A High-Efficiency Super-Resolution Reconstruction Method for Ultrasound Microvascular Imaging
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Super-Resolution Imaging Process
2.2. Microbubble Localization
2.2.1. PSF Models
2.2.2. Data Approximation
2.2.3. Localization Uncertainty
2.3. Trajectory Tracking
2.4. In Vivo Experiment Setups
2.4.1. In Vivo Ultrafast Imaging
2.4.2. Radio-Frequency (RF) Data Processing and Wall Filtering
2.4.3. Methods for Comparison
2.4.4. Parameters Measurement
3. Experimental Results
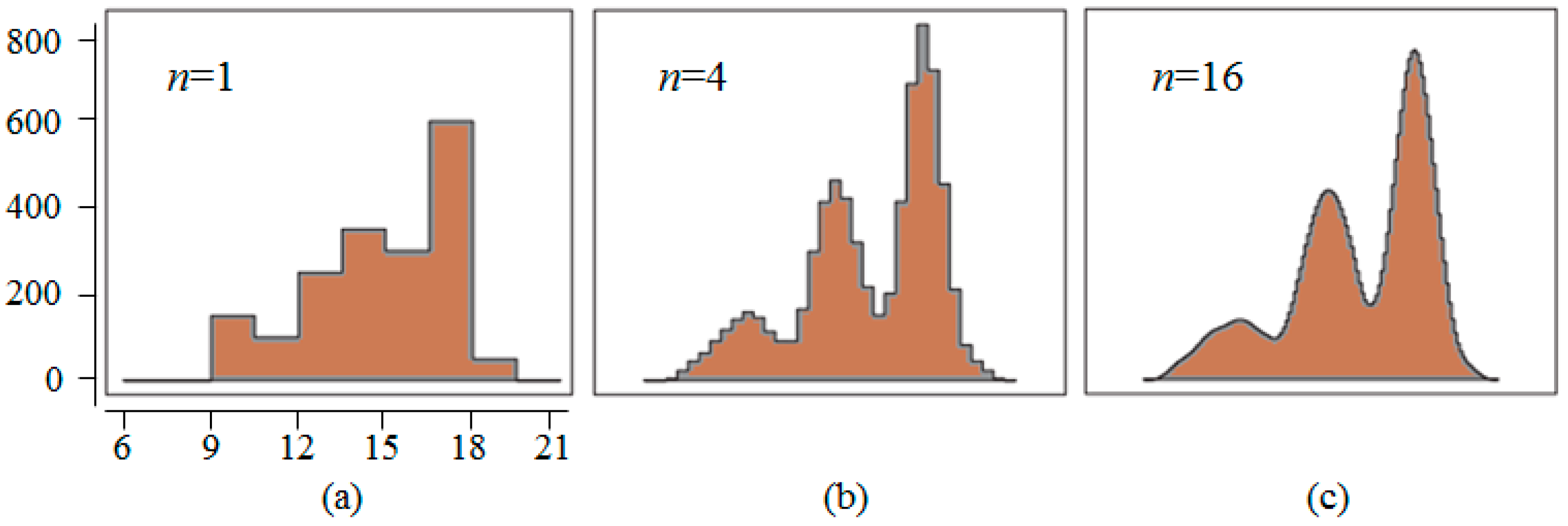
3.1. Preprocess
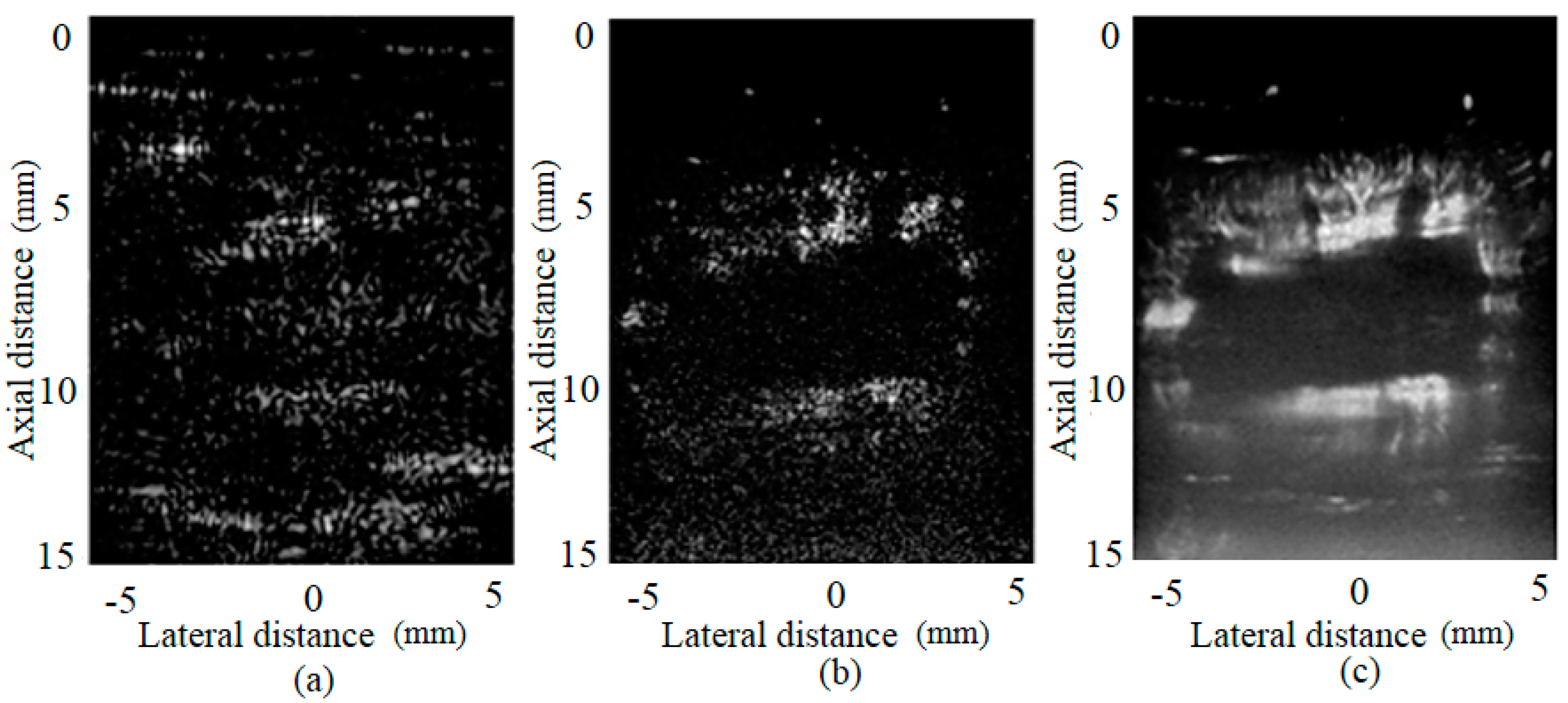
3.2. Results of Different Methods
4. Discussion
5. Conclusions
Authors Contributions
Funding
Conflicts of Interest
References
- Cootney, R.W. Ultrasound imaging: Principles and applications in rodent research. Ilar J. 2001, 42, 233–247. [Google Scholar] [CrossRef]
- Jain, R.K. Normalization of tumor vasculature: An emerging concept in antiangiogenic therapy. Science 2005, 307, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Errico, C.; Pierre, J.; Pezet, S.; Desailly, Y.; Lenkei, Z.; Couture, O.; Tanter, M. Ultrafast ultrasound localization microscopy for deep super-resolution vascular imaging. Nature 2015, 527, 499–502. [Google Scholar] [CrossRef] [PubMed]
- Rust, M.J.; Bates, M.; Zhuang, X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Method 2006, 3, 793–796. [Google Scholar] [CrossRef] [PubMed]
- Betzig, E.; Patterson, G.H.; Sougrat, R.; Lindwasser, O.W.; Olenych, S.; Bonifacino, J.S.; Davidson, M.W.; Lippincott-Schwartz, J.; Hess, H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313, 1642–1645. [Google Scholar] [CrossRef] [PubMed]
- Hess, S.T.; Girirajan, T.P.K.; Mason, M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006, 91, 4258–4272. [Google Scholar] [CrossRef] [PubMed]
- Errico, C.; Couture, O.; Tanter, M. Ultrafast ultrasound localization microscopy. J. Acoust. Soc. Am. 2017, 141, 3951. [Google Scholar] [CrossRef]
- Ghosh, D.; Xiong, F.; Sirsi, S.R.; Mattrey, R.; Brekken, R.; Kim, J.W.; Hoyt, K. Monitoring early tumor response to vascular targeted therapy using super-resolution ultrasound imaging. In Proceedings of the 2017 IEEE International Ultrasonics Symposium (IUS), Washington, DC, USA, 6–9 September 2017. [Google Scholar]
- Lin, F.; Tsuruta, J.K.; Rojas, J.D.; Dayton, P.A. Optimizing sensitivity of ultrasound contrast-enhanced super-resolution imaging by tailoring size distribution of microbubble contrast agent. Ultrasound Med. Biol. 2017, 43, 2488–2493. [Google Scholar] [CrossRef] [PubMed]
- Yu, A.; Lovstakken, L. Eigen-based clutter filter design for ultrasound color flow imaging: A review. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2010, 57, 1096–1111. [Google Scholar] [CrossRef] [PubMed]
- Gallippi, C.M.; Trahey, G.E. Adaptive clutter filtering via blind source separation for two-dimensional ultrasonic blood velocity measurement. Ultrasonic Imaging 2002, 24, 193–214. [Google Scholar] [CrossRef] [PubMed]
- Thompson, R.E.; Larson, D.R.; Webb, W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002, 82, 2775–2783. [Google Scholar] [CrossRef]
- Stallinga, S.; Rieger, B. Accuracy of the Gaussian point spread function model in 2D localization microscopy. Opt. Express 2010, 18, 24461–24476. [Google Scholar] [CrossRef] [PubMed]
- Baddeley, D.; Cannell, M.B.; Soeller, C. Visualization of localization microscopy data. Microsc. Microanal. 2010, 16, 64–72. [Google Scholar] [CrossRef] [PubMed]
- Rix, A.; Lederle, W.; Theek, B.; Lammers, T.; Moonen, C.; Schmitz, G.; Kiessling, F. Advanced ultrasound technologies for diagnosis and therapy. J. Nucl. Med. 2018, 59, 740–746. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Schwartz, S.L.; Byars, J.M.; Lidke, K.A. Simultaneous multiple emitter fitting for single molecule super-resolution imaging. Biomed. Opt. Express 2011, 2, 1377–1393. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.W. Averaged shifted histograms: Effective nonparametric density estimators in several dimensions. Ann. Stat. 1985, 13, 1024–1040. [Google Scholar] [CrossRef]
- Tanter, M.; Fink, M. Ultrafast imaging in biomedical ultrasound. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2014, 61, 102–119. [Google Scholar] [CrossRef] [PubMed]
- Tiran, E.; Deffieux, T.; Correia, M.; Maresca, D.; Osmanski, B.F.; Sieu, L.A.; Bergel, A.; Cohen, I.; Pernot, M.; Tanter, M. Multiplane wave imaging increases signal-to-noise ratio in ultrafast ultrasound imaging. Phys. Med. Biol. 2015, 60, 8549–8566. [Google Scholar] [CrossRef] [PubMed]
- Cox, B.; Beard, P. Imaging techniques: Super-resolution ultrasound. Nature 2015, 527, 451–452. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.S.; Joseph, N.; Rieger, B.; Lidke, K.A. Fast, single-molecule localization that achieves theoretically minimum uncertainty. Nat. Method 2010, 7, 373–375. [Google Scholar] [CrossRef] [PubMed]
- Kendall, M.; Stuart, A. The Advanced Theory of Statistics, 2nd ed.; Charles Griffin: London, UK, 1946; pp. 29–48. ISBN 9780340814932. [Google Scholar]
- Mortensen, K.I.; Churchman, L.S.; Spudich, J.A.; Flyvbjerg, H. Optimized localization analysis for single-molecule tracking and super-resolution microscopy. Nat. Method 2010, 7, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Bevington, P.R.; Robinson, D.K.; Blair, J.M.; Mallinckrodt, A.J.; McKay, S. Data reduction and error analysis for the physical sciences. Comput. Phys. 1993, 7, 415–416. [Google Scholar] [CrossRef]
- Math, C. The Apache Commons Mathematics Library, Version 3.2. Available online: http://commons.apache.org/math/ (accessed on 4 February 2018).
- Křížek, P.; Raška, I.; Hagen, G.M. Minimizing detection errors in single molecule localization microscopy. Opt. Express 2011, 19, 3226–3235. [Google Scholar] [CrossRef] [PubMed]
- Quan, T.; Zeng, S.; Huang, Z.L. Localization capability and limitation of electron-multiplying charge-coupled, scientific complementary metal-oxide semiconductor, and charge-coupled devices for superresolution imaging. J. Biomed. Opt. 2010, 15, 066005. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.W. Multivariate Density Estimation: Theory, Practice, and Visualization, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2015; pp. 242–245. ISBN 9780471697558. [Google Scholar]
- Schneider, M. Sonovue, a new ultrasound contrast agent. Eur. Radiol. 1999, 9, S347–S348. [Google Scholar] [CrossRef] [PubMed]
- Demené, C.; Deffieux, T.; Pernot, M.; Osmanski, B.F.; Biran, V.; Gennisson, J.L.; Sieu, L.; Bergel, A.; Franqui, S.; Correas, J.M.; et al. Spatiotemporal clutter filtering of ultrafast ultrasound data highly increases doppler and fultrasound sensitivity. IEEE Trans. Med. Imaging 2015, 34, 2271–2285. [Google Scholar] [CrossRef] [PubMed]
- Bar-Zion, A.; Tremblay-Darveau, C.; Solomon, O.; Adam, D.; Eldar, Y.C. Fast vascular ultrasound imaging with enhanced spatial resolution and background rejection. IEEE Trans. Med. Imaging 2017, 36, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of image analysis. Nat. Method 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Ovesný, M.; Křížek, P.; Borkovec, J.; Švindrych, Z.; Hagen, G.M. ThunderSTORM: A comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics 2014, 30, 2389–2390. [Google Scholar] [CrossRef] [PubMed]
- Viessmann, O.M.; Eckersley, R.J.; Christensenjeffries, K.; Tang, M.X.; Dunsby, C. Acoustic super-resolution with ultrasound and microbubbles. Phys. Med. Biol. 2013, 58, 6447–6458. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, D.; Schmitz, G. Detection and tracking of multiple microbubbles in ultrasound B-mode images. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2015, 63, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Christensenjeffries, K.; Browning, R.; Tang, M.X.; Dunsby, C.; Eckersley, R.J. In vivo acoustic super-resolution and super-resolved velocity mapping using microbubbles. IEEE Trans. Med. Imaging 2014, 34, 433–440. [Google Scholar] [CrossRef]



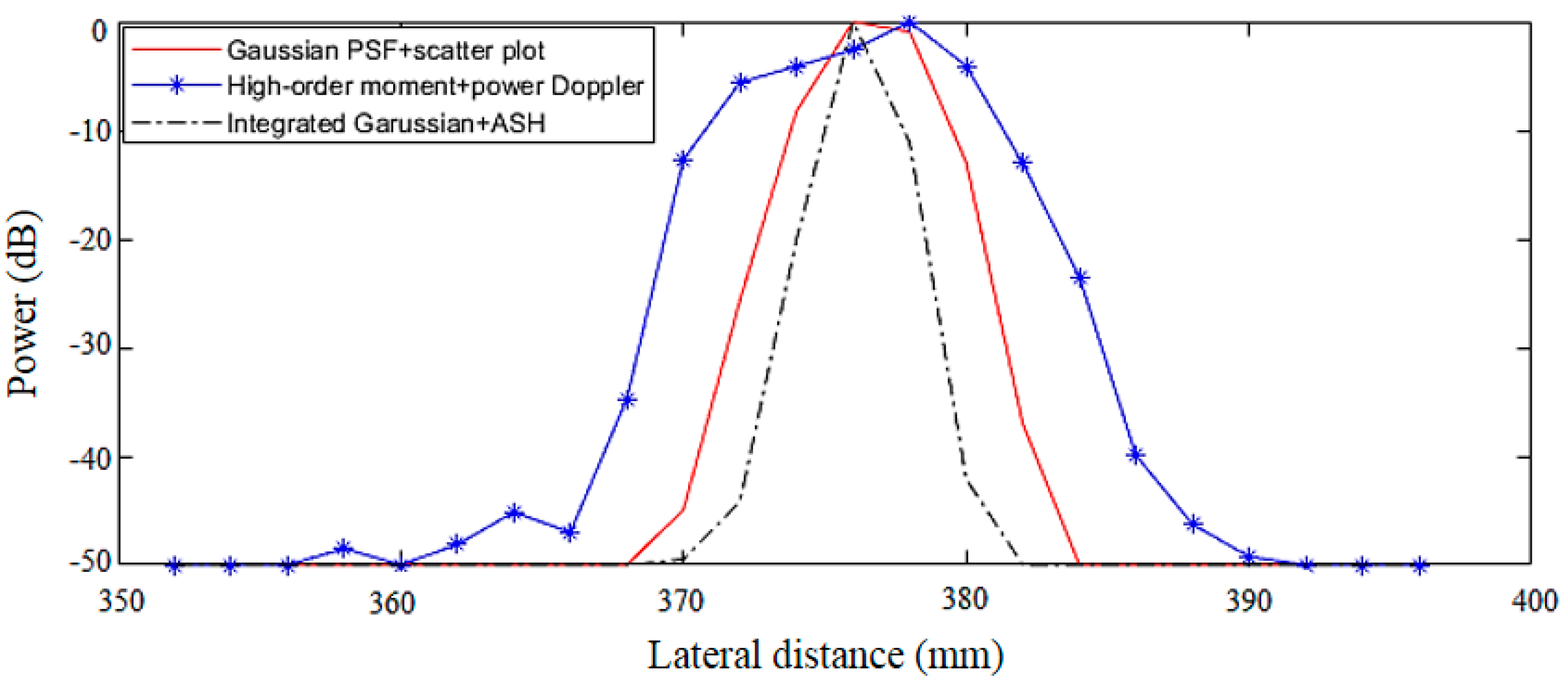
| Method | FWHM (μm) | PSL (dB) | Run Time (s) |
|---|---|---|---|
| Gaussian PSF deconvolution + scatter plot | 7.72 | −37.22 | 14.20 |
| High-order moment + power Doppler integration | 13.44 | −26.46 | 9.27 |
| Integral form of Gaussian PSF deconvolution + ASH | 3.88 | −42.35 | 17.11 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, W.; Tong, Y.; Huang, Y.; Wang, Y.; Yu, J. A High-Efficiency Super-Resolution Reconstruction Method for Ultrasound Microvascular Imaging. Appl. Sci. 2018, 8, 1143. https://doi.org/10.3390/app8071143
Guo W, Tong Y, Huang Y, Wang Y, Yu J. A High-Efficiency Super-Resolution Reconstruction Method for Ultrasound Microvascular Imaging. Applied Sciences. 2018; 8(7):1143. https://doi.org/10.3390/app8071143
Chicago/Turabian StyleGuo, Wei, Yusheng Tong, Yurong Huang, Yuanyuan Wang, and Jinhua Yu. 2018. "A High-Efficiency Super-Resolution Reconstruction Method for Ultrasound Microvascular Imaging" Applied Sciences 8, no. 7: 1143. https://doi.org/10.3390/app8071143
APA StyleGuo, W., Tong, Y., Huang, Y., Wang, Y., & Yu, J. (2018). A High-Efficiency Super-Resolution Reconstruction Method for Ultrasound Microvascular Imaging. Applied Sciences, 8(7), 1143. https://doi.org/10.3390/app8071143




