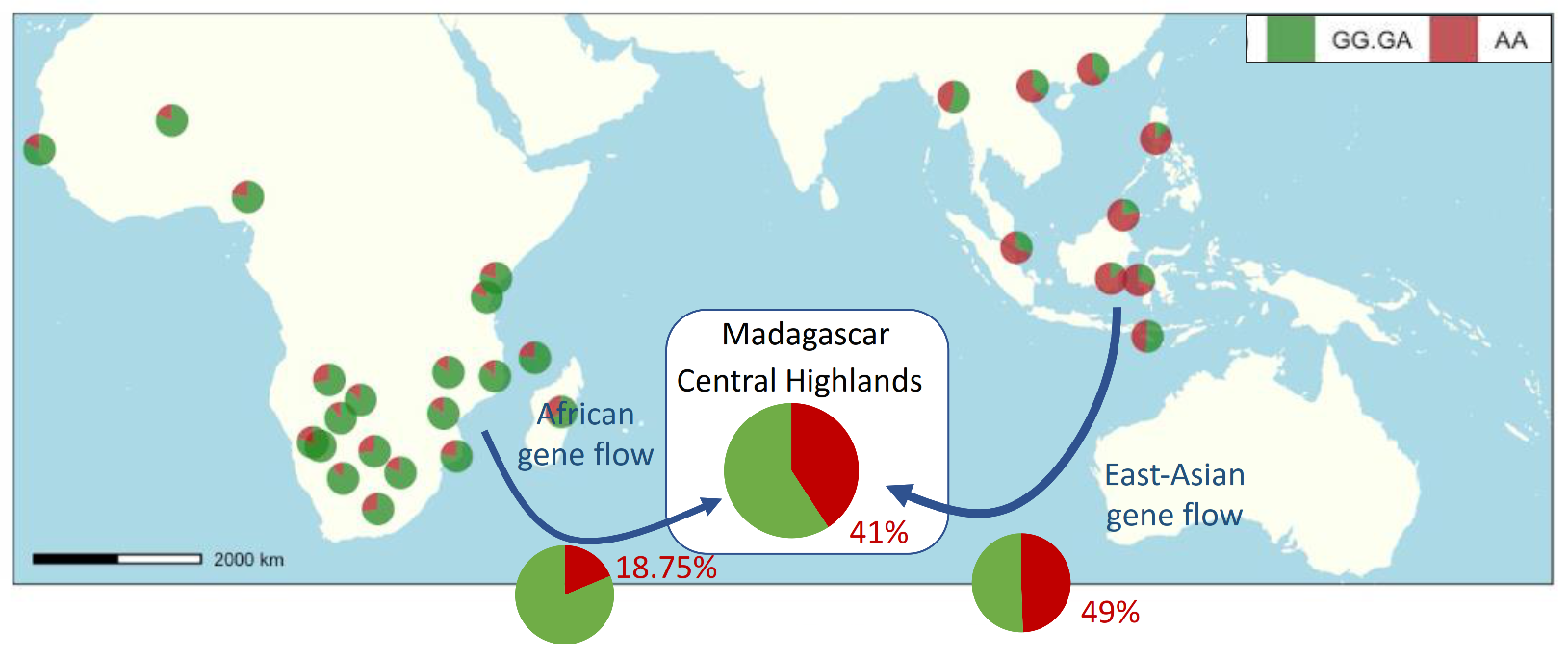
4.2. Relationship between Insensitivity to βI and Local Ancestry
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lunde, K.; Egelandsdal, B.; Skuterud, E.; Mainland, J.D.; Lea, T.; Hersleth, M.; Matsunami, H. Genetic variation of an odorant receptor OR7D4 and sensory perception of cooked meat containing androstenone. PLoS ONE 2012, 7, e35259. [Google Scholar] [CrossRef] [PubMed]
- Keller, A.; Hempstead, M.; Gomez, I.A.; Gilbert, A.N.; Vosshall, L.B. An olfactory demography of a diverse metropolitan population. BMC Neurosci. 2012, 13, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amoore, J.E. A plan to identify most of the primary odors. In Olfaction and Taste III; The Rockefeller University Press: New York, NY, USA, 1969; pp. 158–171. [Google Scholar]
- Pause, B.M. Are androgen steroids acting as pheromones in humans? Physiol. Behav. 2004, 83, 21–29. [Google Scholar] [CrossRef]
- Blanch, M.; Panella-Riera, N.; Chevillon, P.; Furnols, M.F.; Gil, M.; Gil, J.M.; Kallas, Z.; Oliver, M.A. Impact of consumer’s sensitivity to androstenone on acceptability of meat from entire male pigs in three European countries: France, Spain and United Kingdom. Meat Sci. 2012, 90, 572–578. [Google Scholar] [CrossRef]
- Ayabe-Kanamura, S.; Schicker, I.; Laska, M.; Hudson, R.; Distel, H.; Kobayakawa, T.; Saito, S. Differences in perception of everyday odors: A Japanese-German cross-cultural study. Chem. Senses 1998, 23, 31–38. [Google Scholar] [CrossRef] [Green Version]
- Chrea, C.; Valentin, D.; Sulmont-Rossé, C.; Ly Mai, H.; Hoang Nguyen, D.; Abdi, H. Culture and odor categorization: Agreement between cultures depends upon the odors. Food Qual. Prefer. 2004, 15, 669–679. [Google Scholar] [CrossRef] [Green Version]
- Ferdenzi, C.; Roberts, S.C.; Schirmer, A.; Delplanque, S.; Cekic, S.; Porcherot, C.; Cayeux, I.; Sander, D.; Grandjean, D. Variability of affective responses to odors: Culture, gender, and olfactory knowledge. Chem. Senses 2013, 38, 175–186. [Google Scholar] [CrossRef]
- Joussain, P.; Thevenet, M.; Rouby, C.; Bensafi, M. Effect of Aging on Hedonic Appreciation of Pleasant and Unpleasant Odors. PLoS ONE 2013, 8, e61376. [Google Scholar] [CrossRef] [Green Version]
- Doty, R.L. Odor-guided behavior in mammals. Experientia 1986, 42, 257–271. [Google Scholar] [CrossRef]
- Keller, A.; Zhuang, H.; Chi, Q.; Vosshall, L.B.; Matsunami, H. Genetic variation in a human odorant receptor alters odour perception. Nature 2007, 449, 468–472. [Google Scholar] [CrossRef]
- Menashe, I.; Abaffy, T.; Hasin, Y.; Goshen, S.; Yahalom, V.; Luetje, C.W.; Lancet, D. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS Biol. 2007, 5, e284. [Google Scholar] [CrossRef]
- Carty, C.L.; Spencer, K.L.; Setiawan, V.W.; Fernandez-Rhodes, L.; Malinowski, J.; Buyske, S.; Young, A.; Jorgensen, N.W.; Cheng, I.; Carlson, C.S.; et al. Replication of genetic loci for ages at menarche and menopause in the multi-ethnic Population Architecture using Genomics and Epidemiology (PAGE) study. Hum. Reprod. 2013, 28, 1695–1706. [Google Scholar] [CrossRef] [Green Version]
- Jaeger, S.R.; McRae, J.F.; Bava, C.M.; Beresford, M.K.; Hunter, D.; Jia, Y.; Chheang, S.L.; Jin, D.; Peng, M.; Gamble, J.C.; et al. A mendelian trait for olfactory sensitivity affects odor experience and food selection. Curr. Biol. 2013, 23, 1601–1605. [Google Scholar] [CrossRef] [Green Version]
- Sollai, G.; Melis, M.; Magri, S.; Usai, P.; Hummel, T.; Tomassini Barbarossa, I.; Crnjar, R. Association between the rs2590498 polymorphism of Odorant Binding Protein (OBPIIa) gene and olfactory performance in healthy subjects. Behav. Brain Res. 2019, 372, 112030. [Google Scholar] [CrossRef]
- Settle, R.G.; Meehan, K.; Williams, G.R.; Doty, R.L.; Sisley, A.C. Chemosensory properties of sour tastants. Physiol. Behav. 1986, 36, 619–623. [Google Scholar] [CrossRef]
- Doty, R.L.; Gregor, T.; Monroe, C. Quantitative assessment of olfactory function in an industrial setting. J. Occup. Med. 1986, 28, 457–460. [Google Scholar] [CrossRef]
- Jaeger, S.R.; Reinbach, H.C.; Roigard, C.M.; McRae, J.F.; Pineau, B.; Chheang, S.L.; Beresford, M.K.; Rouse, S.A.; Jin, D.; Paisley, A.G.; et al. Sensory characterisation of food and beverage stimuli containing β-ionone and differences between individuals by genotype for rs6591536. Food Res. Int. 2014, 62, 205–214. [Google Scholar] [CrossRef]
- Auton, A.; Abecasis, G.R.; Altshuler, D.M.; Durbin, R.M.; Bentley, D.R.; Chakravarti, A.; Clark, A.G.; Donnelly, P.; Eichler, E.E.; Flicek, P.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar]
- Heiske, M.; Alva, O.; Pereda-Loth, V.; van Schalkwyk, M.; Radimilahy, C.; Letellier, T.; Rakotarisoa, J.A.; Pierron, D. Genetic evidence and historical theories of the Asian and African origins of the present Malagasy population. Hum. Mol. Genet. 2021, 30, R72–R78. [Google Scholar] [CrossRef]
- Fernández-Rhodes, L.; Gong, J.; Haessler, J.; Franceschini, N.; Graff, M.; Nishimura, K.K.; Wang, Y.; Highland, H.M.; Yoneyama, S.; Bush, W.S.; et al. Trans-ethnic fine-mapping of genetic loci for body mass index in the diverse ancestral populations of the Population Architecture using Genomics and Epidemiology (PAGE) Study reveals evidence for multiple signals at established loci. Hum. Genet. 2017, 136, 771–800. [Google Scholar] [CrossRef]
- Hayes, B. Overview of statistical methods for genome-wide association studies (GWAS). Methods Mol. Biol. 2013, 1019, 149–169. [Google Scholar] [CrossRef]
- Menashe, I.; Man, O.; Lancet, D.; Gilad, Y. Different noses for different people. Nat. Genet. 2003, 34, 143–144. [Google Scholar] [CrossRef]
- Menashe, I.; Man, O.; Lancet, D.; Gilad, Y. Population differences in haplotype structure within a human olfactory receptor gene cluster. Hum. Mol. Genet. 2002, 11, 1381–1390. [Google Scholar] [CrossRef]
- Olender, T.; Waszak, S.M.; Viavant, M.; Khen, M.; Ben-Asher, E.; Reyes, A.; Nativ, N.; Wysocki, C.J.; Ge, D.; Lancet, D. Personal receptor repertoires: Olfaction as a model. BMC Genomics 2012, 13, 414. [Google Scholar] [CrossRef] [Green Version]
- Manichaikul, A.; Mychaleckyj, J.C.; Rich, S.S.; Daly, K.; Sale, M.; Chen, W.M. Robust relationship inference in genome-wide association studies. Bioinformatics 2010, 26, 2867–2873. [Google Scholar] [CrossRef] [Green Version]
- Rouby, C.; Thomas-Danguin, T.; Vigouroux, M.; Ciuperca, G.; Jiang, T.; Alexanian, J.; Barges, M.; Gallice, I.; Degraix, J.-L.; Sicard, G. The Lyon Clinical Olfactory Test: Validation and Measurement of Hyposmia and Anosmia in Healthy and Diseased Populations. Int. J. Otolaryngol. 2011, 2011, 203805. [Google Scholar] [CrossRef]
- Mazzatenta, A.; Cellerino, A.; Origlia, N.; Barloscio, D.; Sartucci, F.; Di Giulio, C.; Domenici, L. Olfactory phenotypic expression unveils human aging. Oncotarget 2016, 7, 19193–19200. [Google Scholar] [CrossRef] [Green Version]
- Manesse, C. Le Déficit Olfactif: Le Comprendre, le Diagnostiquer, et Compenser ses Effets sur la Qualité de Vie; Université de Lyon: Lyon, France, 2018. [Google Scholar]
- Marees, A.T.; de Kluiver, H.; Stringer, S.; Vorspan, F.; Curis, E.; Marie-Claire, C.; Derks, E.M. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int. J. Methods Psychiatr. Res. 2018, 27, e1608. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [Green Version]
- Olender, T.; Nativ, N.; Lancet, D. HORDE: Comprehensive resource for olfactory receptor genomics. Methods Mol. Biol. 2013, 1003, 23–38. [Google Scholar] [CrossRef]
- Boivin, N.; Crowther, A.; Prendergast, M.; Fuller, D.Q. Indian Ocean Food Globalisation and Africa. Afr. Archaeol. Rev. 2014, 31, 547–581. [Google Scholar] [CrossRef]
- Plotto, A.; Barnes, K.W.; Goodner, K.L. Specific anosmia observed for β-ionone, but not for α-ionone: Significance for flavor research. J. Food Sci. 2006, 71, S401–S406. [Google Scholar] [CrossRef]
- Pierron, D.; Razafindrazaka, H.; Pagani, L.; Ricaut, F.X.; Antao, T.; Capredon, M.; Sambo, C.; Radimilahy, C.; Rakotoarisoa, J.A.; Blench, R.M.; et al. Genome-wide evidence of austronesian-bantu admixture and cultural reversion in a hunter-gatherer group of Madagascar. Proc. Natl. Acad. Sci. USA 2014, 111, 936–941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Razafindrazaka, H.; Monnereau, A.; Razafindrazaka, D.; Tonasso, L.; Schiavinato, S.; Rakotoarisoa, J.A.; Radimilahy, C.; Letellier, T.; Pierron, D. Genetic admixture and flavor preferences: Androstenone sensitivity in Malagasy populations. Hum. Biol. 2015, 87, 59–70. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez-Kristeller, D.C.; do Nascimento, J.B.P.; Galante, P.A.F.; Malnic, B. Identification of agonists for a group of human odorant receptors. Front. Pharmacol. 2015, 6, 35. [Google Scholar] [CrossRef] [Green Version]
- Sanz, G.; Schlegel, C.; Pernollet, J.C.; Briand, L. Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chem. Senses 2005, 30, 69–80. [Google Scholar] [CrossRef]
- Sorokowska, A.; Drechsler, E.; Karwowski, M.; Hummel, T. Effects of olfactory training: A meta-analysis. Rhinology 2017, 55, 17–26. [Google Scholar] [CrossRef]
- Buttery, R.G.; Ling, L.C.; Stern, D.J. Studies on Popcorn Aroma and Flavor Volatiles. J. Agric. Food Chem. 1997, 45, 837–843. [Google Scholar] [CrossRef]
- Larsen, M.; Poll, L.; Callesen, O.; Lewis, M. Relations Between the Content of Aroma Compounds and the Sensory Evaluation of 10 Raspberry Varieties (Rubus Idaeus L.). Acta Agric. Scand. 1991, 41, 447–454. [Google Scholar] [CrossRef]
- Mahattanatawee, K.; Rouseff, R.; Valim, M.F.; Naim, M. Identification and aroma impact of norisoprenoids in orange juice. J. Agric. Food Chem. 2005, 53, 393–397. [Google Scholar] [CrossRef]
- Etievant, P.X.; Issanchou, S.N.; Bayonove, C.L. The flavour of muscat wine: The sensory contribution of some volatile compounds. J. Sci. Food Agric. 1983, 34, 497–504. [Google Scholar] [CrossRef]
- Pierron, D.; Heiske, M.; Razafindrazaka, H.; Pereda-Loth, V.; Sanchez, J.; Alva, O.; Arachiche, A.; Boland, A.; Olaso, R.; Deleuze, J.F.; et al. Strong selection during the last millennium for African ancestry in the admixed population of Madagascar. Nat. Commun. 2018, 9, 932. [Google Scholar] [CrossRef]
- Fuller, D.Q.; Boivin, N.; Hoogervorst, T.; Allaby, R. Across the Indian Ocean: The prehistoric movement of plants and animals. Antiquity 2011, 85, 544–558. [Google Scholar] [CrossRef] [Green Version]
- Boivin, N.; Fuller, D.Q. Shell middens, ships and seeds: Exploring coastal subsistence, maritime trade and the dispersal of domesticates in and around the ancient Arabian Peninsula. J. World Prehistory 2009, 22, 113–180. [Google Scholar] [CrossRef]
- Beaujard, P. The first migrants to Madagascar and their introduction of plants: Linguistic and ethnological evidence. Azania 2011, 46, 169–189. [Google Scholar] [CrossRef] [Green Version]
- Adelaar, S. Towards an integrated theory about the Indonesian migrations to Madagascar. In Ancient Huma Migrations: A Multidisciplinary Approach; University of Utah Press: Salt Lake City, UT, USA, 2009; pp. 149–172. [Google Scholar]
- Blench, R. The Austronesians in Madagascar and Their Interaction with the Bantu of the East African Coast: Surveying the Linguistic Evidence for Domestic and Translocated Animals. Stud. Phiippine Lang. Cult. 2008, 18, 18–43. [Google Scholar]
- Jordan, I.K. The Columbian Exchange as a source of adaptive introgression in human populations. Biol. Direct 2016, 11, 17. [Google Scholar] [CrossRef] [Green Version]
- Adhikari, K.; Mendoza-Revilla, J.; Chacón-Duque, J.C.; Fuentes-Guajardo, M.; Ruiz-Linares, A. Admixture in Latin America. Curr. Opin. Genet. Dev. 2016, 41, 106–114. [Google Scholar] [CrossRef]
- Brucato, N.; Kusuma, P.; Cox, M.P.; Pierron, D.; Purnomo, G.A.; Adelaar, A.; Kivisild, T.; Letellier, T.; Sudoyo, H.; Ricaut, F.X. Malagasy Genetic Ancestry Comes from an Historical Malay Trading Post in Southeast Borneo. Mol. Biol. Evol. 2016, 33, 2396–2400. [Google Scholar] [CrossRef] [Green Version]
- Busby, G.B.; Band, G.; Si Le, Q.; Jallow, M.; Bougama, E.; Mangano, V.D.; Amenga-Etego, L.N.; Enimil, A.; Apinjoh, T.; Ndila, C.M.; et al. Admixture into and within sub-Saharan Africa. eLife 2016, 5, e15266. [Google Scholar] [CrossRef]
- Mörseburg, A.; Pagani, L.; Ricaut, F.X.; Yngvadottir, B.; Harney, E.; Castillo, C.; Hoogervorst, T.; Antao, T.; Kusuma, P.; Brucato, N.; et al. Multi-layered population structure in Island Southeast Asians. Eur. J. Hum. Genet. 2016, 24, 1605–1611. [Google Scholar] [CrossRef]
- Brucato, N.; Fernandes, V.; Mazières, S.; Kusuma, P.; Cox, M.P.; Ng’ang’a, J.W.; Omar, M.; Simeone-Senelle, M.C.; Frassati, C.; Alshamali, F.; et al. The Comoros Show the Earliest Austronesian Gene Flow into the Swahili Corridor. Am. J. Hum. Genet. 2018, 102, 58–68. [Google Scholar] [CrossRef] [Green Version]
- Schlebusch, C.M.; Skoglund, P.; Sjödin, P.; Gattepaille, L.M.; Hernandez, D.; Jay, F.; Li, S.; de Jongh, M.; Singleton, A.; Blum, M.G.B.; et al. Genomic variation in seven Khoe-San groups reveals adaptation and complex African history. Science 2012, 338, 374–379. [Google Scholar] [CrossRef] [Green Version]
- Semo, A.; Gayà-Vidal, M.; Fortes-Lima, C.; Alard, B.; Oliveira, S.; Almeida, J.; Prista, A.; Damasceno, A.; Fehn, A.M.; Schlebusch, C.; et al. Along the Indian Ocean Coast: Genomic Variation in Mozambique Provides New Insights into the Bantu Expansion. Mol. Biol. Evol. 2020, 37, 406–416. [Google Scholar] [CrossRef] [Green Version]
- Teo, Y.Y.; Sim, X.; Ong, R.T.H.; Tan, A.K.S.; Chen, J.; Tantoso, E.; Small, K.S.; Ku, C.S.; Lee, E.J.D.; Seielstad, M.; et al. Singapore Genome Variation Project: A haplotype map of three Southeast Asian populations. Genome Res. 2009, 19, 2154–2162. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y. Detecting structure of haplotypes and local ancestry. Genetics 2014, 196, 625–642. [Google Scholar] [CrossRef] [Green Version]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [Green Version]
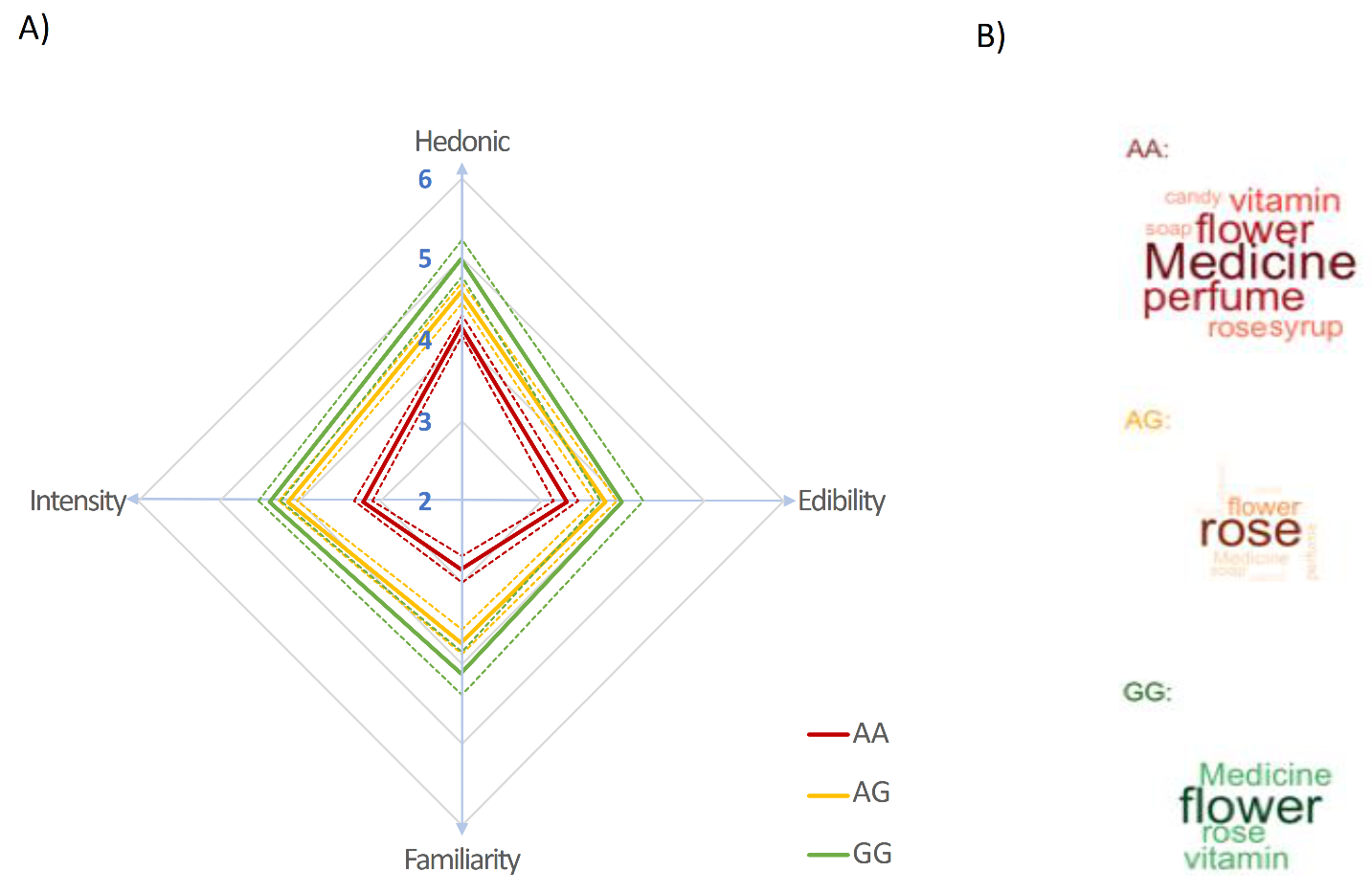

| Words | AA | AG | GG | Total |
|---|---|---|---|---|
| rose | 6 | 19 | 3 | 28 |
| flower | 9 | 9 | 5 | 23 |
| medicine | 11 | 6 | 3 | 20 |
| perfume | 9 | 5 | 2 | 16 |
| vitamin | 7 | 4 | 3 | 14 |
| soap | 4 | 5 | 2 | 11 |
| candy | 4 | 3 | 7 | |
| syrup | 6 | 1 | 7 | |
| fruits | 2 | 3 | 5 |
| Genotype Frequency | |||||
|---|---|---|---|---|---|
| AA | GA | GG | Total | ||
| Locus ancestry | AF/AF | 18.75% (3) | 62.5% (10) | 18.75% (3) | 16 |
| AS/AF | 36.08% (35) | 46.39% (45) | 17.53% (17) | 97 | |
| AS/AS | 48.94% (46) | 42.55% (40) | 8.51% (8) | 93 | |
| Total | 40.58% (84) | 45.89% (94) | 13.53% (28) | 206 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Razafindrazaka, H.; Pereda-Loth, V.; Ferdenzi, C.; Heiske, M.; Alva, O.; Randriamialisoa, M.; Costedoat, C.; Signoli, M.; Talou, T.; Courtade-Saidi, M.; et al. African Gene Flow Reduces Beta-Ionone Anosmia/Hyposmia Prevalence in Admixed Malagasy Populations. Brain Sci. 2021, 11, 1405. https://doi.org/10.3390/brainsci11111405
Razafindrazaka H, Pereda-Loth V, Ferdenzi C, Heiske M, Alva O, Randriamialisoa M, Costedoat C, Signoli M, Talou T, Courtade-Saidi M, et al. African Gene Flow Reduces Beta-Ionone Anosmia/Hyposmia Prevalence in Admixed Malagasy Populations. Brain Sciences. 2021; 11(11):1405. https://doi.org/10.3390/brainsci11111405
Chicago/Turabian StyleRazafindrazaka, Harilanto, Veronica Pereda-Loth, Camille Ferdenzi, Margit Heiske, Omar Alva, Minah Randriamialisoa, Caroline Costedoat, Michel Signoli, Thierry Talou, Monique Courtade-Saidi, and et al. 2021. "African Gene Flow Reduces Beta-Ionone Anosmia/Hyposmia Prevalence in Admixed Malagasy Populations" Brain Sciences 11, no. 11: 1405. https://doi.org/10.3390/brainsci11111405
APA StyleRazafindrazaka, H., Pereda-Loth, V., Ferdenzi, C., Heiske, M., Alva, O., Randriamialisoa, M., Costedoat, C., Signoli, M., Talou, T., Courtade-Saidi, M., Boland, A., Deleuze, J.-F., Rouby, C., Radimilahy, C., Letellier, T., Bensafi, M., & Pierron, D. (2021). African Gene Flow Reduces Beta-Ionone Anosmia/Hyposmia Prevalence in Admixed Malagasy Populations. Brain Sciences, 11(11), 1405. https://doi.org/10.3390/brainsci11111405







