Featured Application
In the biomedical field, accessing data by classical methods is getting more difficult day by day, as it is in any other field, due to the data growth rate. Different methods are needed to access the desired data more quickly. In particular, more specific methods need to be developed for question answering systems. In this study, a model is proposed for the document retrieval and answer extraction modules which are a part of biomedical question answering systems.
Abstract
In this paper, we describe our biomedical document retrieval system and answers extraction module, which is part of the biomedical question answering system. Approximately 26.5 million PubMed articles are indexed as a corpus with the Apache Lucene text search engine. Our proposed system consists of three parts. The first part is the question analysis module, which analyzes the question and enriches it with biomedical concepts related to its wording. The second part of the system is the document retrieval module. In this step, the proposed system is tested using different information retrieval models, like the Vector Space Model, Okapi BM25, and Query Likelihood. The third part is the document re-ranking module, which is responsible for re-arranging the documents retrieved in the previous step. For this study, we tested our proposed system with 6B training questions from the BioASQ challenge task. We obtained the best MAP score on the document retrieval phase when we used Query Likelihood with the Dirichlet Smoothing model. We used the sequential dependence model at the re-rank phase, but this model produced a worse MAP score than the previous phase. In similarity calculation, we included the Named Entity Recognition (NER), UMLS Concept Unique Identifiers (CUI), and UMLS Semantic Types of the words in the question to find the sentences containing the answer. Using this approach, we observed a performance enhancement of roughly 25% for the top 20 outcomes, surpassing another method employed in this study, which relies solely on textual similarity.
1. Introduction
This paper describes our document retrieval system and answers extraction module, which is part of the BioASQ question answering task. In general, Question Answering (QA) is an automatic process capable of understanding questions asked in a natural language, such as English, and responding exactly with the requested information. An “ideal” QA system has a very complicated architecture because it has to determine the desired information in the question, find the requested information from suitable sources, extract it, and then create the right answer. Users prefer that a QA system finds precise answers to questions, rather than acquires all the documents relevant to their search query. Studies on systems that can automatically answer questions asked in a natural language started in the 1960s. However, this research area became extremely popular within the information retrieval community in 1999 after the Text Retrieval Conference (TREC) [1]. Compared to open domain QA systems, far fewer researchers are working on the medical branch of domain-specific question answering. Rinaldi and his colleagues [2] have customized an open-domain QA system to construct answers for questions related to the genome, focusing on defining term relations based on a linguistic-rich full-parser. Due to the continuous increase in information produced in the biomedical field, there is also an increasing need for biomedical QA, especially for the public, medical students, healthcare professionals, and biomedical researchers [3]. Biomedical QA is one of the most important applications of real-world biomedical systems, and there are many initiatives to promote research in this field [1].
The BioASQ challenge is an organization that has been focusing on the advancement of contemporary biomedical semantic indexing and QA at large-scale since 2013 [4]. The organizers have undertaken the task of evaluating existing solutions to various QA sub-tasks. Within this organization, various benchmarks have been provided to assess researchers in the field of QA systems [5]. The BioASQ challenge consists of two tasks: large-scale biomedical semantic indexing and QA. Several systems have been developed within the scope of this challenge.
To begin, the NCBI framework [6] is one of these. It uses a PubMed search to return relevant documents. It employs cosine similarity to calculate the likeness between the question and sentence. The highest-scored sentence in the abstract is to be returned as a snippet. A modified dictionary search algorithm is being used with MetaMap [7] to identify biomedical concepts. BioPortal services are also employed to expand the query. For example, for the question: “Is Rheumatoid Arthritis more common in men or women?” the synonyms extracted are “arthropathy” for “arthritis” and “female”, “femme”, and “adult” for “women”. The system presented in [8] depends on the SAP HANA Database, which has text analysis features, like tokenization, sentence splitting, named-entity recognition, full-text indexing, and approximate text matching, for text processing. Another example is the CMU OAQA system, which has a hierarchical retrieval architecture. In this system, each query is completed in three steps. In the first step, all stop words are removed from the query. The Dirichlet smoothing retrieval model is used for scoring documents, and only the top 10,000 are retrieved. The Negative Query Generation (NQG) model is used to re-rank documents in the second step. After re-ranking, only the top 100 documents are to be retrieved. In the third step, in-depth analysis is performed on documents, and pre-trained Learning to Rank (LETOR) algorithms are used to score documents, and then only the top 10 documents are retrieved [9]. Ref. [10] has built a generic retrieval model based on the Sequential Dependence Model, Word Embedding, and the Ranking Model for document retrieval. In this generic model, titles have special significance, and top-K results are re-ranked according to meaningful nouns in their titles. The Fudan team uses a statistical language model and query likelihood model to retrieve relevant documents. They have employed the Indri search engine to build their system. According to this, nouns in the query have higher significance. Retrieved documents are re-ranked according to the presence of the keywords in the query in the document. If the document contains all the keywords in the query, it will score higher than others [11]. The IIIT-Hyderabad team has used chunking, stop words removal, and query formulation techniques to retrieve documents. The most relevant phrases are collected as snippets from the top documents. Cosine similarity and noun chunk identification techniques have been used to produce exact and ideal answers from the snippets [12]. Pubmed articles are indexed by the Lucene search engine. Various models have been employed to retrieve documents in this system. They include cosine similarity, sequential dependence, and semantic concept-enriched dependence models. This system uses UMLS concepts in the query as additional criteria for ranking the documents [13]. Another team [14] has used the Indri search engine for document retrieval. A unigram language model with Dirichlet prior smoothing is used as a retrieval method. Generally, the sequential dependence model (SDM) and semantic concept-enriched dependence model (SCDM) perform better than the method used in the baseline query likelihood model (QL). KSAnswer is a biomedical QA system that returns the most relevant documents and snippets. Candidate snippets are retrieved by using a cluster-based language model. Five independent similarity models are used to re-rank the retrieved top-N snippets [15]. The “AUTH” team has tried approaches based on BioASQ search services and ElasticSearch for the document retrieval task by querying the top 10 documents, with a combination of words in each question [16]. The AUEB team has developed their BioASQ6 document retrieval system, which they have modified to give a relevance score for each sentence, and experimented with BERT and PACRR for this task [17]. The Google team has used different models in their system, such as the BM25 retrieval model, BioBERT, Synthetic Query Generation (QGen), a retrieval model based on BM25, and a neural model [18]. Additionally, the BERT model was used to re-rank the results [19]. The University of Aveiro’s “bioinfo” team has created a retrieval process consisting of two stages. Initially, they utilized the traditional BM25 model in the first stage. Subsequently, the second stage incorporated neural re-ranking models based on transformers, specifically sourced from Pub-MedBERT and monoT5 checkpoints [20]. The University of Regensburg’s team has employed “UR-gpt” systems that harnessed two commercial iterations of the GPT Large Language Model (LLM). Their approach involved experimenting with both GPT-3.5-turbo and GPT-4 models. Zero-shot learning for query expansion, query re-formulation, and re-ranking was utilized in this system [21]. The MindLab team’s approach in the realm of document retrieval involved utilizing the BM25 scoring function coupled with a re-ranking strategy based on semantic similarity [22]. Fudan University’s “dmiip” team has employed a two-stage approach in their systems. In the retrieval stage, they utilized both BM25 and GPT. They implemented a cross-encoder ranker for the ranking stage to leverage various biomedical Pre-trained Language Models (PLMs), including PubMedBERT, BioBERT, BioLinkBERT, and ELECTRA [23]. The A&Q team has employed a multi-stage approach in their systems, involving a bi-encoder model for the retrieval stage and a cross-encoder model for the subsequent re-ranking stage. They have implemented a hybrid retriever combining dense and sparse methods. The dense retrieval aspect utilized the bi-encoder, while the sparse retrieval used BM25. Both encoders were initialized with Pub-MedBERT and underwent additional training using PubMed query-article search logs [24].
In this study, we tested ranking algorithms under different scenarios and found the ranking algorithm that performs well in the depicted scenarios. We examined the impact of query expansion techniques in various scenarios (Pos tagger, UMLS Services) on biomedical document retrieval performance. We also employed NER, UMLS CUI, Semantic Type, and Semantic Group features for answer extraction, and this method has resulted in performance improvement.
2. Materials and Methods
This study covers two main tasks. The first task is to find the document containing the answer to the question, and the second is to find the specific sentences that can be the answer. We developed a question analysis module to find the relevant document for the first task. This module, which is responsible for improving the query, enriches the query by adding biomedical concepts and nouns related to the terminology in the question. Known ranking algorithms were used for the first task. The answer extraction module was supposed to find sentences that might be the answer to the question. For this purpose, the similarity between each sentence and the question was measured, and the sentences with the highest similarity were taken as the answer. We included semantic similarity in the calculation, along with textual similarity, to increase the system’s performance. The similarity algorithm consists of the Named Entity Recognition (NER), UMLS Concept Unique Identifiers (CUI), UMLS Semantic Type, and UMLS Semantic Group features of the question and sentence.
All application modules are written in Python.
2.1. Ranking Models
Commonly used models for ranking document similarity are as follows.
2.1.1. Vector Space Model
The vector space model is an algebraic model for representing text documents as vectors [25]. In this model, documents and queries are represented as vectors. Each separate term is represented as a dimension. The term’s value in the vector differs from zero if the document contains the term. Several ways have been developed to calculate the weight of the term. The most popular term weight calculation method is tf-idf. The cosine similarity between two vectors is a measure that calculates the cosine of the angle between the vectors. The cosine value of the angle between the two vectors can be between 0 and 1. Using the Euclidean point product formula, the cosine of two vectors can be calculated as shown in Equation (1).
Hence, the similarity of two documents (a and b) is calculated as shown in Equation (2).
2.1.2. Okapi BM25
Okapi BM25 (BM stands for Best Matching) is based on the probabilistic retrieval framework [26]. The BM25 ranks the documents according to the fact that the terms in the query are found within them, regardless of their relationships. It has slightly different scoring functions. One of the most common uses of the function is as in Equation (3).
Given a query Q, containing terms q1…, qn, the BM25 score of a document D is:
where f(qi,D) is the term frequency of the qi in the document D, |D| is the length of the document according to the number of words it contains, and avgdl is the average document length in the text collection. k1 and b are free parameters. The parameter k1 is usually selected in the range (1.2,2.0), and parameter b is chosen as 0.75. IDF(qi) is the inverse document frequency (IDF) weight of the query term qi. It is generally calculated as shown in Equation (4).
where N denotes the total number of documents in the collection and n(qi) denotes the number of documents containing the term qi.
2.1.3. Bayesian Smoothing with Dirichlet Priors
This model produced the best performance in our document retrieval system. It is also described by [27] and is defined in Equation (5).
where c(w;d) is the frequency of presence of the word in the document, and µ is the smoothing parameter. p(w|C) is the collection model containing all the documents.
2.1.4. Sequential Dependence Model
In [28], Metzler and Croft proposed a sequential dependence model (SDM). In this model, the adjacent query term affects the similarity score. There are three features in the SDM to be considered: single-word features (a collection consisting of single-word, QT), ordered bi-words phrase features (the two words in a phrase appearing in order, QO), and unordered window features (one or several words can be allowed, appearing between the two words, QU). A score for documents is calculated as shown in Equation (6).
where Q is a sequence of keywords extracted from a user query, D is a candidate document, and qi is the i-th query keyword of Q. fT(qi,D) denotes the frequency of a term in the document D, fO(qi,qi+1,D) denotes the frequency of the exact phase qi..qi+1 appearing as ordered in the document D, and fU(qi,qi+1,D) denotes the frequency of phase qi..qi+1 ordered or unordered within window N terms in document D. λT, λO, and λU are weighting parameters. Setting these parameters as λT = 0.85, λO = 0.10, and λU = 0.05 is recommended.
2.2. QA System Components
Typically, an automated QA system has three components:
- Question Analysis
- Document Retrieval
- Answer Extraction
2.2.1. Question Analysis
One of the difficulties in question analysis is creating query terms from a question asked in a natural language [5]. In the procedure for preprocessing queries, we performed a sequence of tasks to extract the keywords from user queries. This is how we planned our system. At the first step of the question analysis, the question is tokenized, and stop words and unrelated symbols are removed. Then, nouns, noun phrases, and other word classes are determined using the nltk pos tagger package [29]. Since the nouns and noun phrases are more significant and distinctive words in terms of text similarity, the query is enriched by adding these words. There are some other important points in terms of question analysis. For example, biomedical concepts play a crucial role in identifying synonym terms and semantic analysis. Since similar concepts are written differently in the document, ranking algorithms that calculate scores based on word similarity cannot include these words in the similarity score. At this step, our system determines UMLS concept unique identifiers (CUI) in query terms using MetaMap18 [30]. After determining biomedical concepts, the QA module enriches the question with biomedical concepts. A description of each step is provided:
- Tokenization: involves breaking down a sequence of text into smaller units, known as tokens.
- Stemming: a process that substitutes all word variations with the word’s single stem or root. This typically includes removing any attached suffixes and prefixes from words. For instance, the words “reading”, “reader”, and “reads” are transformed into the root “read”.
- Stop-words are a series of commonly used words in a language. They can be safely ignored without compromising the meaning of the sentence. Examples of stop words in English include “a”, “the”, “is”, “our”, etc. Eliminating these terms helps increase the accuracy of the findings.
The Question Analysis System Architecture is shown in Figure 1.

Figure 1.
Question Analysis System Architecture. The modified query was used in the document retrieval phase. We employed 6B training questions from the BioASQ challenge task as the question set of our study.
2.2.2. Document Retrieval
The task of this component is to find documents containing the answer for the question. We employed the MEDLINE database as the corpus. Documents in MEDLINE contain miscellaneous information, such as journal names, contents of the title, author, abstract, publication date, chemical codes, Medical Subject Heading (MeSH) terms [31], and MeSH codes. After analyzing these files, we selected the fields for title, abstract, publication date, chemical codes, MeSH terms, and MeSH codes to index with the Apache Lucene search engine. Approximately 26.5 million PubMed articles were indexed as a corpus with the Apache Lucene text search engine for this study. For ranking documents, four models were used: Vector Space Model, Okapi BM25, Query Likelihood with Dirichlet Smoothing, and the Jelinek–Mercer Smoothing Model. The search process consisted of four steps. The system performance was measured according to the results of each separate step in the search process.
Lucene Step: This is the first step in the search process. Similar documents are retrieved by sending a modified query to the lucene search engine.
Stemming Step: This is the second step in the search process. In this step, the modified query is expanded by adding root forms of the modified query words. The new query obtained is sent to the lucene search engine.
Feedback Step: This is the third step in the search process. In this step, the system retrieves the documents from the previous step and receives the five most frequently repeated mesh terms in those documents. The new query is obtained by adding these mesh terms to the modified query. The new query obtained is sent to the lucene search engine.
Re-rank Step: This is the last step in the search process. The 1000 most relevant documents are retrieved from the previous step. These documents are split into trigram units [15]. A trigram unit is generated by the sliding window technique. In this method, the first trigram unit is generated using the first, second, and third sentences. The second trigram unit is generated using the second, third, and fourth sentences. The third trigram unit is generated by using the third, fourth, and fifth sentences, and so on. This method aims to find the most relevant sentences that might include the answer. Two similarity scores are used to obtain the similarity between the question and documents. These are document similarity and trigram unit similarity scores. These similarity scores are calculated as shown in Equation (7).
denotes the similarity score of the trigram unit, and denotes the similarity score of the documents. The weighting parameter has a value between 0 and 1. To give more weight to the trigram unit, α parameter is set as 0.65.
According to results obtained from the re-ranking operation, the top-ranked 20 documents are retrieved and returned as an output of the system. Our proposed document retrieval system architecture is shown in Figure 2.
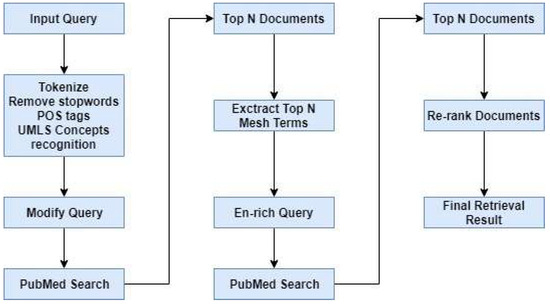
Figure 2.
Document Retrieval System Architecture.
2.2.3. Answer Extraction
The biomedical question answering task of the BioASQ challenge has four types of questions: “yes/no”, “factoid”, “list” and “summary”. The answer extraction module is designed to produce answers only for the factoid and list type questions. In this task, articles containing the answer to the question are provided by the BioASQ challenge organization. We have employed the dataset prepared within the scope of the BioASQ challenge as the training data for our study. The answer extraction architecture is shown in Figure 3.
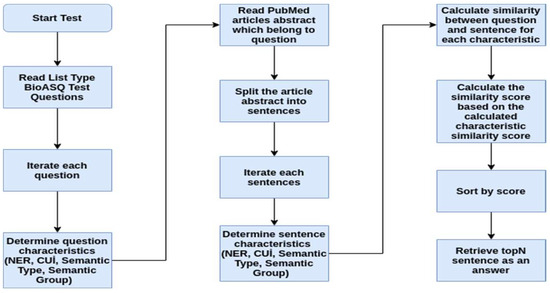
Figure 3.
Answer Extraction Architecture.
Named Entity Recognition (NER), UMLS Concept Unique Identifiers (CUIs), UMLS Semantic Types, and UMLS Semantic Group features have been employed to find sentences that might be the answer. In this way, both text similarity and semantic similarity are included in the score.
Named Entity Recognition (NER) plays a crucial role in information extraction, especially in the biomedical domain. The primary objective of the NER task is to identify and categorize specific chunks of text that refer to entities of interest, such as gene names, protein names, drug names, and disease names. Various NER systems have been developed for biomedical purposes, employing diverse approaches and techniques. These can be broadly categorized into three main types: rule-based, dictionary matching, and machine learning.
Processing biomedical and clinical texts is a crucial domain within natural language processing, and finding robust and practical models can be challenging. We use scispaCy [32], a Python library and set of models for practical biomedical/scientific text processing, which heavily leverages the spaCy [33] library. The scispaCy has four packages: en_ner_{bc5cdr|craft |jnlpba|bionlp13cg}_md with finer-grained NER models trained on BC5CDR (for chemicals and diseases), CRAFT (for cell types, chemicals, proteins, and genes), JNLPBA (for cell lines, cell types, DNAs, RNAs, and proteins) and BioNLP13CG (for cancer genetics), respectively [18]. NER packages are shown in Table 1.

Table 1.
NER packages.
Named Entity Recognition (NER)
Four NER packages are used to identify a specific entity in the sentence. Each result is combined with others to obtain the NER feature group. For example, named entities of the query “Which are the different isoforms of the mammalian Notch receptor?” are shown in Table 2.

Table 2.
Sample NER output.
We compose a new sentence by combining the named entity labels occurring in the question. For example, the sentence combined with the named entities in this question is “SO, TAXON, PROTEIN, GENE_OR_GENE_PRODUCT”. Sentences containing the same type of NER as the question are more likely to contain the answers. Even when there is no textual similarity, when NER is included in the similarity check, it is easier to find the sentence that can be the answer.
UMLS Concept Unique Identifiers (CUI)
One of the main objectives of the UMLS concept is to link diverse terms referring to identical concepts across numerous vocabularies. To include the relevant terms in the similarity calculation, we employed the Metamap tool to determine the conceptual words both in the sentence and the question. For example, the CUI of the question above is shown in Table 3.

Table 3.
Sample CUI output.
Semantic Types and Groups
The UMLS’s Semantic Network provides a consistent categorization of all concepts related to medical studies and establishes a set of valuable relationships among these concepts. The Metamap tool can be used to determine the semantic types of a text. Thus, we used it to determine the semantic types of the sentence. Then, we combined all semantic types of the sentence and obtained its semantic features.
- Semantic Type Sentence: mamm, aapp, qlco, bsoj, aapp, rcpt
- Semantic Group Sentence: LIVB, CHEM, CONC, ANAT, CHEM, CHEM
Similarity Calculation
Finally, we obtained five sentences created with different features of a single sentence. One is the original sentence, whereas the other four are a NER sentence, CUI sentence, Semantic Type sentence, and Semantic Groups sentence. The question above is shown as an example below.
- Query: Which are the different isoforms of the mammalian Notch receptor?
- NER Sentence: SO, TAXON, PROTEIN, GENE_OR_GENE_PRODUCT
- CUI Sentence: C0024660, C0597298, C1705242, C1235660, C0597357
- Semantic Type Sentence: mamm, aapp, qlco, bsoj, aapp, rcpt
- Semantic Group Sentence: LIVB, CHEM, CONC, ANAT, CHEM, CHEM
We used these five sentences in the similarity score calculation. Similarity Score is calculated as shown in Equation (8).
where Simi(q,s) is the ith similarity model among Simsentence(q,s), and Simner(q,s), Simcui(q,s), Simsemtype(q,s), Simsemgroup(q,s). Simsentence(q,s), Simner(q,s), Simcui(q,s), Simsemtype(q,s), Simsemgroup(q,s) denote that similarity between the named entities, UMLS concept unique identifiers (CUI), UMLS semantic types, and UMLS semantic groups of the query and sentence in the article, respectively. Simsentence(q,s) refers to the textual similarity between the question and the individual sentences in the article that contains the answer. Simner(q,s) denotes the similarity between the named entities of the question and the named entities of the individual sentences in the article. Simcui(q,s) denotes the similarity between the UMLS Concept Unique Identifiers (CUI) of the question and the UMLS Concept Unique Identifiers (CUI) of the individual sentences in the article.
We have used two different score calculation models. In the first model, the similarity score is calculated by the textual similarity between the question and the sentence. In the second model, the similarity score is calculated by the textual similarity, named entity similarity, cui similarity, semantic type similarity, and semantic group similarity score. Table 4 and Table 5 show the Top10 and Top20 evaluation results, respectively.

Table 4.
Top10 evaluation results.

Table 5.
Top20 evaluation results.
Calculation of the similarity according to five different features of the query and the sentence has provided an improvement in the performance score of our system.
In the second proposed model, we assigned 0.2 as the weight of each one of the five features. At this point, we concluded that the performance of the system can be increased if the weight values are optimized. Therefore, we used different methods to optimize the weight values.
As the first method, the F1 score of each feature was used in calculating their weight value. Hence, the F1 score of each feature (sentence, ner, cui, semgroup, semtype) was calculated separately. Fifty-seven sample questions were used for this evaluation. Table 6 and Table 7 present the Top 10 and Top 20 evaluation results, respectively.

Table 6.
Top 10 evaluation results of each feature.

Table 7.
Top 20 evaluation results of each feature.
The weight values were calculated by normalizing the weighted averages of the calculated F1 scores to 1. Table 8 and Table 9 show the calculated weight scores.

Table 8.
Top 10 normalized weights of each characteristic.

Table 9.
Top 20 normalized weights of each characteristic.
The final weight score was calculated using the arithmetic mean of the top10 and top20 evaluation weights. The final weight is presented in Table 10.

Table 10.
Weight of each characteristic.
The same method was used as the second weight calculation method. As an exception, two categories with low performances were not included in the calculation. The similarity score was calculated as follows:
where . Simi(q,s) is the ith similarity model among Simsentence(q,s), Simcui(q,s), and Simsemtype(q,s).
The sentence, cui, and semtype weighting scores were calculated as 0.3528, 0.3416, and 0.3056, respectively.
As the third method, we added the NER filter to our system. The NER filter functions so as to compare the question and the sentences in the articles in terms of their named entities. It eliminates sentences not including the same named entities as the question and only allows sentences including the same named entity as the question as candidate answer sentences.
3. Results
3.1. Document Retrieval Component Evaluation
In general, we obtained the following results based on the test results.
- Dirichlet similarity method with expanding query with MESH terms produces the best performance for 100 questions and MAP@20.
- Expanded query with noun/noun phrases increases the performance.
- Expanded query with mesh terms increases the performance.
- Expanded query with stemming terms has a better performance than expanded query with UMLS concept.
- In general, expanded query with the UMLS concept decreases the system performance.
Figure 4 shows the evaluation results of the expanding query with MESH terms.
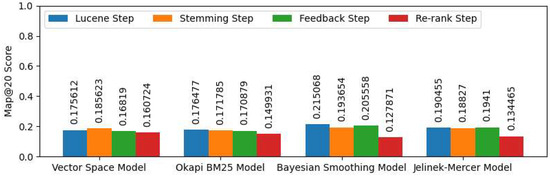
Figure 4.
Expanding query with MESH terms Map@20 Score.

Table 11.
Performance of the Vector Space Model.

Table 12.
Performance of the Okapi BM25 Model.

Table 13.
Performance of the Bayesian Smoothing with Dirichlet Priors Model.

Table 14.
Performance of the Jelinek–Mercer Smoothing Model.
When we compare the results of our study with others which have employed similar classical methods, it is clear that we have achieved a partial improvement in performance. To improve performance, embedding/transformer-based algorithms can be used in the document re-ranking stage. Table 15 shows comparative performance scores of certain studies along with ours ranked according to their MAP scores.

Table 15.
MAP score of document retrieval system.
Table 16 [34] and Table 17 [35] show the names and performance results of the systems participating in the BioASQ challenge held in 2022 and 2023 for test batch 1, respectively. When we compare our MAP score with the systems that have participated in the challenge, it is clear that our score is not very compatible. That is why we propose suggestions to improve our score in the coming section.

Table 16.
2022 Task 10b Document Retrieval Phase Test Batch 1 Result.

Table 17.
2023 Task 10b Document Retrieval Phase Test Batch 1 Result.
3.2. Answer Extraction Evaluation
To test the system’s performance, 101 questions were used. We took the top 10 and top 20 sentences with the highest similarity score as answers for each question. Table 18 and Table 19 present the similarity score evaluation results calculated with different weights.

Table 18.
Top 10 evaluation score.

Table 19.
Top 20 evaluation score.
As can be seen from the results, the performance levels of the methods are close to one another. In these methods, we aimed to find the answer after combining all abstracts of all relevant articles. Alternatively, we applied the same methods on the sentences in each article to score them within themselves, which we call the “One-by-One” technique. After evaluating the sentences in individual articles, we ranked all the sentences in all articles according to their similarity scores. We created an answer set by combining the sentences with the highest scores. The evaluation results of these methods are shown in Table 20.

Table 20.
One-by-One evaluation score.
The One-by-One method has a better performance than the previous one. Performance improvements by top 10, top 20, and all answers (One-by-One method) are approximately 20%, 25%, and 32%, respectively.
4. Discussion
Commonly used ranking algorithms cannot measure words’ semantic similarity or their similarity/closeness to other words. These algorithms work on a dictionary basis. Generally, the similarity score between the query and the documents is calculated based on the number of common words. Although this gives fast and good results in searches based on word similarity, it is insufficient for queries based on comprehension. In our study, we tried to eliminate the disadvantages of ranking algorithms by using information retrieval techniques and textual and semantic similarity. Therefore, we used UMLS services to provide semantically similar concepts to enrich the query. At the same time, we added important words (nouns) to the query and thus increased the effect of these words in similarity calculations, since nouns are more significant and distinctive for similarity calculations. These applied techniques provided a general increase in performance, although it remains below the desired level. The performance did not reach a certain level because the similarity score calculation was carried out using the dictionary-based bag-of-words method. To overcome this problem, it is necessary to use embedding/transformer-based methods that capture the meaning of words in the relevant context, calculate the semantic similarity between words, and can be trained according to the specific domains.
As for the answer extraction module, we obtained five sentences created with different features of the same sentence. One is the original sentence, whereas the other four consist of an NER sentence, CUI sentence, Semantic Type sentence, and Semantic Groups sentence. We have employed two distinct score calculation methodologies. In the initial model, the similarity score is determined based on the textual resemblance between the question and the sentence. In the second model, the similarity score is computed using various factors, including textual similarity, named entity similarity, CUI similarity, semantic type similarity, and semantic group similarity scores. We achieved approximately a 25% performance increase for the top 20 results using the second model compared to the first. To obtain even better results, we need deep learning algorithms that can understand the question and sentences in their context.
Future work can be improved and extended as follows. At the first stage, dictionary-based ranking algorithms (like Okapi BM25) can be used to quickly retrieve a certain number of relevant documents. The similarity score between the documents and the query can be calculated using embedding/transformer-based models at the second stage. For this purpose, the BERT model, which is pre-trained for the general domain, can be trained using PubMed abstracts to learn the biomedical terminology and the similarity between words related to one another in the biomedical domain. Subsequently, fine-tuning may be performed for both document retrieval and answer extraction.
5. Conclusions
We developed a document retrieval system for this study. We tested the Vector Space Model, Okapi BM25, and Query Likelihood with Dirichlet Smoothing models to find the best ranking model for the biomedical document retrieval system. We used a variety of methods, such as adding the root form of the question words, to enrich the query. As a result of the tests, we achieved a better system performance by adding word roots, nouns, noun phrases, and MESH terms to the query terms.
To choose the statements that might be the answer for the question, we calculated the similarity score according to five different features, namely the textual similarity, named entity similarity, cui similarity, semantic type similarity, and semantic group similarity. We propose that the “One-by-One” method evaluates similarities between the query and the sentences within individual articles separately. Then, all sentences across articles are ranked based on their scores. The highest-scoring sentences are then combined to form an answer set, likely to contain the most relevant information from the articles. We achieved an approximately 25% performance increase for the top 20 results using this method, compared to another method we used in this study, which is based on only textual similarity. However, calculating the similarity score according to these five features is a much longer procedure.
Author Contributions
Data Collection, H.B.; methodology, H.B. and B.Ş.; software, H.B.; formal analysis, H.B.; writing—original draft preparation, H.B.; writing—review and editing, B.Ş. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this article and are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Athenikos, S.J.; Han, H. Biomedical question answering: A survey. Comput. Methods Programs Biomed. 2010, 99, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Rinaldi, F.; Dowdall, J.; Schneider, G.; Persidis, A. Answering questions in the genomics domain. In Proceedings of the ACL 2004 Workshop on Question Answering in Restricted Domains, Barcelona, Spain, 25 July 2004; pp. 46–53. [Google Scholar]
- Zweigenbaum, P. Question answering in biomedicine. In Proceedings of the Workshop on Natural Language Processing for Question Answering, Budapest, Hungary, 14 April 2003. [Google Scholar]
- Tsatsaronis, G.; Balikas, G.; Malakasiotis, P.; Partalas, I.; Zschunke, M.; Alvers, M.R.; Weissenborn, D.; Krithara, A.; Petridis, S.; Polychronopoulos, D.; et al. An overview of the BIOASQ large-scale biomedical semantic indexing and question answering competition. BMC Bioinform. 2015, 16, 138. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.-X.; Zhang, B.-W.; Fang, F.; Zhang, L.-L.; Yin, X.-C. A Multi-strategy Query Processing Approach for Biomedical Question Answering: USTB_PRIR at BioASQ 2017 Task 5B. BioNLP 2017, 2017, 373–380. [Google Scholar]
- Mao, Y.; Wei, C.H.; Lu, Z. NCBI at the 2014 BioASQ challenge task: Large-scale biomedical semantic indexing and question answering. CEUR Workshop Proc. 2014, 1180, 1319–1327. [Google Scholar]
- Aronson, R.; Lang, F.-M. An overview of MetaMap: Historical perspective and recent advances. J. Am. Med. Inform. Assoc. 2010, 13, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Neves, M. HPI in-memory-based database system in Task 2b of BioASQ. In Proceedings of the CEUR Workshop Proceedings, Sheffield, UK, 15–18 September 2014. [Google Scholar]
- Yang, Z.; Gupta, N.; Sun, X.; Xu, D.; Zhang, C.; Nyberg, E. Learning to answer biomedical factoid & list questions: OAQA at BioASQ 3B. In Proceedings of the CEUR Workshop Proceedings, Toulouse, France, 8–11 September 2015. [Google Scholar]
- Zhang, Z.J.; Liu, T.T.; Zhang, B.W.; Li, Y.; Zhao, C.H.; Feng, S.H.; Yin, X.C.; Zhou, F. A generic retrieval system for biomedical literatures: USTB at BioASQ2015 Question Answering Task. In Proceedings of the CEUR Workshop Proceedings, Toulouse, France, 8–11 September 2015. [Google Scholar]
- Peng, S.; You, R.; Xie, Z.; Wang, B.; Zhang, Y.; Zhu, S. The Fudan participation in the 2015 BioASQ Challenge: Large-scale biomedical semantic indexing and question answering. In Proceedings of the CEUR Workshop Proceedings, Toulouse, France, 8–11 September 2015. [Google Scholar]
- Yenala, H.; Kamineni, A.; Shrivastava, M.; Chinnakotla, M. IIITH at BioASQ challange 2015 task 3b: Bio-medical question answering system. In Proceedings of the CEUR Workshop Proceedings, Toulouse, France, 8–11 September 2015. [Google Scholar]
- Choi, S.; Choi, J. Classification and retrieval of biomedical literatures: SNUMedinfo at CLEF QA track BioASQ 2014. In Proceedings of the Question Answering Lab at CLEF, Sheffield, UK, 15–18 September 2014; pp. 1283–1295. [Google Scholar]
- Choi, S. SNUMedinfo at CLEF QA track BioASQ 2015. In Proceedings of the CEUR Workshop Proceedings, Toulouse, France, 8–11 September 2015. [Google Scholar]
- Lee, H.-G.; Kim, M.; Kim, H.; Kim, J.; Kwon, S.; Seo, J.; Choi, J.; Kim, Y.-R. KSAnswer: Question-answering System of Kangwon National University and Sogang University in the 2016 BioASQ Challenge. In Proceedings of the Fourth BioASQ Workshop, Berlin, Germany, 12–13 August 2016; pp. 45–49. [Google Scholar]
- Dimitriadis, D.; Tsoumakas, G. Word embeddings and external resources for answer processing in biomedical factoid question answering. J. Biomed. Inform. 2019, 92, 103–118. [Google Scholar] [CrossRef] [PubMed]
- Brokos, G.; Liosis, P.; McDonald, R.; Pappas, D.; Ion, A. AUEB at BioASQ 6: Document and Snippet Retrieval. In Proceedings of the 6th BioASQ Workshop A Challenge on Large-Scale Biomedical Semantic Indexing and Question Answering, Brussels, Belgium, 1 November 2018; pp. 30–39. [Google Scholar]
- Ma, J.; Korotkov, I.; Yang, Y.; Hall, K.B.; McDonald, R.T. Zero-shot Neural Passage Retrieval via Domain-targeted Synthetic Question Generation. arXiv 2020, arXiv:2004.14503. [Google Scholar]
- Pappas, D.; McDonald, R.; Brokos, G.-I.; Androutsopoulos, I. AUEB at BioASQ 7: Document and Snippet Retrieval. In Proceedings of the Seventh BioASQ Workshop: A Challenge on Large-Scale Biomedical Semantic Indexing and Question Answering, Würzburg, Germany, 20 September 2019. [Google Scholar]
- Almeida, T.; Jonker, R.; Poudel, R.; Silva, J.; Matos, S. Two-stage IR with synthetic training and zero-shot answer generation at BioASQ 11. In Proceedings of the CLEF2023: Conference and Labs of the Evaluation Forum, Thessaloniki, Greece, 18–21 September 2023. [Google Scholar]
- Ateia, S.; Kruschwitz, U. Is ChatGPT a Biomedical Expert? In Proceedings of the BioASQWorkshop at CLEF 2023, Thessaloniki, Greece, 18–21 September 2023.
- Rosso-Mateus, A.; Muñoz-Serna, L.A.; Montes-y-Gómez, M.; González, F.A. Deep Metric Learning for Effective Passage Retrieval in the BioASQ Challenge. In Proceedings of the CLEF2023: Conference and Labs of the Evaluation Forum, Thessaloniki, Greece, 18–21 September 2023. [Google Scholar]
- Nentidis, A.; Katsimpras, G.; Krithara, A.; Lima López, S.; Farr, E.; Gasco, L.; Krallinger, M.; Paliouras, G. Overview of bioasq 2023: The eleventh bioasq challenge on large-scale biomedical semantic indexing and question answering. In Proceedings of the International Conference of the Cross-Language Evaluation Forum for European Languages, Thessaloniki, Greece, 8–21 September 2023. [Google Scholar]
- Shin, A.; Jin, Q.; Lu, Z. Multi-stage Literature Retrieval System Trained by PubMed Search Logs for Biomedical Question Answering. In Proceedings of the CLEF2023: Conference and Labs of the Evaluation Forum, Thessaloniki, Greece, 18–21 September 2023. [Google Scholar]
- Salton, G.; Wong, A.; Yang, C.S. A Vector Space Model for Automatic Indexing. Commun. ACM 1975, 18, 613–620. [Google Scholar] [CrossRef]
- Robertson, S.; Jones, K.S. Simple, Proven Approaches to Text Retrieval; University of Cambridge, Computer Laboratory: Cambridge, UK, 1994. [Google Scholar]
- Zhai, C.; Lafferty, J. A study of smoothing methods for language models applied to Ad Hoc information retrieval. In Proceedings of the 24th Annual İnternational ACM SIGIR Conference on Research and Development in İnformation Retrieval—SIGIR ‘01, New Orleans, LA, USA, 9–13 September 2001; pp. 334–342. [Google Scholar]
- Metzler, D.; Croft, W.B. A Markov random field model for term dependencies. In Proceedings of the 28th Annual İnternational ACM SIGIR Conference on Research and Development in İnformation Retrieval SIGIR 05, Salvador, Brazil, 15–19 August 2005; p. 472. [Google Scholar]
- Natural Language Toolkit. Available online: https://www.nltk.org/ (accessed on 13 February 2024).
- Aronson, R. Effective mapping of biomedical text to the UMLS Metathesaurus: The MetaMap program. In Proceedings of the AMIA Symposium, Washington, DC, USA, 3–7 November 2001; p. 17. [Google Scholar]
- Medical Subject Headings. Available online: https://www.nlm.nih.gov/mesh/meshhome.html (accessed on 13 February 2024).
- scispaCy. Available online: https://spacy.io/universe/project/scispacy (accessed on 13 February 2024).
- Industrial-Strength Natural Language Processing. Available online: https://spacy.io/ (accessed on 13 February 2024).
- BioASQ Participants Area Task 10b: Test Results of Phase A. Available online: http://participants-area.bioasq.org/results/10b/phaseA/ (accessed on 13 February 2024).
- BioASQ Participants Area Task 11b: Test Results of Phase A. Available online: http://participants-area.bioasq.org/results/11b/phaseA/ (accessed on 13 February 2024).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).