1. Introduction
Neurological control through muscle and sensing is part of almost every human activity. It is so natural that we do not even realize it. This is also the case for speech production. Even though the process itself is complex, its functioning is taken for granted. Unfortunately, neurological diseases, infections, tissue changes, and injuries can negatively affect speech production. Impaired speech production is frequently represented by dysphonia or dysphonic voice. Dysphonia is a perceptual quality of the voice that indicates that some negative changes have occurred in the phonation organs [
1]. The relationship between voice pathology and acoustic voice features has been clinically established and confirmed both quantitatively and subjectively by speech experts [
2,
3,
4].
As indicated above, pathological voice is an indication of health-related problems. However, recognizing dysphonic voice at an early stage is not an easy task. Usually, a trained expert is required, and a series of speech exercises need to be completed. Automatized speech evaluation can allow for time- and cost-effective speech analysis that, in turn, enables the screening of a wider population. Since some diseases, such as Parkinson’s disease, manifest themselves early in speech disruption, early discovery through screening can lead to earlier treatment and to improved treatment results. The main motivation for the realization of this work is the utilization of artificial intelligence for the diagnosis of different diseases. This can lead to significant improvements in diagnosis and healthcare and, in turn, to the improvement of human life [
5,
6]. Several diseases include a speech disorder as an early symptom. This is usually due to damage of the neurological system or caused directly by damage to some part of the vocal tract, such as the vocal cords [
7]. Frequently, a speech disorder leads to a secondary symptom, and early detection may reveal many high-risk illnesses [
1,
8].
The ultimate goal of this research is to develop a decision support system that provides an accurate, objective, and time-efficient diagnosis and helps medical personnel provide the correct diagnostic decision and treatment. In this work, we focused on the detection of pathological speech based on features obtained from voice samples from multiple subjects. It is a noninvasive way of examining a voice and only digital voice recordings are needed. Voice data were obtained from the publicly available Saarbrucken Voice Database (SVD). We exported 194 samples from SVD, of which 94 samples originated from patients with dysphonia and 100 samples came from healthy ones. In order to detect pathological speech, it is necessary to build and train a classification model. For this purpose, three state-of-the-art classification algorithms were utilized: support vector machine (SVM), random forest classifier (RFC), and K-nearest neighbors (KNN).
The paper is organized as follows. In the next section, we provide a brief overview of similar works in the area of pathological speech processing. Then, we describe the preprocessing of the dataset and provide a brief overview of classification algorithms. Later, we propose a decision support model and present the results of numerical experiments. Conclusions are drawn in the last section.
2. Related Work
Speech processing is a very active area of research. There are many contributions that focus on different aspects of speech processing, from feature extraction to decision support systems based on speech analysis. We a provide a brief overview of some recent findings related to our research topic.
There are several existing solutions in the field of pathological speech detection [
9,
10]. For example, Al-Nasheri et al. in their work [
11] concentrated on developing feature extraction for the detection and classification of voice pathologies by investigating different frequency bands using autocorrelation and entropy. The voice impairment cases studied were caused by vocal cysts, vocal polyps, and vocal paralysis. They found that the most contributive frequency bands in both detection and classification were between 1000 and 8000 Hz. Each voice sample consisted of the sustained vowel /
a/, and a support vector machine was used as a classifier. The highest obtained accuracies in the case of detection were 99.69%, 92.79%, and 99.79% for Massachusetts Eye and Ear Infirmary (MEEI), the Saarbrucken Voice Database (SVD), and the Arabic Voice Pathology Database (AVPD), respectively.
Martinez et al. [
12] in their work presented a set of experiments on pathological voice detection with the SVD by using the MultiFocal toolkit for discriminative calibration and fusion. Results were compared with the MEEI database. Since they used the data from the SVD dataset, sustained vowel recordings of /
a/, /
i/, and /
u/ were analyzed. The samples were not differentiated according to the diagnosis, but they used all samples in SVD and distinguished only between healthy and pathological ones. Samples of 650 subjects were healthy and 1320 samples were pathological. Extracted features included mel-cepstral coefficients (MFCCs), harmonics-to-noise ratio (HNR), normalized noise energy (NNE), and glottal-to-noise excitation ratio (GNE), and they mostly measured the quality of the voice. A Gaussian mixture model was used as a classifier. In the case of the vowel /
a/, they reached an accuracy of 80.4%; with the vowel /
i/, it was 78.3%; and with the vowel /
u/, it was 79.9%. For all vowel fusions, they reached an accuracy of 87.9%. Using the MEEI dataset, they achieved an accuracy of 94.3%, which is 6.6% more than with the SVD dataset.
Little et al. [
13,
14] focused on discriminating healthy subjects from subjects with Parkinson’s disease by detecting dysphonia. They introduced a new measure of dysphonia: pitch period entropy (PPE). The utilized data consisted of 195 sustained vowels from 31 subjects, of which 23 were diagnosed with Parkinson’s disease. The extracted features included pitch period entropy, shimmer, jitter, fundamental frequency, pitch marks, HNR, etc. They found that the combination of the features HNR, PPE, detrended fluctuation analysis, and recurrence period density entropy led to quite an accurate classification of the subjects with Parkinson’s disease from healthy subjects. A classification performance of 91.4%, using a kernel support vector machine, was achieved.
Other authors have also investigated the effect of Parkinson’s disease on speech [
15] or various aspects of speech deterioration caused by Alzheimer’s disease [
16], dysphagia [
8], or impaired speech [
17].
Speech signal processing and machine learning are being increasingly explored in the developmental disorder domain, where the methods range from supervised classification to knowledge-based data mining of highly subjective constructs of psychological states [
18,
19].
3. Data and Preprocessing
3.1. Dataset
We used the publicly available Saarbrucken Voice Database [
20]. It is a collection of voice recordings where one subject’s samples consist of recordings of the vowels /
a/, /
i/, and /
u/ produced at a normal, high, low, and low-high-low pitch. The length of the recordings with sustained vowels is from 1–2 s. We exported 194 samples from this database, of which 94 samples belong to patients with dysphonia (41 men, 53 women) and 100 samples belong to healthy ones. The age of all subjects is over 18 years.
3.2. Speech Feature Extraction
To obtain some representative characteristics of speech, it was necessary to implement feature extraction from individual samples and to build a feature matrix. For each intonation of each vowel, 130 features were extracted. This means that 520 features were extracted for all intonations of a particular vowel. For one subject and all corresponding vowel recordings of /a/, /i/, and /u/, the number of features is 1560. The features consist of the following specific types of parameters: energy, low-short time energy ratio, zero crossing rate, Teager–Kaiser energy operator, entropy of energy, Hurst’s coefficient, fundamental frequency, mel-cepstral coefficients, formants, jitter, shimmer, spectral centroid, spectral roll-off, spectral flux, spectral flatness, spectral entropy, spectral spread, linear prediction coefficients, harmonics-to-noise ratio, power spectral density, and phonatory frequency range. Some of the features contain multiple subtypes (e.g., shimmer:local, shimmer:apq3, shimmer:apq5, etc.) and some of the features were extracted from smaller time frames of the recording. In this case, several statistical functionals (median, average, minimum, maximum, and standard deviation) were determined.
3.3. Feature Selection
It was previously shown that feature selection (FS) can improve prediction performance in some areas [
21]. We applied feature selection to find the optimal subset of features for better classification between healthy and pathological samples. We selected simple filter FS to get
k best features. Filter FS is a computationally effective approach that provides results competitive with more complex methods. Mutual information for discrete target variables was used to estimate the score of features. This function is based on entropy estimation from
k-nearest neighbor distances. Mutual information between two random variables is a non-negative value, which measures the dependency between variables. If the variable is independent, it is zero, and the higher the value, the greater the dependence.
3.4. Principal Component Analysis
The purpose of conducting principal component analysis is similar to the purpose of feature selection, i.e., to find a smaller subset of features to improve prediction performance. Unlike the FS method, dimensionality reduction does not preserve the original features but instead transform features from high-dimensional space to new, lower-dimensional space. We implemented dimensionality reduction using the principal component analysis (PCA) method. PCA is a statistical procedure that uses an orthogonal transformation to convert a set of observations of possibly correlated variables into a set of values of linearly uncorrelated variables called principal components. The new feature set is a linear combination of these principal components and is used to build a new feature matrix.
4. Classification Models
Currently, there are many classification algorithms, and there are none that would outperform the others in every scenario [
22]. To improve the robustness of our results, we selected three state-of-the-art classifiers: support vector machine (SVM) with nonlinear kernel, K-nearest neighbors (KNN), and random forests classifier (RFC). All three classifiers are based on different underlying principles and have shown very promising results in many areas.
4.1. Support Vector Machine
SVM uses a hyperplane to classify data samples into two categories. The hyperplane is built in such a way that it allocates the majority of points of the same category on the same side of the hyperplane while trying to maximize the distance of data samples from both categories to this hyperplane. The subset of data samples closest to the separating hyperplane is denoted as the support vectors [
23].
Assume that data samples in the training set
, where
, with class labels
, where
y for
. The optimal hyperplane is defined as [
24]
where
w represents a weight vector and
b represents bias.
The goal is to maximize the margin, which can be achieved through a constrained optimization problem
By introducing slack parameters, the objective function is updated to:
The introduction of kernelization into the SVM allows it to solve a difficult nonlinear task in data mining. For the SVM classifier, we searched through the following parameters:
Kernel: linear, RBF, poly
C: 0.1, 0.2, 0.5, 1, 3, 5, 7, 10 , 20 , 30 , 35 , 40, 45, 50, 70, 100 (only for poly and RBF kernel), 1000 (only for poly and RBF kernel)
Gamma (only RBF): 0.0001, 0.001, 0.01, 0.025, 0.5, 0.1
Polynomial degree (only poly): 1, 2, 3, 4, 5.
4.2. K-Nearest Neighbors
Akbulut et al. [
25] stated that the KNN method is considered to be one of the oldest and the simplest types of nonparametric classifier. A nonparametric model for KNN means that the classification of the test data does not use a function that has been set up in advance based on the learning process on the training dataset. Instead, it uses the memory of this training set and measures the similarity of the new test sample to the original sample based on the distance. The goal is to determine the true class of an undefined test pattern by finding the nearest neighbors within a hypersphere of a predefined radius. The disadvantage is that when choosing a low
k value, the separating boundary is highly adapted to the training data, and over-training occurs. At larger values of
k, the boundary tends to be smoother and achieve better prediction results for new samples. The optimal value of
k needs to be determined experimentally. For the KNN method, we grid-searched through the following parameters:
K: 1, 2, 3, 4, 5, 6, 7, 8 ,9 ,10
Leaf size: 3, 5, 15, 20 , 25 , 30 , 35 , 40 , 45 , 50
Metric: Manhattan, Euclidean
4.3. Random Forest Classifier
RFC is considered a complex classifier, as it comprises a set of decision trees. Each tree is an independent classifier that consists of decision nodes. Each node evaluates a certain condition using the test data from the set {
,
, ⋯,
}. Based on the result, the branch goes onto the next node, up to the tree sheet that holds the classification information [
26,
27].
As stated in [
28], the decision tree process is appropriate for selecting attributes from the next test set that has the greatest information gain to divide the data into two of the most diverse strings. This process is repeated at each node until the tree sheet is obtained. However, this can lead to a large depth of the tree and increase the risk of over-training. For this reason, the maximum depth of the tree is limited in practice. In order to increase classification accuracy, these trees, with weaker prediction capabilities, are grouped to create a more robust and accurate model—RFC. Another drawback associated with decision tree classifiers is their high variance. The random forests should achieve better success with a higher number of decision trees—
k [
26]. The value of
k needs to be found where accuracy is stabilized and will not increase further. The set of parameters for RFC in our experiments was as follows:
Number of estimators: 36, 83, 103, 124
Max. depth of tree: 1, 5, 10, 15, 17, 20, 25
Min. samples for leaf node: 1, 2, 4, 6, 8, 10
The number of features to consider when looking for a split: sqrt(number of features), log2(number of features)
5. Pathological Speech Detection
The main aim of this work is to design a machine learning model that is able to discriminate pathological speech. As was indicated above, from the machine learning point of view, this is a binary classification task. The following list describes the sequence of steps for creating the system for pathological speech detection:
Export of data: Recordings of the vowels /a/, /i/, and /u/ were obtained from the freely available Saarbrucken Voice Database.
Feature extraction: From the exported samples, it was necessary to extract speech features that express voice quality and potential pathological disorders present in the voice. Whole feature extraction was performed in Python. The types of features are described in
Section 3.
Dimensionality reduction: In order to improve the accuracy of the classification, the selection of the features and principal component analysis were performed. Both methods are described in more detail in
Section 3. The most relevant features are depicted in
Table 1.
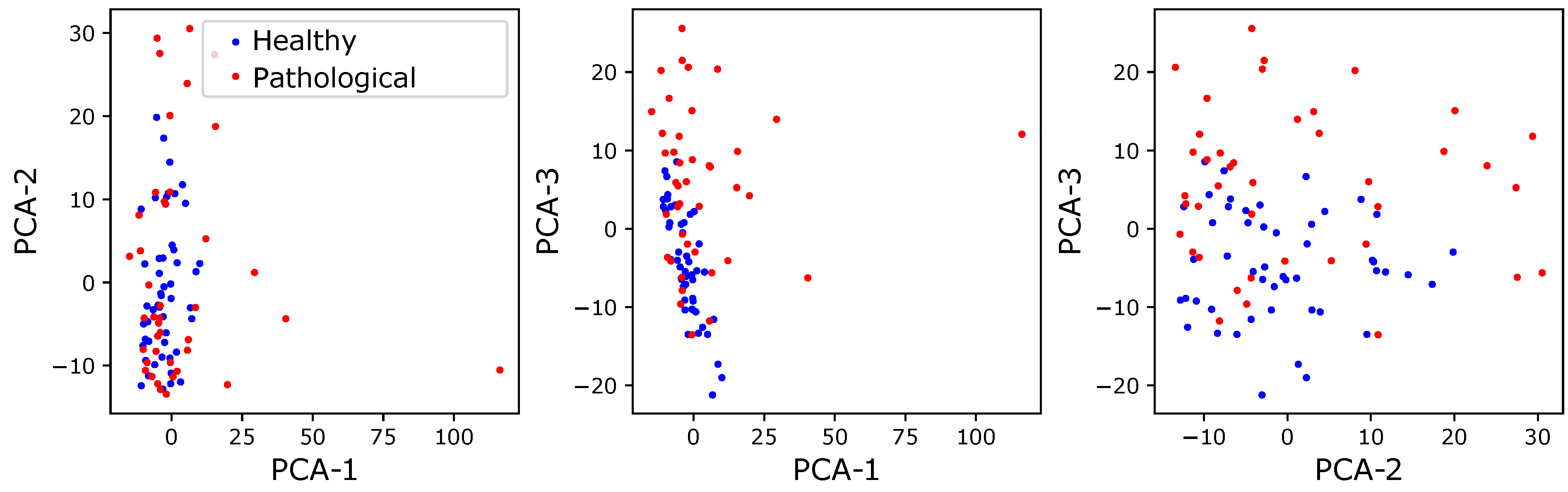
Visualization of features: In order to get a better view of the data structure, we visualized features using the PCA method. These visualizations are shown in
Figure 1,
Figure 2 and
Figure 3. For this visualization, we used the first three principal components from the PCA output.
Model training: Each machine learning model used in this work (SVM, KNN, RFC) which was designed for the classification of samples must be trained prior to classification. A combination of different features and genders was tested. After this step, we should have classifiers with their optimal parameters, because tuning the hyperparameters of the model is also included in this step. The detailed description of the training and cross-validation procedure is provided in the Experimental Results section.
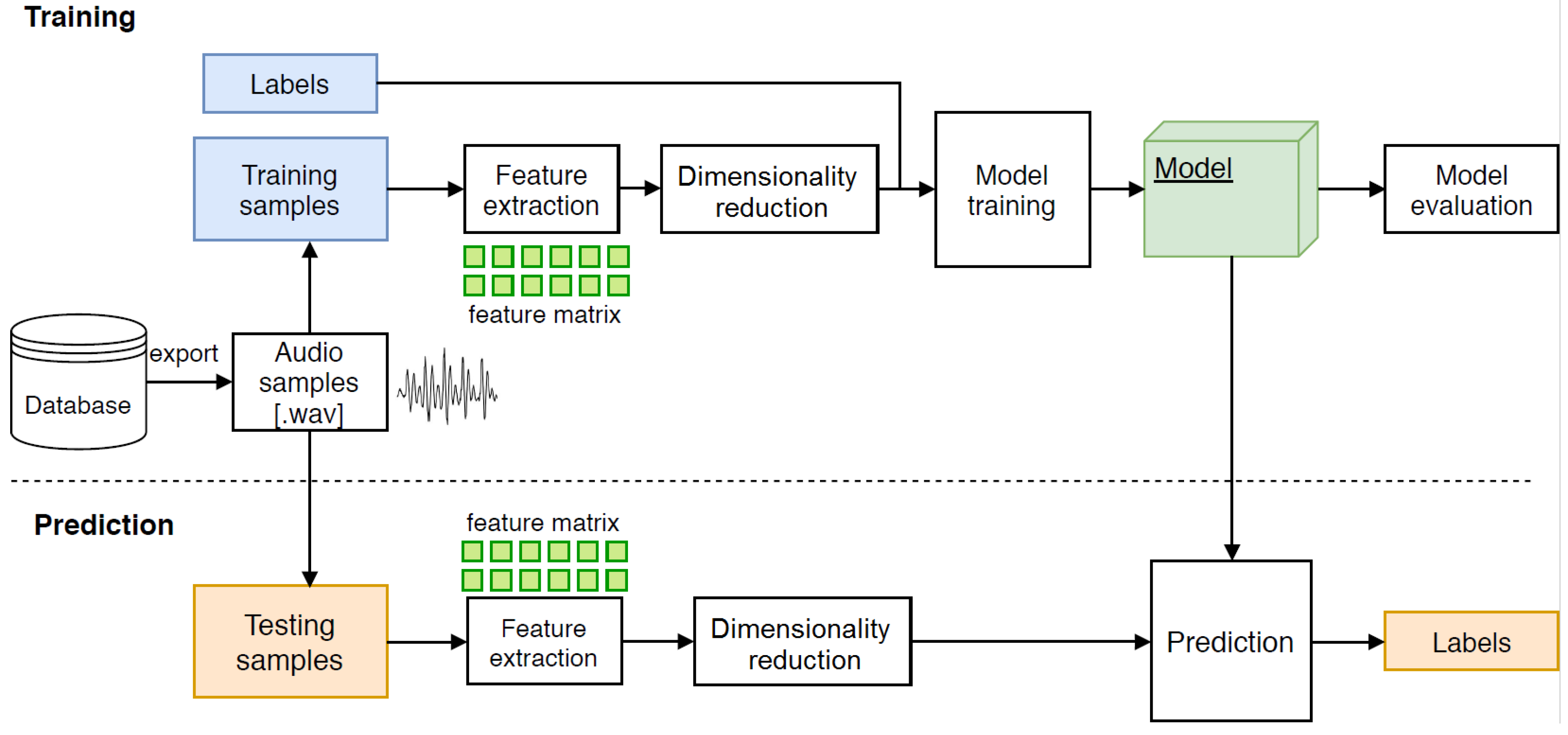
Model evaluation: The created model was tested on new test data, which means that these data were not part of the training process and the classifier did not come into contact with them. The graphical design of the sequence of steps is shown in
Figure 4.
6. Experimental Results
Two types of result comparisons were made for all three classifiers: SVM, KNN, and RFC. In the first case, we compared the influence of dimensionality reduction on classification performance. In the second case, we compared the results by considering the gender and the results yielded by processing different vowels.
Prediction performance is measured by accuracy, defined as
where
tp represents true positive and
tn is true negative. Then,
fp denotes a false positive sample and
fn a false negative sample.
6.1. Influence of Feature Selection on Prediction Performance
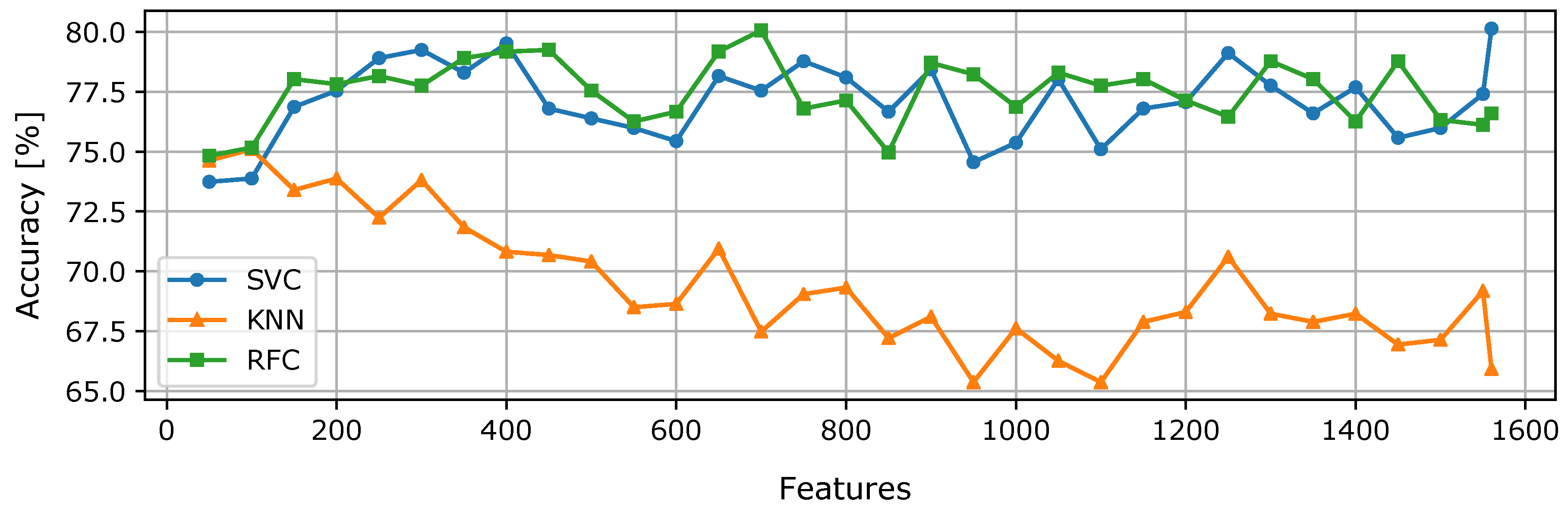
First, we aimed to analyze how the feature selection affects the prediction performance of classifiers. We evaluated the classification accuracy using only selected features; a new subset of
k features was obtained in a loop, where
k = {50, 1560} and, in each iteration,
k was incremented by 50. Then, parameter tuning for the classifier model (SVM, KNN, RFC) was done. The classification performance of the classifiers for each iteration is shown in
Figure 5.
For SVM and RFC, the accuracy increases steadily up to 400 features. After this point, the accuracy decreases and rises repeatedly, and the values usually do not exceed the limit of accuracy that was reached with 400 features. On the other hand, the accuracy of the KNN classifier shows a decreasing tendency with an increasing number of features. This is probably due to the higher dimensionality of the data space since the KNN classifier is known to suffer from higher dimensions. Even though, in this case, the accuracy of KNN dropped in higher dimensional cases, this is not always the case, as can be seen, for example, in [
29].
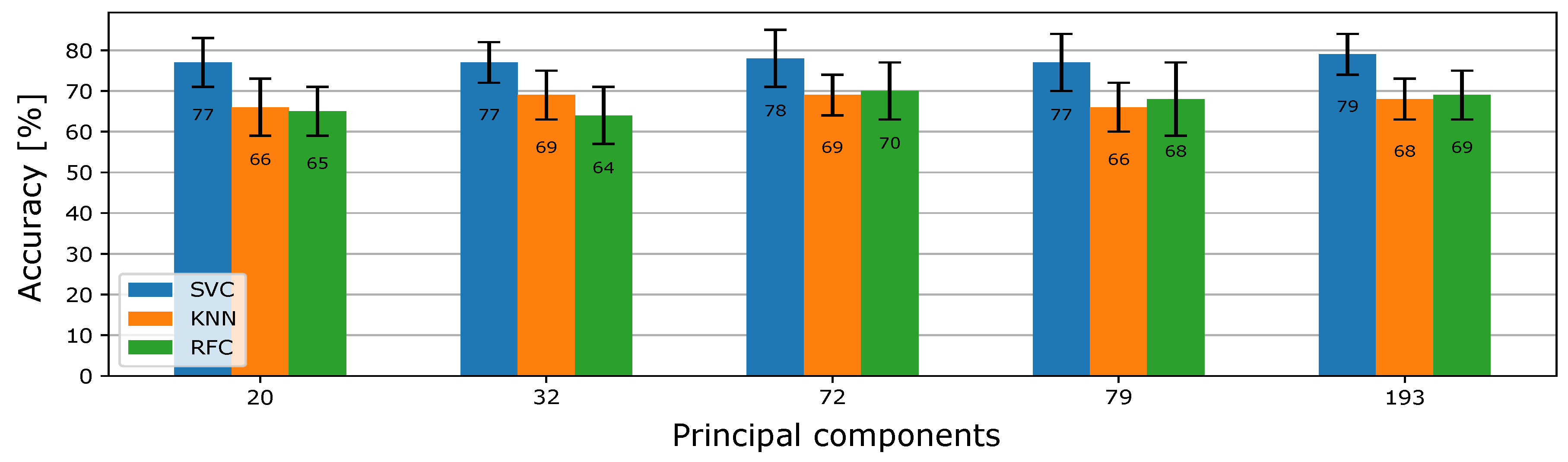
6.2. Influence of PCA on Prediction Performance
The goal of this section is to compare the accuracy of the classifiers with data of reduced dimensions. The first step was to set the number of principal components that make up a new set of training and test data. As with the previous selection method, the hyperparameters of the classification model were tuned with these new data in the training process. An overview of the accuracy of the classification results using the PCA method for different numbers of principal components is shown in
Figure 6.
Using PCA, the classification results did not improve as we expected, so we did not use this method anymore. After this finding, we used only filter feature selection.
6.3. Results by Gender and Classifiers
The previous experiments indicated that the feature selection positively influenced the prediction performance, so we utilized it in further experiments. The number of all features for one subject was 1560, and only 300 features were selected in feature selection.
For each classifier, we applied two types of cross-validation. In the first case, 75% of the data were used for training the model. Out of these training data, 25% were utilized as validation data, employed for hyperparameter tuning. The classifier model with the best parameters found in the previous step was applied to the test data that were not part of the training dataset. The whole process was repeated 10 times and the results were averaged. As a second validation scheme, we used fourfold cross-validation. The dataset was divided into four folds. Then, three folds were used for training (75%) and one fold for testing (25%). The loop proceeded in such a way that each fold was used once for testing and three times for training. This is cross-validation without resampling. The results of testing both cases are shown in
Table 2.
In
Table 2, each column shows the highest value (bold font) achieved within each type of testing method and for each classifier. As can be seen, more bold values are in rows representing results for 300 selected features. Feature selection led to an improvement in overall accuracy. Another finding is that the SVM classifier achieved the best results.
6.4. Results for Individual Vowels
The results of individual classifiers were also compared by evaluating each vowel separately. As in the previous case, we distinguished between male, female, and mixed types of samples, and we employed the same cross-validation approach as before. Each of these sample types was subdivided into four subsets of samples. Each subset contained records of only one kind of vowel. The number of features for each subject was 1560. The best value among the vowels is highlighted with bold font in each column in
Table 3.
Table 3 shows that the vowel /
a/, pronounced in all intonations, achieved significantly better results than the other vowels. In the case of women, the accuracy of each classifier is higher for the vowel /
a/ than for the other vowels. Another finding is that the fusion of all intonations of the particular vowel has a positive influence on the overall accuracy of the classification. The best results for mixed samples were achieved by the SVM classifier, although results are very similar to RFC.
7. Conclusions
In this work, we proposed and implemented a system for pathological speech (dysphonia) detection. For training and testing data, we used recordings of the sustained vowels /a/, /i/, and /u/. Pathological records were from 94 subjects, and the control group was formed by samples from 100 healthy subjects. In order to obtain voice quality information from these recordings, we implemented methods for extracting speech features. In order to design the most optimal classification model, we worked with three types of classifiers based on the following methods: supported vectors machine (SVM), random forests classifier (RFC), and K-nearest neighbors (KNN). The highest accuracy was achieved by the SVM classifier, with the feature set reduced to 300 by the filter FS method from the original 1560 features. There are several other algorithms that can be used for classification, but these are the most frequently used methods that achieve satisfactory performance. A slight boost in prediction accuracy can be achieved by further fine-tuning of hyperparameters. However, further tuning can lead to over-training, so we believe the results provided are representative for the performance of the proposed system. The most representative features are shimmer, jitter, MFCC, and spectral coefficients like spectral roll-off, spectral flux, etc. The overall classification performance with feature selection was 80.3% for mixed samples, 80.6% for female samples, and 86.2% for male samples, which is the best score achieved. The RFC achieved similar results to SVM, but with a lower accuracy for the male and female samples. Lower accuracy could be due to fact that the pathological samples were at different stages of illness and age. Our system is designed to be trained with a new set of recordings (e.g., samples of subjects with different diagnoses), and the graphical interface allows the selection of features that determine whether the sample belongs to a healthy subject or non-healthy subject.
In future work, the system could be extended to classify the stage of disease, and a monitoring function can be added. This would require a larger dataset and the design of a new classification model. Furthermore, there are several other options for further research. Introducing novel features can provide new information for classifiers. More sophisticated features capturing hidden patterns or nonlinear relationships can significantly boost prediction accuracy. Additionally, the majority of studies focus on the diagnosis of a disorder where they differentiate between healthy and non-healthy subjects; however, the more important task is frequently differential diagnosis, where we need to recognize between two or more different diseases. Even though this is a challenging task, it is of crucial importance to move decision support to this level.
Author Contributions
Conceptualization, P.D.; Funding acquisition, P.D. and L.V.; Methodology, P.D. and Z.D.; Project administration, P.D. and L.V.; Resources, L.V.; Software, D.S.; Supervision, P.D. and L.V.; Validation, P.D.; Visualization, D.S.; Writing—original draft, D.S., P.D. and Z.D.; Writing—review & editing, P.D., Z.D. and L.V.
Funding
This work was supported by the Slovak Research and Development Agency under the contract No. APVV-16-0211.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lopez-de-Ipina, K.; Satue-Villar, A.; Faundez-Zanuy, M.; Arreola, V.; Ortega, O.; Clave, P.; Sanz-Cartagena, M.; Mekyska, J.; Calvo, P. Advances in a Multimodal Approach for Dysphagia Analysis Based on Automatic Voice Analysis. In Advances in Neural Networks; Springer International Publishing: Basel, Switzerland, 2016; pp. 201–211. ISBN 978-3-319-33746-3. [Google Scholar]
- Hirano, M. Psycho-Acoustic Evaluation of Voice; Springer: New York, NY, USA, 1981. [Google Scholar]
- Baken, R.J.; Orlikoff, R.F. Clinical Measurement of Speech and Voice; Singular Thomson Learning: San Diego, CA, USA, 2000. [Google Scholar]
- Mekyska, J.; Janousova, E.; Gomez-Vilda, P.; Smekal, Z.; Rektorova, I.; Eliasova, I.; Kostalova, M.; Mrackova, M.; Alonso-Hernandez, J.; Faundez-Zanuy, M.; et al. Robust and complex approach of pathological speech signal analysis. Neurocomputing 2015, 167, 94–111. [Google Scholar] [CrossRef]
- Zheng, K.; Padman, R.; Johnson, M.P.; Diamond, H.S. Understanding technology adoption in clinical care: Clinician adoption behavior of a point-of-care reminder system. Int. J. Med. Inform. 2005, 74, 535–543. [Google Scholar] [CrossRef] [PubMed]
- Sim, I.; Gorman, P.; Greenes, R.A.; Haynes, R.B.; Kaplan, B.; Lehmann, H.; Tang, P.C. Clinical decision support systems for the practice of evidence-based medicine. J. Am. Med. Inform. Assoc. 2001, 8, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Naranjo, L.; Perez, C.J.; Martin, J.; Campos-Roca, Y. A two-stage variable selection and classification approach for Parkinson’s disease detection by using voice recording replications. Comput. Methods Prog. Biomed. 2017, 142, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Lopez-de-Ipina, K.; Calvo, P.; Faundez-Zanuy, M.; Clave, P.; Nascimento, W.; Martinez-de-Lizarduy, U.; Daniel, A.; Viridiana, A.; Ortega, O.; Mekyska, J.; et al. Automatic voice analysis for dysphagia detection. Speech Lang. Hear. 2018, 21, 86–89. [Google Scholar] [CrossRef]
- Gupta, R.; Chaspari, T.; Kim, J.; Kumar, N.; Bone, D.; Narayanan, S. Pathological speech processing: State-of-the-art, current challenges, and future directions. In Proceedings of the 2016 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Shanghai, China, 20–25 March 2016; pp. 6470–6474. [Google Scholar]
- Danubianu, M.; Pentiuc, S.G.; Schipor, O.A.; Tobolcea, I. Advanced Information Technology-support of improved personalized therapy of speech disorders. Int. J. Comput. Commun. Control 2010, 5, 684–692. [Google Scholar] [CrossRef]
- Al-Nasheri, A.; Muhammad, G.; Alsulaiman, M.; Ali, Z.; Malki, K.H.; Mesallam, T.A.; Ibrahim, M.F. Voice Pathology Detection and Classification using Auto-correlation and entropy features in Different Frequency Regions. IEEE Access 2017, 6, 6961–6974. [Google Scholar] [CrossRef]
- Martinez, D. Voice Pathology Detection on the Saarbrücken Voice Database with Calibration and Fusion of Scores Using MultiFocal Toolkit. In Communications in Computer and Information Science; Springer: Berlin, Germany, 2012; Volume 328, ISBN 978-3-642-35291-1. [Google Scholar]
- Little, A.M. Suitability of Dysphonia Measurements for Telemonitoring of Parkinson’s Disease. IEEE Trans. Biomed. Eng. 2009, 56, 1015–1022. [Google Scholar] [CrossRef] [PubMed]
- Tsanas, A.; Little, M.A.; McSharry, P.E.; Ramig, L.O. Nonlinear speech analysis algorithms mapped to a standard metric achieve clinically useful quantification of average Parkinson’s disease symptom severity. J. R. Soc. Interface 2011, 8, 842–855. [Google Scholar] [CrossRef] [PubMed]
- Saldert, C.; Bauer, M. Multifaceted Communication Problems in Everyday Conversations Involving People with Parkinson’s Disease. Brain Sci. 2017, 7, 123. [Google Scholar] [CrossRef] [PubMed]
- Lopez-de-Ipina, K.; Martinez-de-Lizarduy, U.; Calvo, P.M.; Mekyska, J.; Beitia, B.; Barroso, N.; Estanga, A.; Tainta, M.; Ecay-Torres, M. Advances on Automatic Speech Analysis for Early Detection of Alzheimer Disease: A Non-linear Multi-task Approach. Curr. Alzheimer Res. 2018, 15, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Grigore, O.; Velican, V. Self-Organizing Maps For Identifying Impaired Speech. Adv. Electr. Comput. Eng. 2011, 11, 41–48. [Google Scholar] [CrossRef]
- Bone, D.; Bishop, S.L.; Black, M.P.; Goodwin, M.S.; Lord, C.; Narayanan, S.S. Use of machine learning to improve autism screening and diagnostic instruments: Effectiveness, efficiency, and multi-instrument fusion. J. Child Psychol. Psychiatry 2016, 57, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Bone, D.; Gibson, J.; Chaspari, T.; Can, D.; Narayanan, S. Speech and language processing for mental health research and care. In Proceedings of the 2016 50th Conference on Signals, Systems and Computers, Pacific Grove, CA, USA, 6–9 November 2016; pp. 831–835. [Google Scholar]
- Barry, B. Saarbruecken Voice Database. Institute of Phonetics; Saarland University. Available online: http://stimmdb.coli.uni-saarland.de/ (accessed on 23 February 2017).
- Cai, J.; Luo, J.; Wang, S.; Yang, S. Feature selection in machine learning: A new perspective. Neurocomputing 2018, 300, 70–79. [Google Scholar] [CrossRef]
- Drotar, P.; Smekal, Z. Comparative study of machine learning techniques for supervised classification of biomedical data. Acta Electrotech. Inform. 2014, 14, 5–11. [Google Scholar] [CrossRef]
- Suykens, J.A.; Vandewalle, J. Least squares support vector machine classifiers. Neural Process. Lett. 1999, 9, 293–300. [Google Scholar] [CrossRef]
- Vapnik, V. Statistical Learning Theory; Willey-Interscience: New York, NY, USA, 1998. [Google Scholar]
- Akbulut, Y.; Sengur, A.; Guo, Y.; Smarandache, F. NS-k-NN: Neutrosophic Set-Based k-Nearest Neighbors Classifier. Symmetry 2017, 9, 179. [Google Scholar] [CrossRef]
- Abellan, J.; Mantas, C.J.; Castellano, J.G. A random forest approach using imprecise probabilities. Knowl.-Based Syst. 2017, 134, 72–84. [Google Scholar] [CrossRef]
- Boulesteix, A.L.; Janitza, S.; Kruppa, J.; König, I.R. Overview of random forest methodology and practical guidance with emphasis on computational biology and bioinformatics. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2012, 2, 496. [Google Scholar] [CrossRef]
- Raschka, S. Python Machine Learning; Packt Publishing Ltd.: Birmingham, UK, 2015; p. 90. ISBN 978-1-78355-513-0. [Google Scholar]
- Zhu, G.; Li, Y.; Wen, P.; Wang, S. Analysis of alcoholic EEG signals based on horizontal visibility graph entropy. Brain Inform. 2014, 1, 19–25. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).