Characterization of Four Liver-Expressed Antimicrobial Peptides from Antarctic Fish and Their Antibacterial Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatic Analysis and Peptide Synthesis
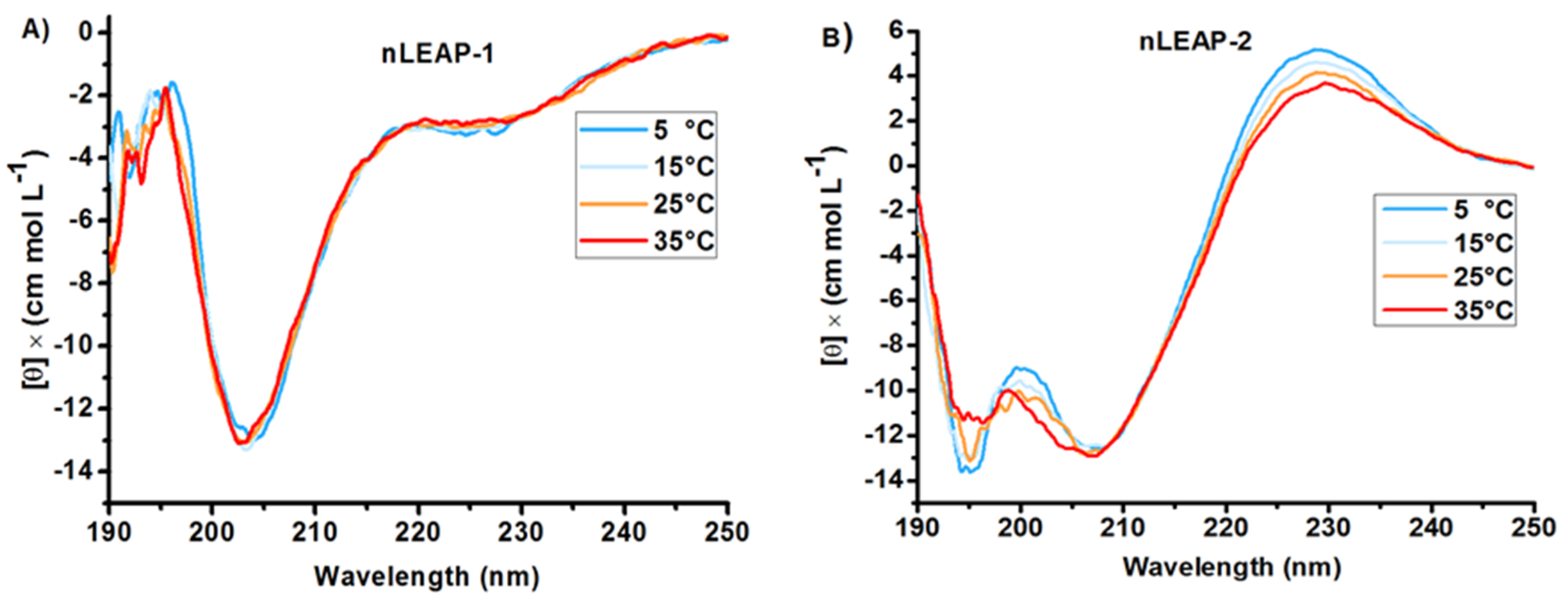
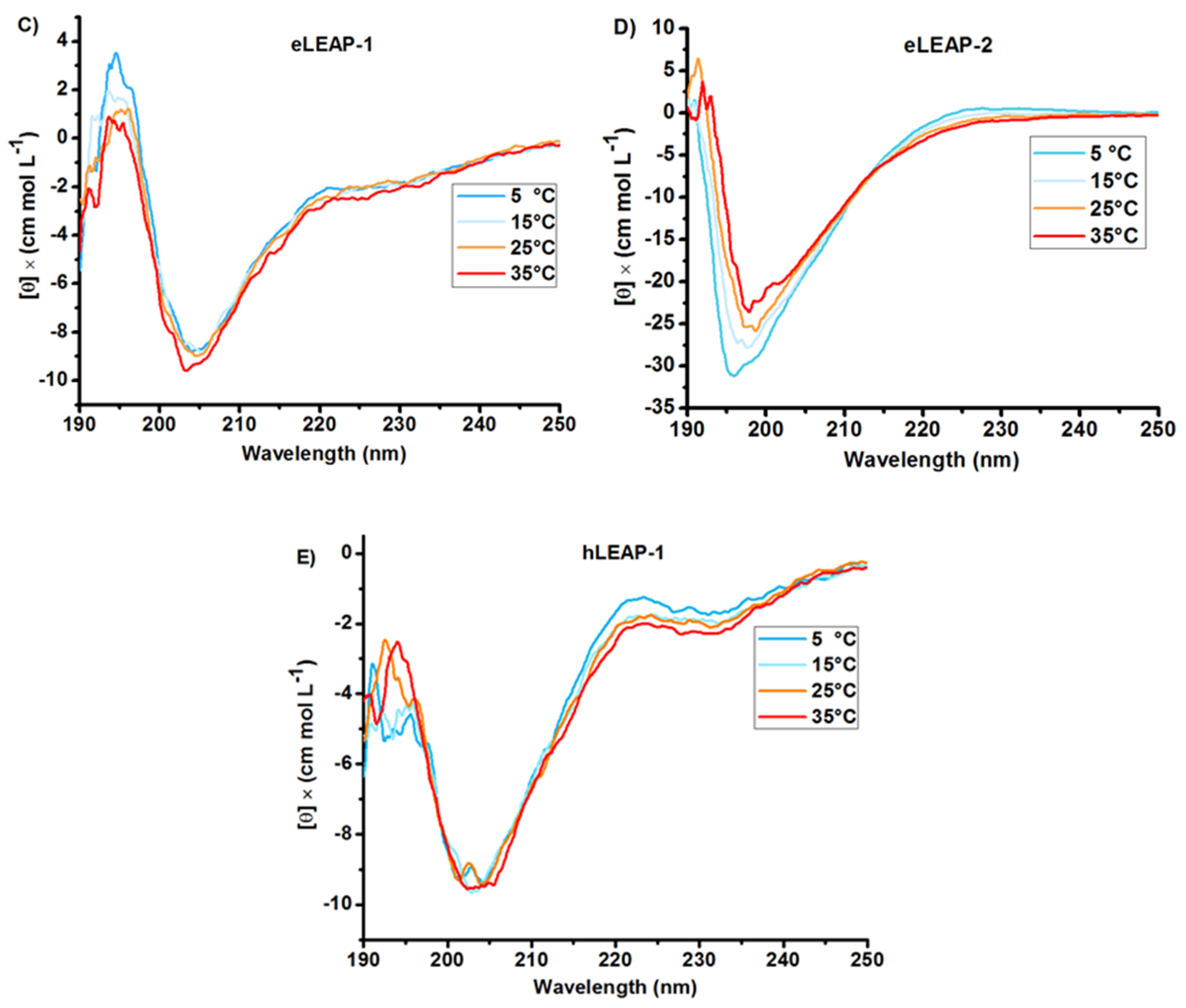
2.2. Circular Dichroism (CD) Spectroscopy
2.3. Antibacterial Assay
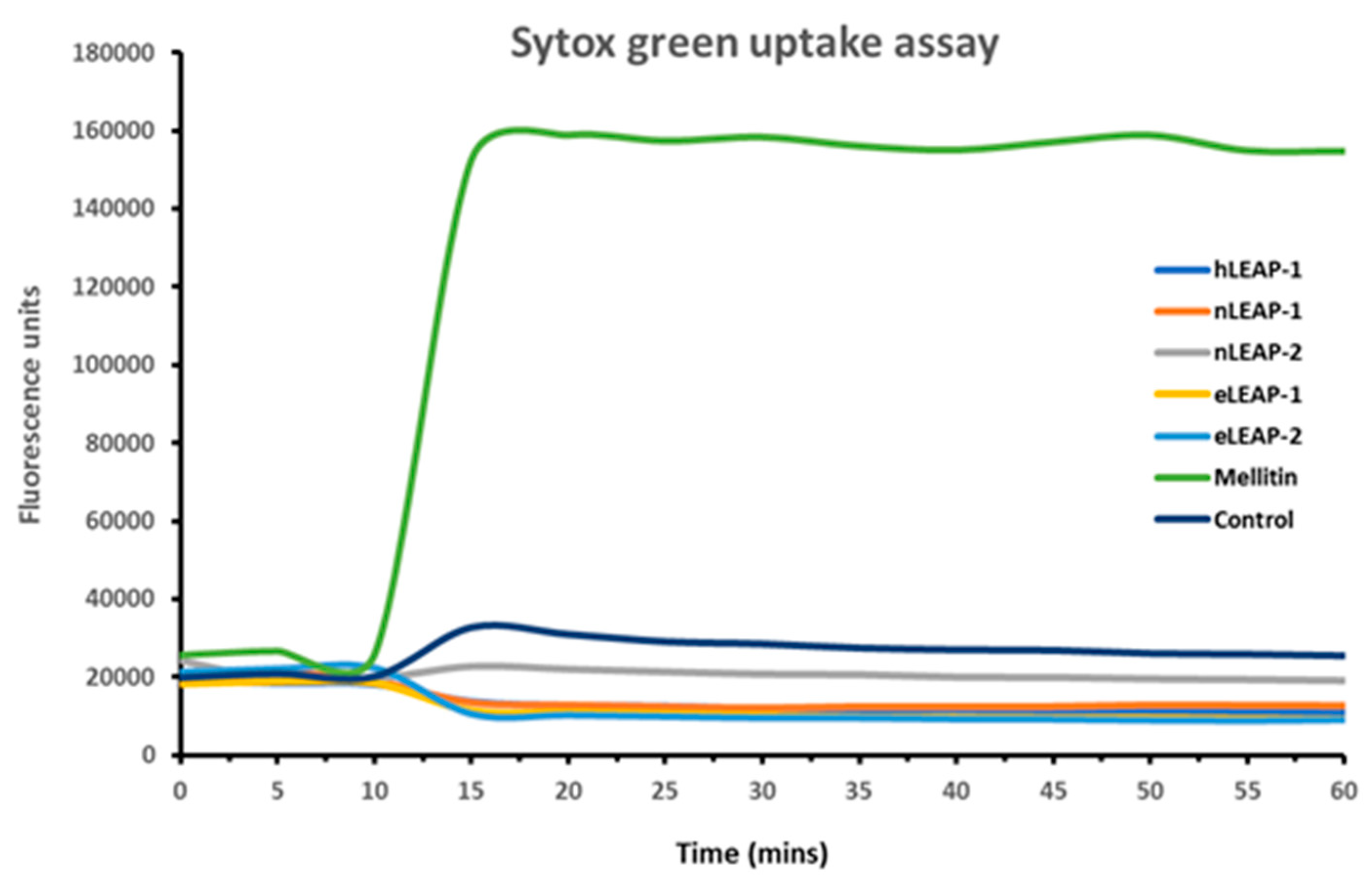
2.4. SYTOX Green Uptake Assay
2.5. Competitive Dye Displacement Assay
3. Results
3.1. Bioinformatics and CD Analysis of LEAPs
3.2. Antibacterial Activity
3.3. Mechanism of Antibacterial Action
3.4. Competitive Dye Displacement Assay
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Park, C.H.; Valore, E.V.; Waring, A.J.; Ganz, T. Hepcidin, a Urinary Antimicrobial Peptide Synthesized in the Liver. J. Biol. Chem. 2001, 276, 7806–7810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krause, A.; Neitz, S.; Schulz, A.; Forssmann, W.; Schulz-knappe, P.; Adermann, K. LEAP-1, a novel highly disul ¢ de-bonded human peptide, exhibits antimicrobial activity 1. FEBS Lett. 2000, 480, 147–150. [Google Scholar] [CrossRef]
- Krause, A. Isolation and biochemical characterization of LEAP-2, a novel blood peptide expressed in the liver. Protein Sci. 2003, 12, 143–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordan, J.B.; Poppe, L.; Haniu, M.; Arvedson, T.; Syed, R.; Li, V.; Kohno, H.; Kim, H.; Schnier, P.D.; Harvey, T.S.; et al. Hepcidin revisited, disulfide connectivity, dynamics, and structure. J. Biol. Chem. 2009, 284, 24155–24167. [Google Scholar] [CrossRef] [PubMed]
- A’lvarez, C.A.; Guzma´n, F.; Cárdenas, C.; Marshall, S.H.; Mercado, L. Antimicrobial activity of trout hepcidin. Fish Shellfish Immunol. 2014, 41, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Harford, C.; Sarkar, B. Amino Terminal Cu(II)- and Ni(II)-Binding (ATCUN) Motif of Proteins and Peptides: Metal Binding, DNA Cleavage, and Other Properties †. Acc. Chem. Res. 1997, 30, 123–130. [Google Scholar] [CrossRef]
- Libardo, M.D.J.; Paul, T.J.; Prabhakar, R.; Angeles-Boza, A.M. Hybrid peptide ATCUN-sh-Buforin: Influence of the ATCUN charge and stereochemistry on antimicrobial activity. Biochimie 2015, 113, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Shike, H.; Lauth, X.; Westerman, M.E.; Ostland, V.E.; Carlberg, J.M.; Van Olst, J.C.; Shimizu, C.; Bulet, P.; Burns, J.C. Bass hepcidin is a novel antimicrobial peptide induced by bacterial challenge. Eur. J. Biochem. 2002, 269, 2232–2237. [Google Scholar] [CrossRef]
- Townes, C.L.; Michailidis, G.; Hall, J. The interaction of the antimicrobial peptide cLEAP-2 and the bacterial membrane. Biochem. Biophys. Res. Commun. 2009, 387, 500–503. [Google Scholar] [CrossRef]
- Liu, D.; Pu, Y.; Xiong, H.; Wang, Y.; Du, H. Porcine hepcidin exerts an iron-independent bacteriostatic activity against pathogenic bacteria. Int. J. Pept. Res. Ther. 2015, 21, 229–236. [Google Scholar] [CrossRef]
- Lou, D.Q.; Nicolas, G.; Lesbordes, J.C.; Viatte, L.; Grimber, G.; Szajnert, M.F.; Kahn, A.; Vaulont, S. Functional differences between hepcidin 1 and 2 in transgenic mice. Blood 2004, 103, 2816–2821. [Google Scholar] [CrossRef] [PubMed]
- Locke, J.B.; Colvin, K.M.; Varki, N.; Vicknair, M.R.; Nizet, V.; Buchanan, J.T. Streptococcus iniae β-hemolysin streptolysin S is a virulence factor in fish infection. Dis. Aquat. Organ. 2007, 76, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.J.; Cai, J.J.; Cai, L.; Qu, H.D.; Yang, M.; Zhang, M. Cloning and expression of a hepcidin gene from a marine fish (Pseudosciaena crocea) and the antimicrobial activity of its synthetic peptide. Peptides 2009, 30, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Lauth, X.; Babon, J.J.; Stannard, J.A.; Singh, S.; Nizet, V.; Carlberg, J.M.; Ostland, V.E.; Pennington, M.W.; Norton, R.S.; Westerman, M.E. Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J. Biol. Chem. 2005, 280, 9272–9282. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.H.; Chen, J.Y.; Kuo, C.M. Three different hepcidins from tilapia, Oreochromis mossambicus: Analysis of their expressions and biological functions. Mol. Immunol. 2007, 44, 1922–1934. [Google Scholar] [CrossRef] [PubMed]
- Ke, F.; Wang, Y.; Yang, C.S.; Xu, C. Molecular cloning and antibacterial activity of hepcidin from chinese rare minnow (Gobiocypris rarus). Electron. J. Biotechnol. 2015, 18, 169–174. [Google Scholar] [CrossRef]
- Yang, M.; Chen, B.; Cai, J.J.; Peng, H.; Ling-Cai; Yuan, J.J.; Wang, K.J. Molecular characterization of hepcidin AS-hepc2 and AS-hepc6 in black porgy (Acanthopagrus schlegelii): Expression pattern responded to bacterial challenge and in vitro antimicrobial activity. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2011, 158, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Cheng, C.H.C.; Hu, P.; Ye, H.; Chen, Z.; Cao, L.; Chen, L.L.; Shen, Y.; Chen, L.L. Adaptive evolution of hepcidin genes in antarctic notothenioid fishes. Mol. Biol. Evol. 2008, 25, 1099–1112. [Google Scholar] [CrossRef] [PubMed]
- Rajanbabu, V.; Chen, J.Y. Applications of antimicrobial peptides from fish and perspectives for the future. Peptides 2011, 32, 415–420. [Google Scholar] [CrossRef] [PubMed]
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.G.; Wei, J.G.; Xu, D.; Cui, H.C.; Yan, Y.; Ou-Yang, Z.L.; Huang, X.H.; Huang, Y.H.; Qin, Q.W. Molecular cloning and characterization of two novel hepcidins from orange-spotted grouper, Epinephelus coioides. Fish Shellfish Immunol. 2011, 30, 559–568. [Google Scholar] [CrossRef] [PubMed]
- Neves, J.V.; Ramos, M.F.; Moreira, A.C.; Silva, T.; Gomes, M.S.; Rodrigues, P.N.S. Hamp1 but not Hamp2 regulates ferroportin in fish with two functionally distinct hepcidin types. Sci. Rep. 2017, 7, 14793. [Google Scholar] [CrossRef] [PubMed]
- Neves, J.V.; Caldas, C.; Vieira, I.; Ramos, M.F.; Rodrigues, P.N.S. Multiple Hepcidins in a Teleost Fish, Dicentrarchus labrax: Different Hepcidins for Different Roles. J. Immunol. 2015, 195, 2696–2709. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.C.; Ahn, I.H.; Ahn, D.H.; Lee, Y.M.; Lee, J.; Lee, J.H.; Kim, H.W.; Park, H. Characterization of two antimicrobial peptides from antarctic fishes (Notothenia coriiceps and parachaenichthys charcoti). PLoS ONE 2017, 12, e0170821. [Google Scholar] [CrossRef] [PubMed]
- Czuprynski, C.J.; Brown, J.F.; Roll, J.T. Growth at reduced temperatures increases the virulence of Listeria monocytogenes for intravenously but not intragastrically inoculated mice. Microb. Pathog. 1989, 7, 213–223. [Google Scholar] [CrossRef]
- Schmid, B.; Klumpp, J.; Raimann, E.; Loessner, M.J.; Stephan, R.; Tasara, T. Role of cold shock proteins in growth of Listeria monocytogenes under cold and osmotic stress conditions. Appl. Environ. Microbiol. 2009, 75, 1621–1627. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Rajasekaran, G.; Shin, S.Y. LL-37-derived short antimicrobial peptide KR-12-a5 and its D-amino acid substituted analogs with cell selectivity, anti-biofilm activity, synergistic effect with conventional antibiotics, and anti-inflammatory activity. Eur. J. Med. Chem. 2017, 136, 428–441. [Google Scholar] [CrossRef] [PubMed]
- Cutrona, K.J.; Kaufman, B.A.; Figueroa, D.M.; Elmore, D.E. Role of arginine and lysine in the antimicrobial mechanism of histone-derived antimicrobial peptides. FEBS Lett. 2015, 589, 3915–3920. [Google Scholar] [CrossRef] [Green Version]
- Maisetta, G.; Petruzzelli, R.; Brancatisano, F.L.; Esin, S.; Vitali, A.; Campa, M.; Batoni, G. Antimicrobial activity of human hepcidin 20 and 25 against clinically relevant bacterial strains: Effect of copper and acidic pH. Peptides 2010, 31, 1995–2002. [Google Scholar] [CrossRef]
- Rajasekaran, G.; Kim, E.Y.; Shin, S.Y. LL-37-derived membrane-active FK-13 analogs possessing cell selectivity, anti-biofilm activity and synergy with chloramphenicol and anti-inflammatory activity. Biochim. Biophys. Acta Biomembr. 2017, 1859, 722–733. [Google Scholar] [CrossRef]
- Roth, B.L.; Poot, M.; Yue, S.T.; Millard, P.J. Bacterial viability and antibiotic susceptibility testing with SYTOX green nucleic acid stain. Appl. Environ. Microbiol. 1997, 63, 2421–2431. [Google Scholar] [PubMed]
- Papadopoulos, A.N.; Zoi, I.; Raptopoulou, C.P.; Kessissoglou, D.P.; Tarushi, A.; Psycharis, V.; Kakoulidou, C.; Psomas, G. Zinc complexes of diflunisal: Synthesis, characterization, structure, antioxidant activity, and in vitro and in silico study of the interaction with DNA and albumins. J. Inorg. Biochem. 2017, 170, 85–97. [Google Scholar] [Green Version]
- Russo, R.; Riccio, A.; di Prisco, G.; Verde, C.; Giordano, D. Molecular adaptations in Antarctic fish and bacteria. Polar Sci. 2010, 4, 245–256. [Google Scholar] [CrossRef] [Green Version]
- Ellis, A.E. Innate host defense mechanisms of fish against viruses and bacteria. Dev. Comp. Immunol. 2001, 25, 827–839. [Google Scholar] [CrossRef]
- Uribe, C.; Folch, H.; Enriquez, R.; Moran, G. Innate and adaptive immunity in teleost fish: a review. Vet. Med. 2011, 56, 486–503. [Google Scholar] [CrossRef]
- Kemna, E.H.J.M.; Tjalsma, H.; Willems, H.L.; Swinkels, D.W. Hepcidin: From discovery to differential diagnosis. Haematologica 2008, 93, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Lin, W.J.; Lin, T.L. A fish antimicrobial peptide, tilapia hepcidin TH2-3, shows potent antitumor activity against human fibrosarcoma cells. Peptides 2009, 30, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Sun, Y.; Shi, G.; Wang, R. Miiuy Croaker Hepcidin Gene and Comparative Analyses Reveal Evidence for Positive Selection. PLoS ONE 2012, 7, e35449. [Google Scholar] [CrossRef]
- Hilton, K.B.; Lambert, L.A. Molecular evolution and characterization of hepcidin gene products in vertebrates. Gene 2008, 415, 40–48. [Google Scholar] [CrossRef]
- Álvarez, C.A.; Acosta, F.; Montero, D.; Guzmán, F.; Torres, E.; Vega, B.; Mercado, L. Synthetic hepcidin from fish: Uptake and protection against Vibrio anguillarum in sea bass (Dicentrarchus labrax). Fish Shellfish Immunol. 2016, 55, 662–670. [Google Scholar] [CrossRef]
- Qu, H.; Chen, B.; Peng, H.; Wang, K. Molecular Cloning, Recombinant Expression, and Antimicrobial Activity of EC-hepcidin3, a New Four-Cysteine Hepcidin Isoform from Epinephelus coioides. Biosci. Biotechnol. Biochem 2013, 77, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Baharith, L.A.; Al-khouli, A.; Raab, G.M.; Cd, N.; Oa, C.; Ene, C.; Nicolae, I.I.; Fulga, I.; Harford, C.; Sarkar, B.; et al. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Peptides 2011, 30, 1–4. [Google Scholar]
- Tam, J.P.; Lu, Y.A.; Yang, J.L. Design of salt-insensitive glycine-rich antimicrobial peptides with cyclic tricystine structures. Biochemistry 2000, 39, 7159–7169. [Google Scholar] [CrossRef] [PubMed]
- Nemeth, E.; Preza, G.C.; Jung, C.L.; Kaplan, J.; Waring, A.J.; Ganz, T. The N-terminus of hepcidin is essential for its interaction with ferroportin: Structure-function study. Blood 2006, 107, 328–333. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Chapple, D.S. MINIREVIEW Peptide Antibiotics. Antimicrob. Agents Chemother. 1999, 43, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Hancock, R.E.W.; Diamond, G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000, 8, 402–410. [Google Scholar] [CrossRef]
- Findlay, B.; Zhanel, G.G.; Schweizer, F. Cationic amphiphiles, a new generation of antimicrobials inspired by the natural antimicrobial peptide scaffold. Antimicrob. Agents Chemother. 2010, 54, 4049–4058. [Google Scholar] [CrossRef] [PubMed]
- Malanovic, N.; Lohner, K. Antimicrobial peptides targeting Gram-positive bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar] [CrossRef]
- Yang, G.; Guo, H.; Li, H.; Shan, S.; Zhang, X.; Rombout, J.H.W.M.; An, L. Molecular characterization of LEAP-2 cDNA in common carp (Cyprinus carpio L.) and the differential expression upon a Vibrio anguillarum stimulus; indications for a significant immune role in skin. Fish Shellfish Immunol. 2014, 37, 22–29. [Google Scholar] [CrossRef]
- Clarke, A.; Crame, J.A.; Stromberg, J.; Barker, P.F.; Trans, P.; Lond, R.S. The Southern Ocean benthic fauna and climate change: a historical perspective. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2006, 338, 299–309. [Google Scholar]
- Buonocore, F.; Randelli, E.; Casani, D.; Picchietti, S.; Belardinelli, M.C.; de Pascale, D.; De Santi, C.; Scapigliati, G. A piscidin-like antimicrobial peptide from the icefish Chionodraco hamatus (Perciformes: Channichthyidae): Molecular characterization, localization and bactericidal activity. Fish Shellfish Immunol. 2012, 33, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Mathew, B.; Nagaraj, R. Variations in the interaction of human defensins with Escherichia coli: Possible implications in bacterial killing. PLoS ONE 2017, 12, e0175858. [Google Scholar] [CrossRef] [PubMed]
- Tse, W.C.; Boger, D.L. A Fluorescent Intercalator Displacement Assay for Establishing DNA Binding Selectivity and Affinity. Acc. Chem. Res. 2004, 37, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, P. Multifunctional host defense peptides: Intracellular-targeting antimicrobial peptides. FEBS J. 2009, 276, 6483–6496. [Google Scholar] [CrossRef] [PubMed]
- Thundimadathil, J.; Roeske, R.W.; Jiang, H.Y.; Guo, L. Aggregation and porin-like channel activity of a β sheet peptide. Biochemistry 2005, 44, 10259–10270. [Google Scholar] [CrossRef]
- Gupta, K.; Jang, H.; Harlen, K.; Puri, A.; Nussinov, R.; Schneider, J.P.; Blumenthal, R. Mechanism of membrane permeation induced by synthetic β-hairpin peptides. Biophys. J. 2013, 105, 2093–2103. [Google Scholar] [CrossRef]
- Zheng, Z.; Tharmalingam, N.; Liu, Q.; Jayamani, E.; Kim, W.; Fuchs, B.B.; Zhang, R.; Vilcinskas, A.; Mylonakis, E. Synergistic efficacy of Aedes aegypti antimicrobial peptide cecropin A2 and tetracycline against Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2017, 61, 1–15. [Google Scholar] [CrossRef]
- Park, C.B.; Kim, H.S.; Kim, S.C. Mechanism of action of the antimicrobial peptide buforin II: Buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 1998, 244, 253–257. [Google Scholar] [CrossRef]
- Huo, L.; Zhang, K.; Ling, J.; Peng, Z.; Huang, X.; Liu, H.; Gu, L. Antimicrobial and DNA-binding activities of the peptide fragments of human lactoferrin and histatin 5 against Streptococcus mutans. Arch. Oral Biol. 2011, 56, 869–876. [Google Scholar] [CrossRef]
- Li, L.; Shi, Y.; Cheng, X.; Xia, S.; Cheserek, M.J.; Le, G. A cell-penetrating peptide analogue, P7, exerts antimicrobial activity against Escherichia coli ATCC25922 via penetrating cell membrane and targeting intracellular DNA. Food Chem. 2015, 166, 231–239. [Google Scholar] [CrossRef]
- Li, L.; Shi, Y.; Cheserek, M.J.; Su, G.; Le, G. Antibacterial activity and dual mechanisms of peptide analog derived from cell-penetrating peptide against Salmonella typhimurium and Streptococcus pyogenes. Appl. Microbiol. Biotechnol. 2013, 97, 1711–1723. [Google Scholar] [CrossRef] [PubMed]
- Mathew, B.; Nagaraj, R. Antimicrobial activity of human α-defensin 5 and its linear analogs: N-terminal fatty acylation results in enhanced antimicrobial activity of the linear analogs. Peptides 2015, 71, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Khuller, G. DNA as the intracellular secondary target for antibacterial action of human neutrophil peptide-I against Mycobacterium tuberculosis H 37 Ra. Curr. Microbiol. 2001, 43, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Lin, X.; Long, Y.; Wang, Y.; Zhang, Y.; Mi, H.; Yan, H. A peptide fragment derived from the T-cell antigen receptor protein α-chain adopts β-sheet structure and shows potent antimicrobial activity. Peptides 2009, 30, 647–653. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Shi, Y.; Su, G.; Le, G. Selectivity for and destruction of Salmonella typhimurium via a membrane damage mechanism of a cell-penetrating peptide ppTG20 analogue. Int. J. Antimicrob. Agents 2012, 40, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Ulvatne, H.; Samuelsen, Ø.; Haukland, H.H.; Krämer, M.; Vorland, L.H. Lactoferricin B inhibits bacterial macromolecular synthesis in Escherichia coli and Bacillus subtilis. FEMS Microbiol. Lett. 2004, 237, 377–384. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Camus, A.C. Hepcidins in amphibians and fishes: Antimicrobial peptides or iron-regulatory hormones? Dev. Comp. Immunol. 2006, 30, 746–755. [Google Scholar] [CrossRef]
- Boumaiza, M.; Chahed, H.; Ezzine, A.; Jaouan, M.; Gianoncelli, A.; Longhi, G.; Carmona, F.; Arosio, P.; Sari, M.A.; Marzouki, M.N. Recombinant overexpression of camel hepcidin cDNA in Pichia pastoris: purification and characterization of the polyHis-tagged peptide HepcD-His. J. Mol. Recognit. 2017, 30, 1–11. [Google Scholar] [CrossRef]
- Yang, N.; Liu, X.; Teng, D.; Li, Z.; Wang, X.; Mao, R.; Wang, X.; Hao, Y.; Wang, J. Antibacterial and detoxifying activity of NZ17074 analogues with multi-layers of selective antimicrobial actions against Escherichia coli and Salmonella enteritidis. Sci. Rep. 2017, 7, 3392. [Google Scholar] [CrossRef]
- Fields, P.A. Hot spots in cold adaptation: Localized increases in conformational flexibility in lactate dehydrogenase A4 orthologs of Antarctic notothenioid fishes. Proc. Natl. Acad. Sci. USA 1998, 95, 11476–11481. [Google Scholar] [CrossRef]
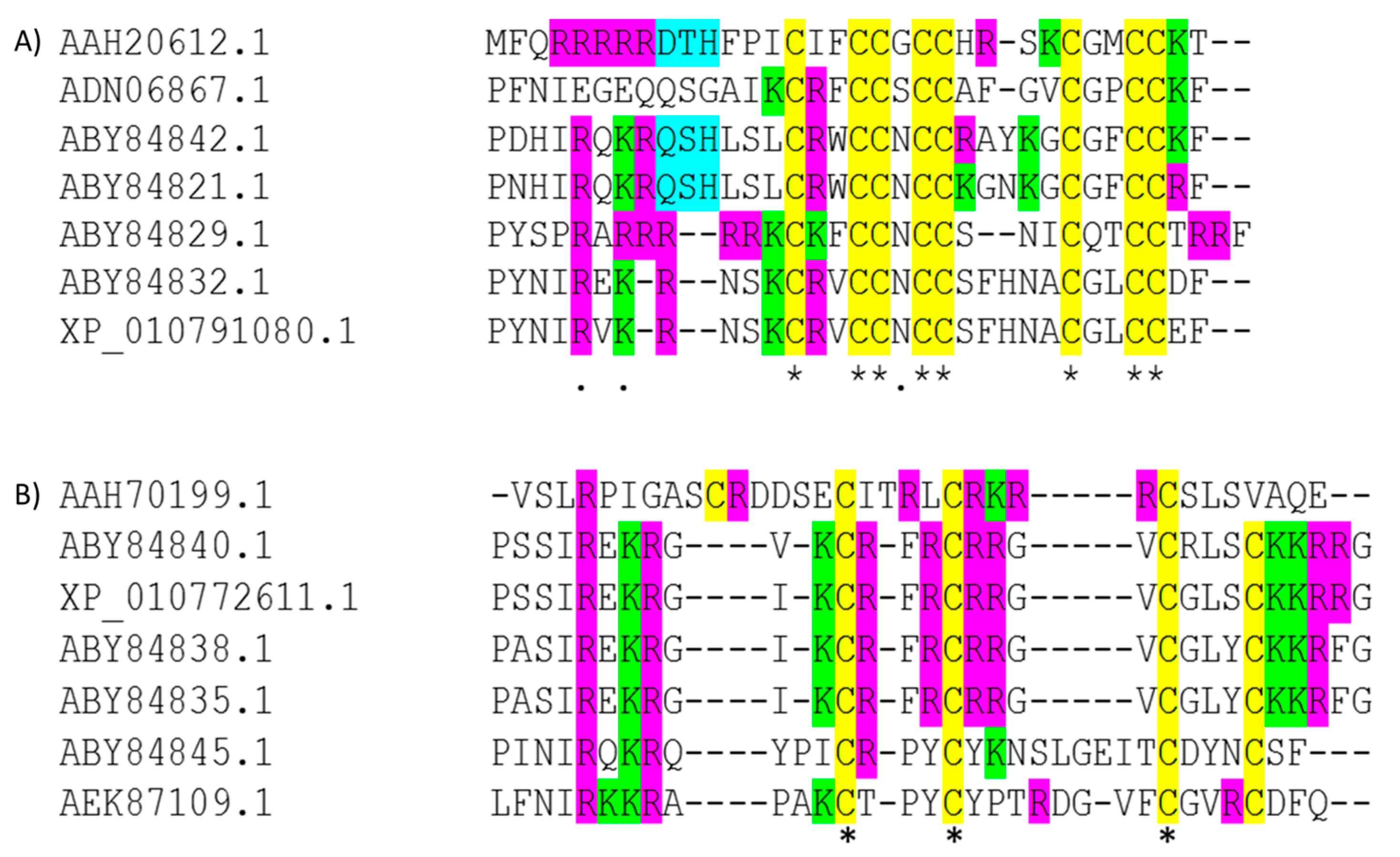
| Peptide | Amino Acid Sequence and Schematic Representation | Disulfide Bonds | Net Charge at pH 7 |
|---|---|---|---|
| hLEAP-1 | NH2-DTHFPICIFCCGCCHRSKCGMCCKT-COOH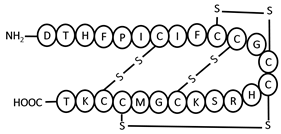 | 7–23, 10–13, 11–19, 14–22 | +2.2 |
| nLEAP-1 | NH2-RRRKCKFCCNCCSNICQTCCTRRF-COOH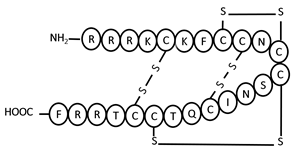 | 5–20, 8–11, 9–16, 12–19 | +7 |
| nLEAP-2 | NH2-GIKCRFRCRRGVCGLYCKKRFG-COOH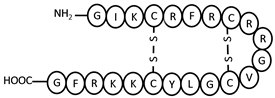 | 4–17, 8–13 | +8 |
| eLEAP-1 | NH2-QSHLSLCRWCCNCCRAYKGCGFCCKF-COOH | 7–24, 10–13, 11–20, 14–23 | +4.1 |
| eLEAP-2 | NH2-QYPICRPYCYKNSLGEITCDYNCSF-COOH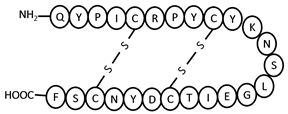 | 5–23, 9–19 | 0 |
| MIC for Gram-Positive Bacteria (µM) | MIC for Gram-Negative Bacteria (µM) | |||||
|---|---|---|---|---|---|---|
| Peptides | E. facaelis | B. subtilis | S. aureus | E. coli | S. typhimurium | V. parahaemolyticus |
| hLEAP-1 | 9 | 4 | 6 | 6 | >9 | 1 |
| nLEAP-1 | 9 | 4 | 6 | 9 | 9 | 1 |
| nLEAP-2 | 1 | 1 | 1 | 1 | 1 | 0.25 |
| eLEAP-1 | 9 | 9 | 9 | >9 | > 9 | 4 |
| eLEAP-2 | 9 | 1 | 1 | >9 | > 9 | 1 |
| MIC * Values (µM) | ||||
|---|---|---|---|---|
| Peptides | 5 °C | 15 °C | 25 °C | 37 °C |
| hLEAP-1 | 4 | 9 | >9 | >9 |
| nLEAP-1 | 9 | >9 | >9 | >9 |
| nLEAP-2 | 0.25 | 1 | >9 | >9 |
| eLEAP-1 | 4 | >9 | >9 | >9 |
| eLEAP-2 | 0.25 | 1 | >9 | >9 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borkar, S.B.; Nandanwar, S.K.; Lee, J.H.; Kim, H.J. Characterization of Four Liver-Expressed Antimicrobial Peptides from Antarctic Fish and Their Antibacterial Activity. Appl. Sci. 2019, 9, 4299. https://doi.org/10.3390/app9204299
Borkar SB, Nandanwar SK, Lee JH, Kim HJ. Characterization of Four Liver-Expressed Antimicrobial Peptides from Antarctic Fish and Their Antibacterial Activity. Applied Sciences. 2019; 9(20):4299. https://doi.org/10.3390/app9204299
Chicago/Turabian StyleBorkar, Shweta Bharat, Sondavid K. Nandanwar, Jun Hyuck Lee, and Hak Jun Kim. 2019. "Characterization of Four Liver-Expressed Antimicrobial Peptides from Antarctic Fish and Their Antibacterial Activity" Applied Sciences 9, no. 20: 4299. https://doi.org/10.3390/app9204299
APA StyleBorkar, S. B., Nandanwar, S. K., Lee, J. H., & Kim, H. J. (2019). Characterization of Four Liver-Expressed Antimicrobial Peptides from Antarctic Fish and Their Antibacterial Activity. Applied Sciences, 9(20), 4299. https://doi.org/10.3390/app9204299






