1. Introduction
With the rapid development of artificial intelligence, an increasing number of studies have focused on applying intelligent algorithms to the healthcare domain [
1,
2,
3]. Traditional Chinese Medicine(TCM), as an essential component of the global healthcare system, plays a crucial role in medical practices worldwide. Chinese herbal medicines, the core elements of TCM, have demonstrated remarkable efficacy in regulating bodily functions, enhancing the immune system, promoting blood circulation, alleviating fatigue symptoms, and preventing chronic diseases. However, the classification of Chinese herbal medicines faces several challenges: the wide variety and morphological diversity of herbs, the existence of multiple subspecies within the same species, high similarity in morphological features among different herbs, and significant morphological variations due to growth stages, processing methods, and storage conditions. Traditional methods of herbal identification rely heavily on the expertise of professionals [
4], which are not only inefficient and costly but also prone to subjectivity and high error rates. Automated classification of Chinese herbal medicines can significantly improve efficiency and reduce the cost of identification, providing robust technical support for the modernization and internationalization of TCM.
Image datasets of Chinese herbal medicines are fundamental to automated classification research. A dataset with a wide variety of species, sufficient samples, diverse morphologies, multiple shooting angles, and complex backgrounds is crucial for building classification models with high generalization capabilities. Previous studies have made some progress in this area. For instance, Miao et al. constructed a dataset containing images of six Chinese herbal medicines [
5]; Mookdarsanit et al. developed a dataset with images of 11 Thai herbs [
6]; Zhang et al. established a dataset with images of 60 types of Chinese herbal slices [
7]. Additionally, Azzez et al. created a dataset with images of five herbs [
8]. In our previous work, we developed the CHMD1 dataset [
9], which includes 16 types of Chinese herbal medicine with 3552 sample images, providing a valuable data foundation for subsequent research.
Convolutional Neural Networks (CNNs) have been widely applied in computer vision due to their ability to automatically extract image features, effectively overcoming the limitations of traditional manual feature extraction [
10,
11,
12,
13]. For example, Lee et al. demonstrated the effectiveness of CNNs in feature extraction through deconvolution network visualization techniques, providing a significant basis for further research [
14]. In the field of herbal image classification, researchers have made notable progress using CNNs. Miao et al. achieved advanced classification performance on a six-class Chinese herb dataset using an improved CNN architecture [
5], and Mookdarsanit et al. obtained similar high-efficiency results [
6]. Furthermore, Zhao et al. employed PointNet to classify seven types of Chinese herbal medicine, further validating the potential of deep learning methods [
15]. Transfer learning has also been widely applied, such as Khalid et al. using the pre-trained ResNet-50 model [
16,
17], and Xing et al. and Azeez et al. employing ImageNet pre-trained CNNs for herb classification, all achieving excellent results [
8,
18]. These studies fully demonstrate the significant application value of CNNs in Chinese herbal image analysis.
Despite the progress in datasets and methods for Chinese herbal classification, two challenges remain. (1) Existing Chinese herbal image datasets generally suffer from small sample sizes, insufficient categories, monotonous backgrounds, and excessive inter-class differences. These issues severely limit the generalization capabilities of models, particularly in handling inter-class similarities and morphological diversity. Moreover, existing datasets often fail to adequately reflect the diversity of Chinese herbal medicine in real-world scenarios, leading to poor model performance in practical applications; (2) current research mainly involves simple improvements to CNNs or direct application of existing pre-trained models, which often struggle to effectively capture multi-scale features in Chinese herbal images, resulting in limited receptive fields and insufficient extraction of multi-level features.
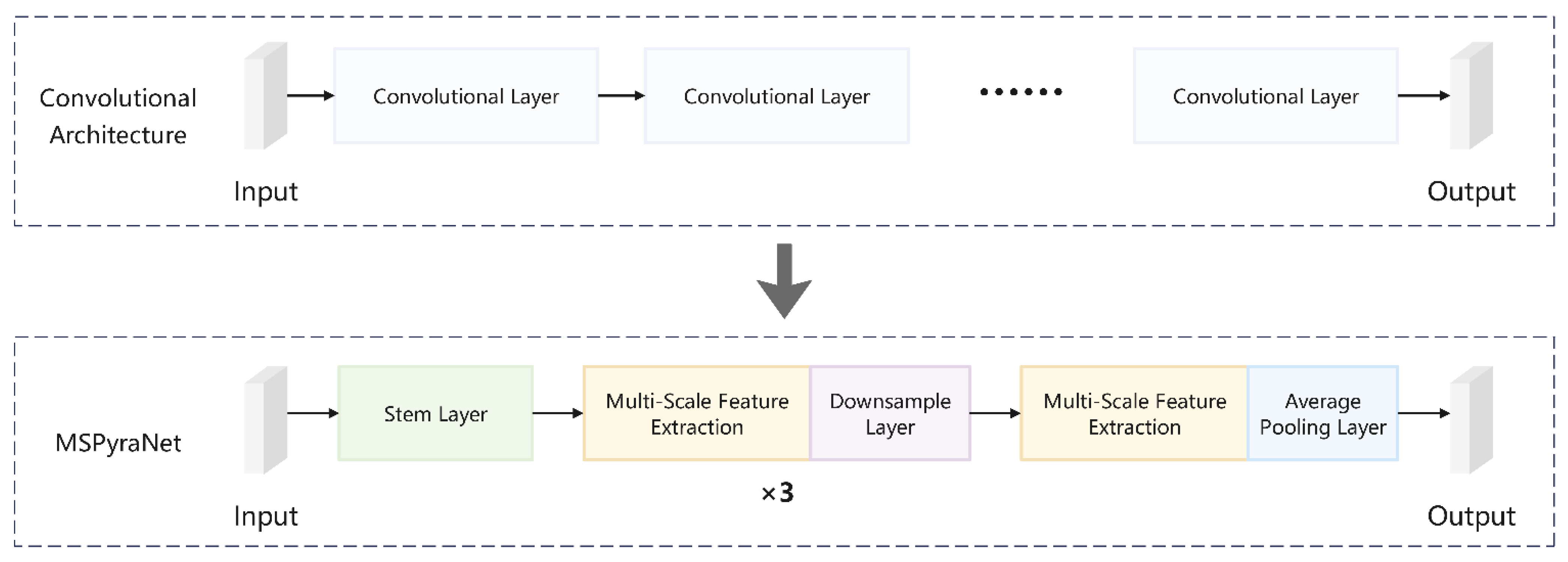
To address these issues, we have made contributions in two key areas. First, in terms of datasets, we expanded the CHMD1 dataset by adding four new types of Chinese herbal medicine and 933 sample images, constructing the CHMD2 dataset. This dataset retains the advantages of CHMD1 and significantly enhances inter-class similarity and morphological diversity, providing richer data support for model training. Second, in terms of model network improvement, we proposed an enhancement based on the ConvNeXtv2 architecture [
19]. Specifically, we replaced the original 7 × 7 large-kernel depthwise separable convolution [
20] with a multi-scale representation module, creating the FACNBlock (Fusion Atrous Convolutional Network Block) unit tailored for Chinese herbal images capable of extracting multi-scale features. The FACNBlock can generate feature maps with diverse receptive fields, and it considers the characteristic that Chinese herbal medicines often appear in stacked forms. FACNBlock enables a more comprehensive and accurate capture of the multi-level features of Chinese herbal medicine images. Additionally, the FACNBlock incorporates Global Response Normalization (GRN) technology [
19], significantly enhancing the expressive power between channels. GRN improves the contrast between feature map channels, alleviating feature collapse and enabling different channels to focus on extracting diverse feature information, thereby enhancing the model’s expressive power. Based on this, we proposed the MSPyraNet (Multi-Scale Pyramid Network), which fuses feature maps of different receptive fields to more comprehensively capture multi-level features of Chinese herbal images, further optimizing the performance of Chinese herbal image classification tasks. The comparison between the MSPyraNet and other convolutional architecture can be seen in
Figure 1. Experimental results show that our method performs well in Chinese herbal image classification tasks.
The main contributions of this study include the following:
- (1)
Dataset Expansion and Improvement: We constructed the CHMD2 dataset, adding four new types of Chinese herbal medicine. This significantly enhanced inter-class similarity and morphological diversity, providing richer data support for model training;
- (2)
Network Architecture Innovation: Proposed the MSPyraNet, significantly improving the model’s classification ability for Chinese herbal images through multi-scale feature extraction tailored to the characteristics of Chinese herbal images;
- (3)
Experimental Results and Analysis: To validate the effectiveness of the proposed method, we conducted comparative experiments on the CHMD1 and CHMD2 datasets. The results show that the MSPyraNet significantly outperforms baseline models.
3. Dataset Construction
Chinese herbal datasets play a crucial role in improving model performance. However, the construction of Chinese herbal image datasets in current research still has shortcomings. In a previous study, we constructed the CHMD1 dataset, which includes 16 types of Chinese herbal medicine with 3552 images [
9]. This dataset has significant advantages over similar datasets: it is specifically designed for Chinese herbal medicine, effectively addressing issues such as limited categories, insufficient sample sizes, single shooting angles, monotonous backgrounds, and uniform morphology within categories. However, the number of Chinese herb types is still insufficient, leading to excessive differences between different types of Chinese herbal medicine and a lack of inter-class similarity, thereby affecting the model’s generalization capabilities. Therefore, we constructed a new Chinese herbal dataset, CHMD2, based on CHMD1.
The CHMD2 dataset incorporates four new types of Chinese herbal medicine images: Raspberry, Poria Cocos, Rhizoma Phragmitis, and Cinnamon. These images were primarily obtained through web scraping, yielding an initial collection of 2496 raw images. After manual curation, 933 high-quality images were retained, including 249 Raspberry images, 199 Poria Cocos images, 245 Cinnamon images, and 240 Rhizoma Phragmitis images. The total number of images in CHMD2 reaches 4485, with the number of types increasing to 20. As shown in
Table 1. The category with the most images has 380 images, while the category with the fewest images has 110 images. The distribution of the number of images for each Chinese herb is shown in
Figure 2. CHMD2 not only retains the advantages of CHMD1, such as complex and diverse morphological expressions, stacked or clustered presentation, single-category images, diverse backgrounds, and multi-angle shooting, but it also increases inter-class similarity and further enhances morphological diversity.
The addition of the four new categories effectively enhances the inter-class similarity of the dataset, thereby improving the model’s generalization capabilities. As shown in
Figure 3, in the original dataset, matrimony vine is the only red Chinese herb category. However, color is an important feature for model recognition, which may limit the model’s ability to recognize red features. Therefore, we introduced raspberry images. Most Raspberries are also red, which will interfere with the model to some extent, thereby improving its generalization capabilities. Similarly, in the original dataset, alumen is the only white block-shaped Chinese herb. Processed poria cocos is also white and block-shaped, and its inclusion helps the model recognize such forms, thereby improving the model’s ability to handle similar forms. The introduction of rhizoma phragmitis enhances the model’s recognition ability due to its similar texture and morphological features to radix stemonae, while the addition of cinnamon expands the model’s recognition range for strip-shaped species.
To further increase the morphological diversity of the dataset, the four new categories include multiple morphological features. As shown in
Figure 4, raspberry presents different forms, colors, and textures, including fresh and processed states, red and black variants, and morphological changes due to different processing techniques. Poria cocos includes processed and unprocessed samples, with processed samples further divided into block and slice forms. Rhizoma phragmitis also has processed and unprocessed distinctions, with different parts showing morphological differences. Cinnamon includes slice and strip forms in different colors. These diverse morphological features provide more comprehensive learning samples for the model, helping to improve its robustness and accuracy in practical applications.
5. Experiments and Results Analysis
5.1. Evaluation Metrics and Experimental Setup
Following previous work, we chose accuracy, precision, recall, and F1 score as evaluation metrics. These metrics are widely used in classification tasks. Accuracy represents the proportion of correctly predicted samples, reflecting overall classification capability; Precision measures the proportion of actual positives among predicted positives, indicating prediction exactness; Recall calculates the proportion of actual positives correctly identified, demonstrating model coverage; F1-Score is the harmonic mean of Precision and Recall, providing a balanced evaluation for imbalanced datasets. The formulas are defined as:
where
denotes True Positives,
denotes True Negatives,
denotes False Positives, and
denotes False Negatives.
We divided the dataset into training, validation, and test sets in a ratio of 7:2:1. The training set contains 3140 images, the validation set contains 897 images, and the test set contains 448 images, ensuring a balanced sample distribution across each category.
To enhance the model’s generalization ability, we performed data augmentation on the dataset. First, each image was rotated by 90, 180, and 270 degrees, expanding the sample size to four times the original. Then, the training set images were uniformly resized to 256 × 256, followed by random horizontal or vertical flipping, and finally randomly cropped to 224 × 224. The validation and test set images were directly randomly cropped to 224 × 224.
To avoid certain features having an excessive impact on model training and to speed up the training process, we perform normalization on the input image tensor. Specifically, for each channel of the image, we subtract the mean and divide it by the standard deviation, ensuring that the mean of each channel is 0 and the variance is 1. The formula is as follows:
where
represents the pixel value in the original image,
represents the normalized result,
denotes the mean of the pixel values in the dataset, and
denotes the standard deviation of the pixel values in the dataset.
The main training parameter settings are as follows: the number of training epochs is 200, the batch size per epoch is 32, and the optimizer used is AdamW.
5.2. Comparative Experiments with Common Models on CHMD1
To validate the effectiveness of MSPyraNet in the task of Chinese herbal medicine classification, we conducted comparative experiments on the CHMD1 dataset. The performance of MSPyraNet was evaluated against several common deep learning models [
17,
21,
22,
28,
29,
30] and CSA [
9] in terms of accuracy, precision, recall, and F1 score. The classification layer of all models utilized a fully connected layer. Previous studies have shown that the CSA encoder outperforms other deep learning classification algorithms on the CHMD1 dataset, albeit with limited improvement. However, MSPyraNet demonstrated significantly superior performance on the CHMD1 dataset compared to other models. The specific experimental results are presented in
Table 2.
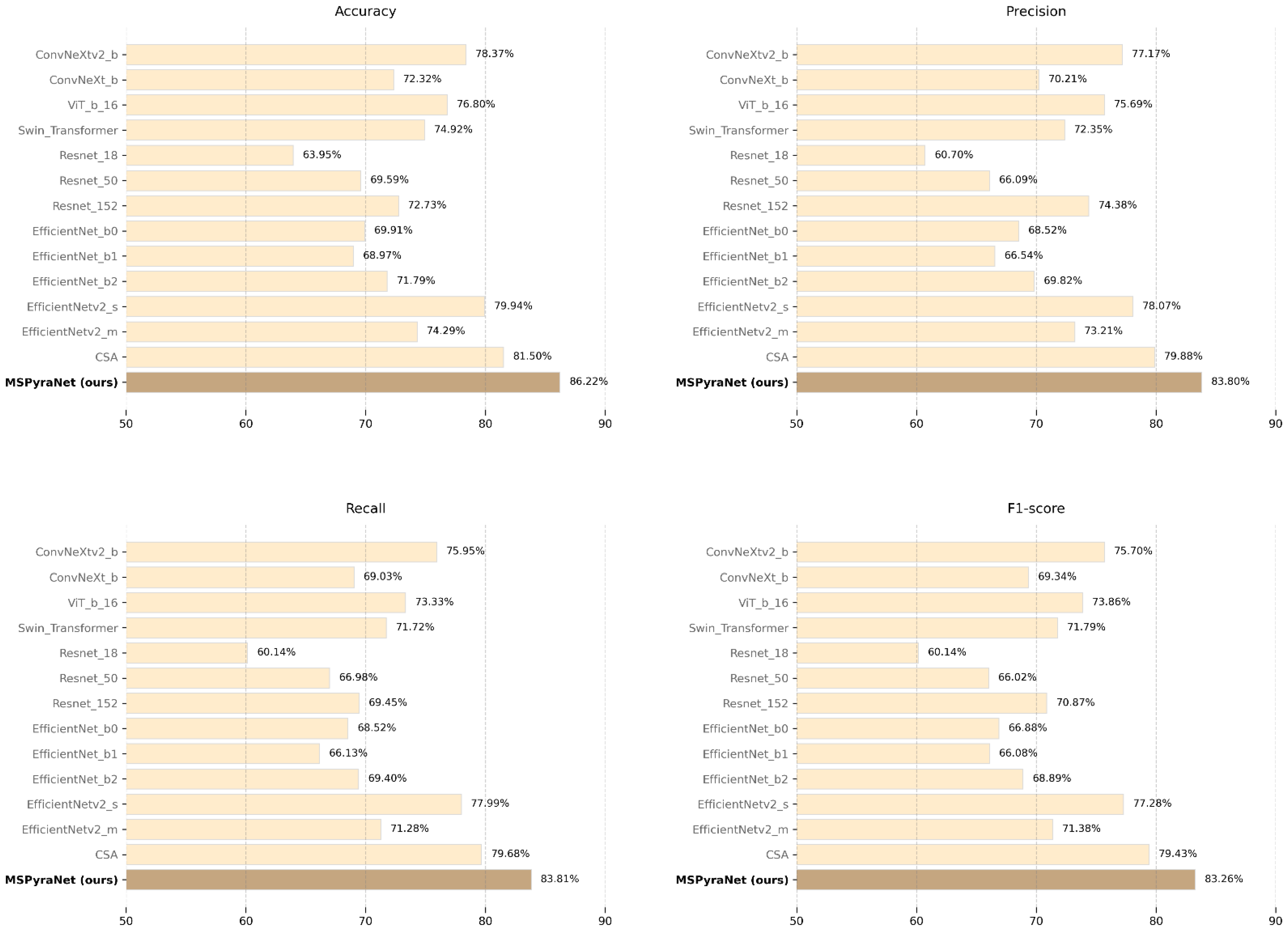
Experimental results demonstrate that MSPyraNet achieves superior performance, with an accuracy of 86.22%, precision of 83.80%, recall of 83.81%, and an F1 score of 83.26%. Compared to the CSA encoder, MSPyraNet shows improvements of 4.72%, 3.92%, 4.13%, and 3.83% in accuracy, precision, recall, and F1 score, respectively. Among all the SOTA models, EfficientNetv2_s performs the best, with scores of 79.94% (accuracy), 78.07% (precision), 77.99% (recall), and 77.28% (F1 score). MSPyraNet outperforms EfficientNetv2_s by 6.28% (accuracy), 5.73% (precision), 5.82% (recall), and 5.98% (F1 score). Within the Resnet family, MSPyraNet surpasses the best-performing model, Resnet-152, by 13.49% (accuracy), 9.42% (precision), 14.36% (recall), and 12.39% (F1 score). Similarly, compared to the best model in the ConvNeXt family, ConvNeXtv2_b, MSPyraNet achieves improvements of 7.85% (accuracy), 6.63% (precision), 7.86% (recall), and 7.56% (F1 score). These results demonstrate that MSPyraNet excels in the task of Chinese herbal medicine classification, marking a significant advancement over previous research.
Figure 6 illustrates the performance comparison of MSPyraNet and SOTA models on the CHMD1 dataset across four evaluation metrics.
5.3. Comparative Experiments with Common Models on CHMD2
To further validate the performance of MSPyraNet on a more complex dataset, in this study, experiments were conducted on the CHMD2 dataset, with comparisons made against the CSA encoder [
9], ConvNeXt v2 [
19] and some other common deep learning models [
17,
29,
30]. The experiments aimed to demonstrate the effectiveness of the MSPyraNet in more complex Chinese herbal medicine classification tasks. The results, presented in
Table 3, show that MSPyraNet maintains commendable performance on the more complex CHMD2 dataset.
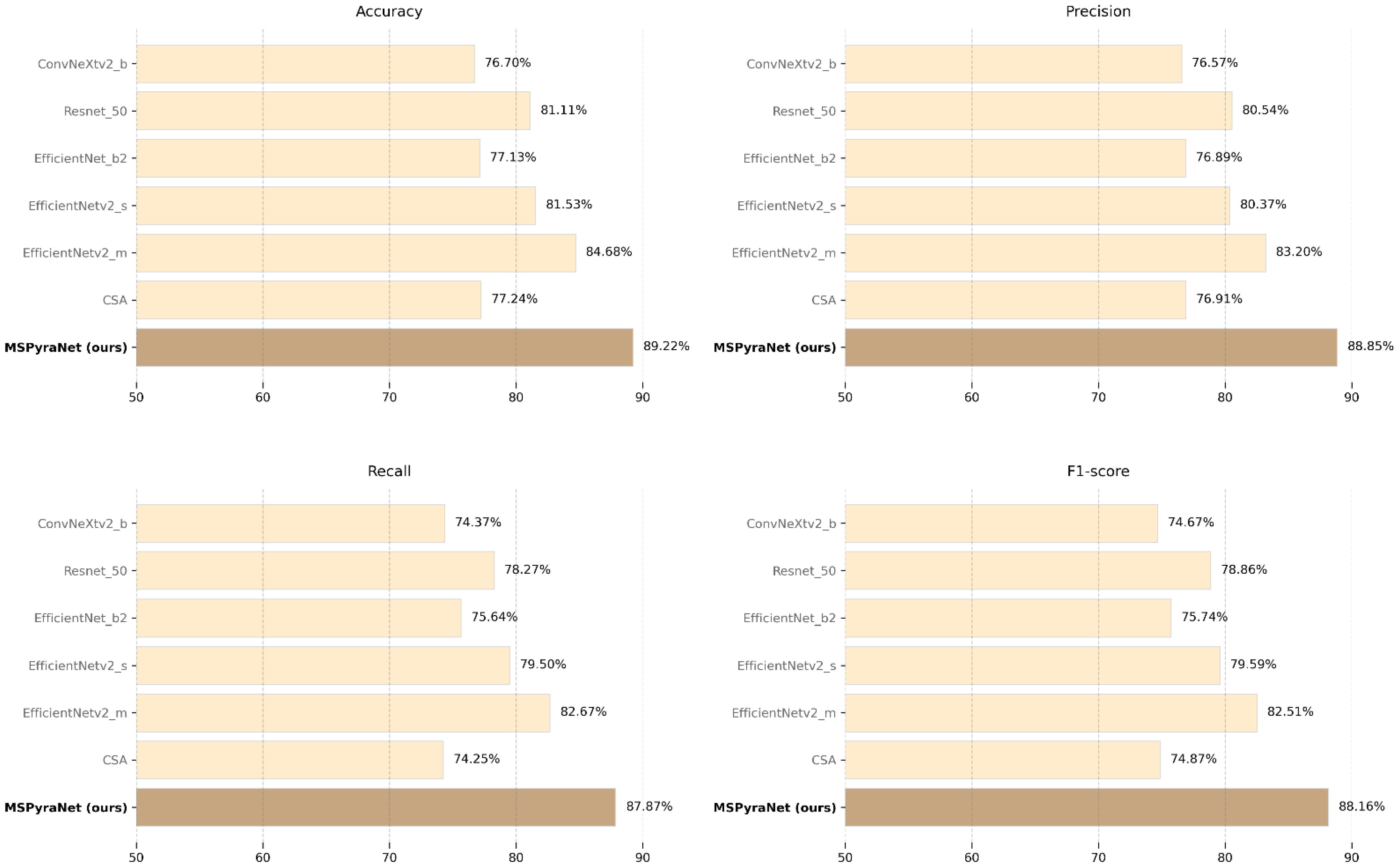
On the CHMD2 dataset, MSPyraNet demonstrates outstanding performance, with an accuracy of 89.22%, precision of 88.85%, recall of 87.87%, and an F1 score of 88.16%. Compared to the best SOTA model, EfficientNetv2_m, MSPyraNet improves accuracy, precision, recall, and F1-score by 4.54%, 5.65%, 5.20%, and 5.65%, respectively. The improvements over the CSA model are even more significant, with gains of 11.98% in accuracy, 11.94% in precision, 13.62% in recall, and 13.29% in F1-score. MSPyraNet also outperforms Resnet_50 by 8.11%, 8.31%, 9.60%, and 9.30% on the four metrics. Compared to ConvNeXtv2_b, the performance gains are 12.52%, 12.28%, 13.50%, and 13.49%, respectively. These results highlight the robustness and effectiveness of MSPyraNet on this classification task.
Figure 7 illustrates the performance comparison of MSPyraNet and SOTA models on the CHMD2 dataset across four evaluation metrics.
5.4. Ablation Experiment Analysis
Ablation experiments were conducted on both the CHMD1 and CHMD2 datasets to systematically evaluate the effectiveness of the multi-scale representation module. By comparing the performance differences between models with and without the multi-scale representation module, the contribution of this module was validated. Specifically, two experimental setups were designed: without the multi-scale representation module and with the multi-scale representation module. The results are presented in
Table 4.
The experimental results reveal that the introduction of the multi-scale representation module significantly enhances the model’s performance on both datasets. Specifically, on the CHMD1 dataset, the model’s accuracy, precision, recall, and F1 score improved by 9.81%, 11.08%, 13.65%, and 12.82%, respectively. On the CHMD2 dataset, the corresponding metrics improved by 12.14%, 11.11%, 13.59%, and 13.08%. These results robustly confirm the importance of the multi-scale representation module in the task of Chinese herbal medicine image classification.
Notably, the MSPyraNet model exhibited superior performance on the more complex CHMD2 dataset. As shown in
Table 3, MSPyraNet’s accuracy, precision, recall, and F1 score on CHMD2 improved by 3%, 5.05%, 4.06%, and 4.9%, respectively, compared to CHMD1. This indicates that the model possesses stronger adaptability and robustness in handling complex Chinese herbal medicine classification tasks.
To validate the suitability of smaller convolutional sizes for feature extraction in scenarios where Chinese herbal medicines are stacked, we conducted experiments on CHMD2 using existing multi-scale fusion pyramid structures [
27] with varying convolutional sizes. The results are presented in
Table 5.
From
Table 4, it is evident that adjusting the convolutional sizes in consideration of the characteristics of Chinese herbal medicines led to a significant improvement in model performance. MSPyraNet achieved improvements of 10.12%, 9.77%, 12.2%, and 11.66% in accuracy, precision, recall, and F1 score, respectively. This demonstrates that the multi-scale representation module in MSPyraNet is particularly well suited for Chinese herbal medicine image classification tasks.
To verify the importance of each convolutional unit in the multi-scale representation module, we conducted ablation experiments on CHMD2 by removing one convolutional unit at a time. The experimental results are shown in
Table 6.
The experimental results indicate that removing any single convolutional unit from the Multi-scale Representation Module leads to a decline in model performance. Among them, the removal of the 3 × 3 convolution has the most significant impact, with accuracy, precision, recall, and F1-score dropping to 82.99%, 82.96%, 80.86%, and 81.42%, respectively. These findings suggest that each convolutional unit plays a distinct and important role within the module.
5.5. Single Chinese Herb Prediction Accuracy Analysis
To showcase MSPyraNet’s capability in identifying individual Chinese herbal medicines, we analyzed the model’s accuracy for each Chinese herb in the CHMD2 dataset. For a more intuitive comparison, the accuracy of CSA [
9] and ConvNeXtv2_b [
19] for each herb in CHMD2 was also presented. The results are shown in
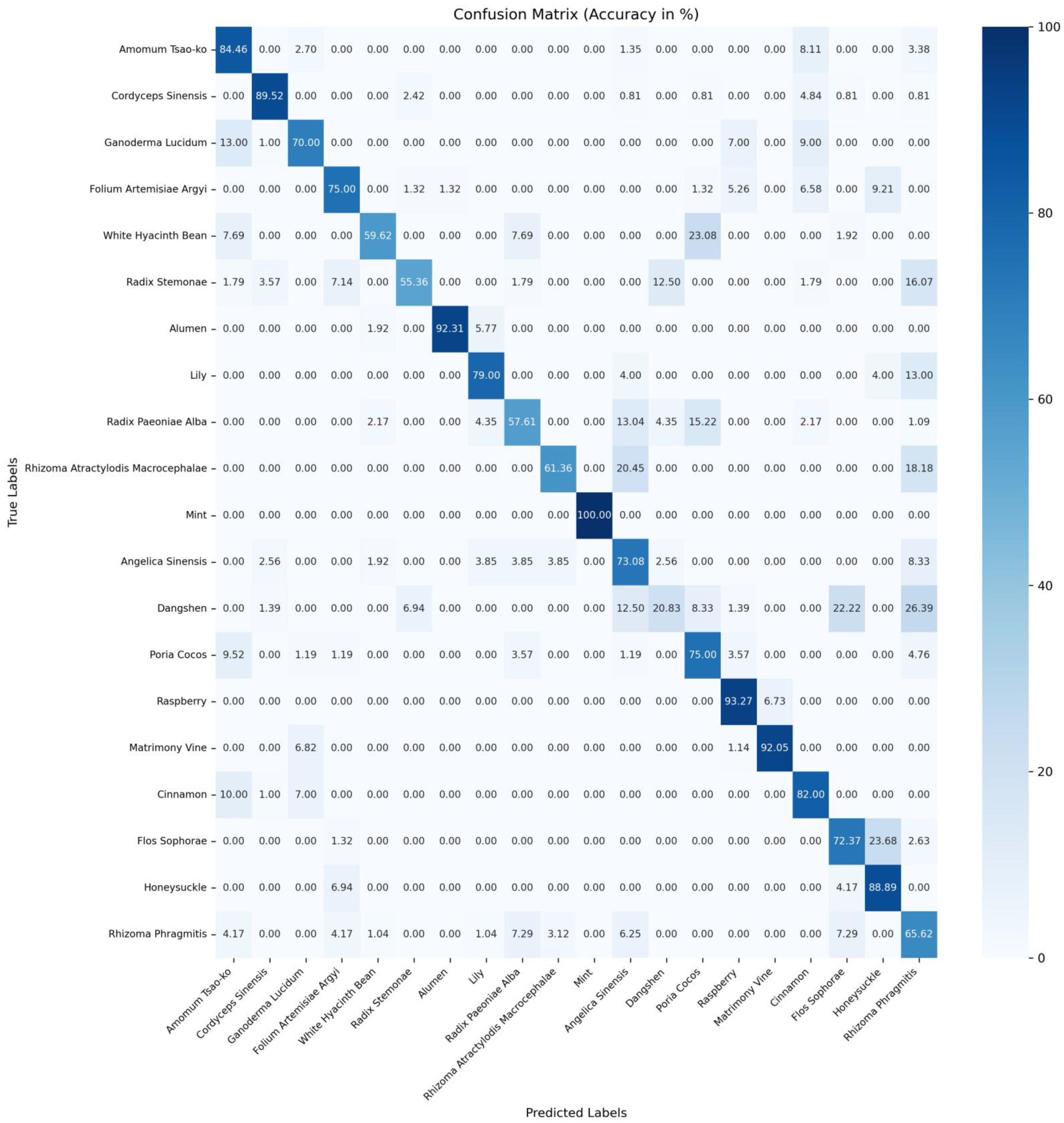
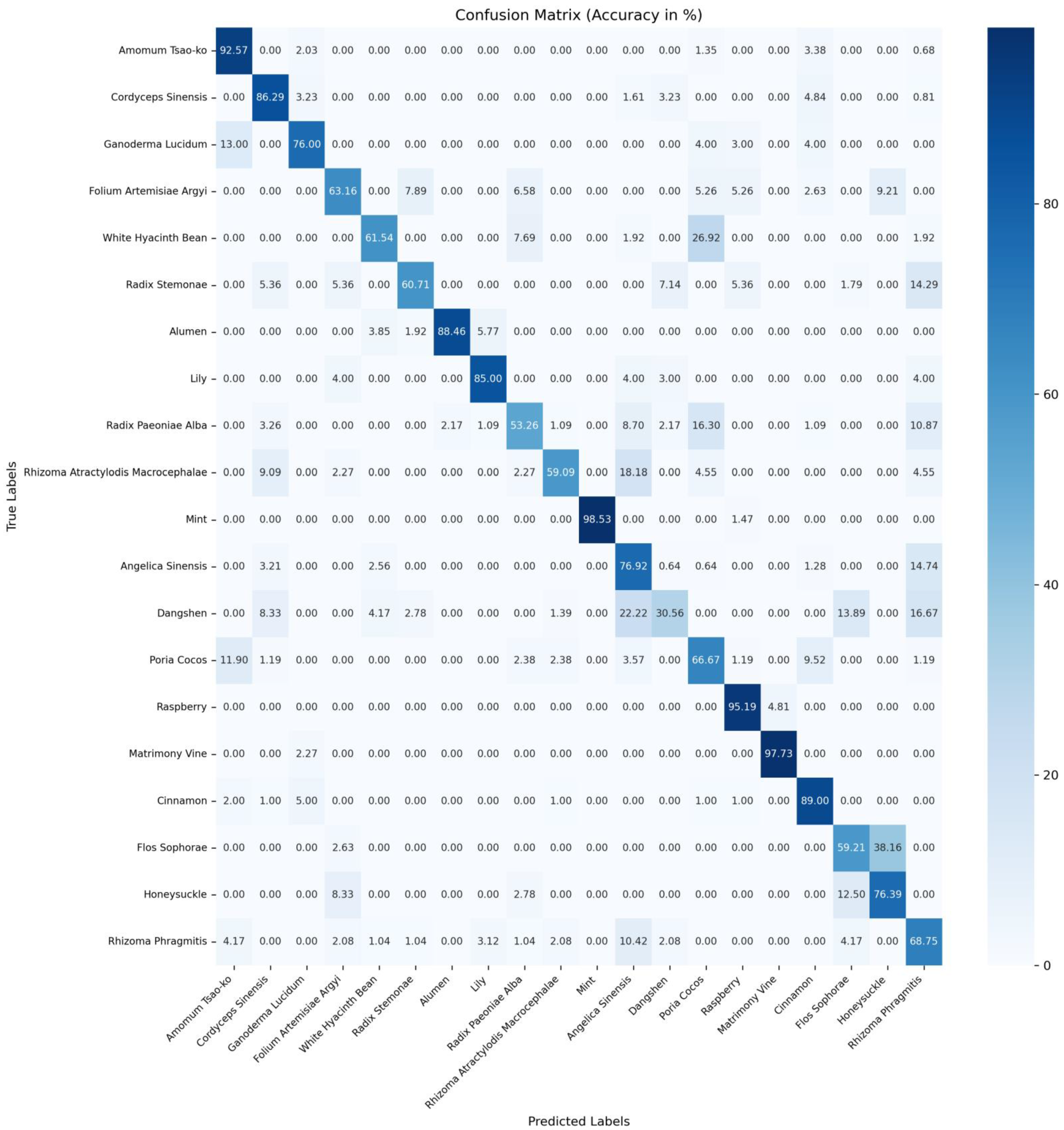
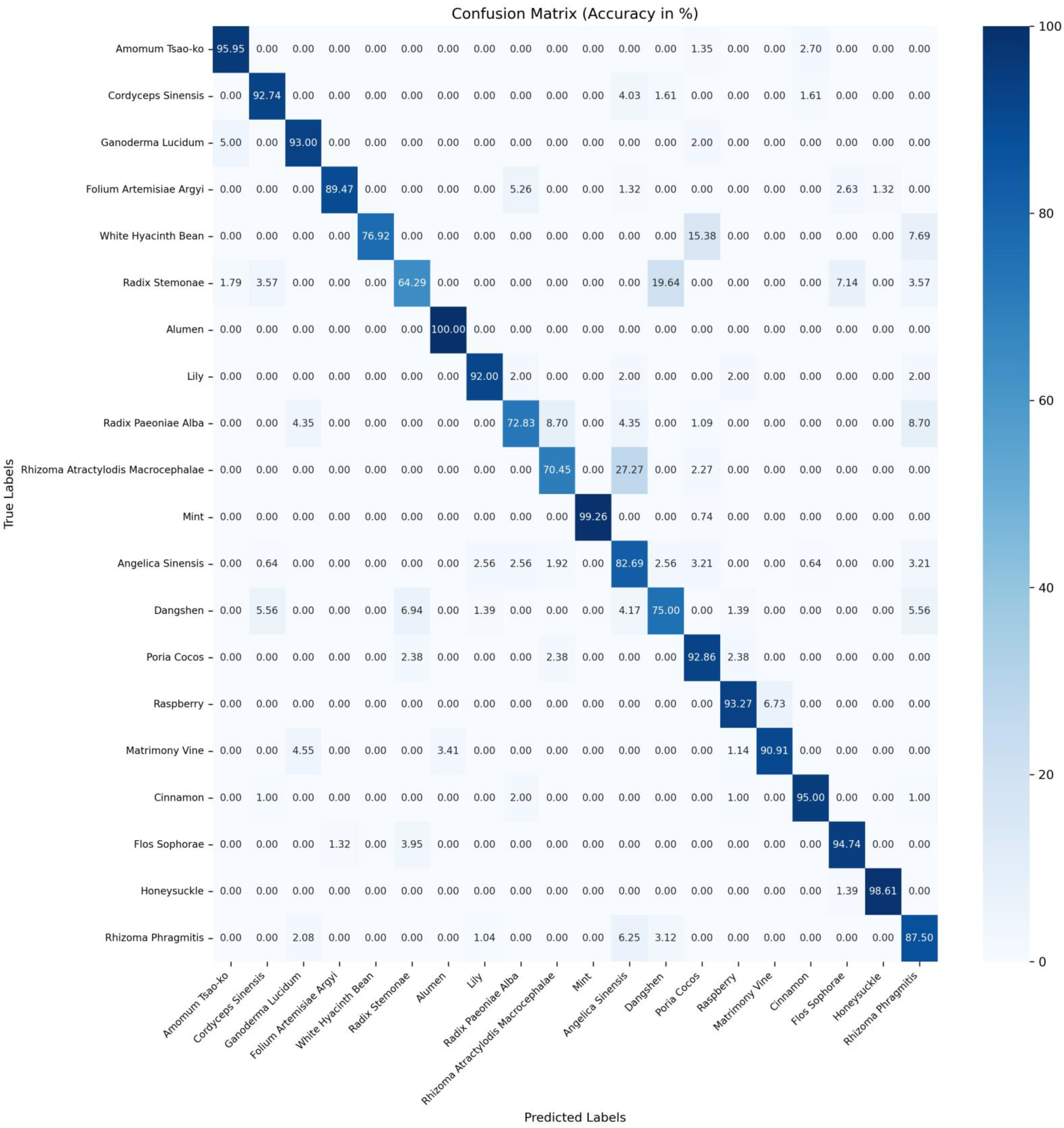
Table 7. The confusion matrices for accuracy of ConvNeXtv2_b [
19], CSA [
9], and MSPyraNet on the CHMD2 dataset can be found in
Figure A1,
Figure A2 and
Figure A3 in
Appendix A, respectively.
The experimental results indicate that MSPyraNet’s accuracy in identifying individual Chinese herbal medicines is significantly superior to that of CSA and ConvNeXtv2_b. Particularly notable improvements were observed for ganoderma lucidum, folium artemisiae argyi, dangshen, poria cocos, and flos sophorae.
To further demonstrate MSPyraNet’s enhanced performance in classifying Chinese herbal medicines with high inter-class similarity, a comparison was conducted of the probabilities of a given Chinese herb being predicted as the true label versus other morphologically similar labels. The results are presented in
Table 8.
The experimental results demonstrate that MSPyraNet exhibits superior performance in distinguishing between morphologically similar Chinese herbal medicines. For instance, flos sophorae, which is morphologically similar to honeysuckle, was frequently misclassified as honeysuckle by ConvNeXtv2_b and CSA. However, MSPyraNet did not misclassify flos sophorae as honeysuckle. Similarly, honeysuckle was often misclassified as folium artemisiae argyi and flos sophorae by ConvNeXtv2_b and CSA, but MSPyraNet did not misclassify honeysuckle as folium artemisiae argyi and only misclassified 1.39% of honeysuckle as flos sophorae. The identification accuracy of dangshen was notably low in ConvNeXtv2_b and CSA, with CSA achieving only 30.56%. However, MSPyraNet achieved an accuracy of 75.00% for dangshen, significantly reducing misclassifications such as angelica dinensis, flos sophorae, and rhizoma rhragmitis. The misclassification rate of white hyacinth bean as poria cocos was also reduced to 15.38%.
5.6. Evaluation Under Different Dataset Splits
To verify the stability and non-randomness of the model’s performance, we conducted training on the CHMD2 dataset by randomly re-shuffling the images in the training, validation, and test sets. The model’s performance was then compared under three different data-splitting strategies. The experimental results are presented in
Table 9.
As can be observed, the performance metrics remain consistently high and exhibit only minor variations across the three different data partitioning strategies. This consistency indicates that the model is not overly dependent on a specific dataset split and confirms its robustness and generalization ability. Therefore, the strong and stable results across all configurations provide solid evidence of the reliability and effectiveness of the proposed model.