Abstract
In recent years, genetic studies have brought new insights into psoriasis, a chronic inflammatory disease with multiple determining and favoring factors. Recent advances in the technology of genetic analysis have enabled the discovery of many loci with causal or susceptibility roles and the finding of correlations related to different types of treatment responses. In this study, genomic deoxyribonucleic acid (DNA) was extracted from 2 mL peripheral blood for the evaluation of rs10204525 for Programmed Cell Death 1 (PDCD1) gene and rs550675 for Collagen Type IX Alpha 1 Chain (COL9A1) gene in 45 psoriasis patients and 43 healthy subjects without a personal pathological history of dermatological diseases. All patients were diagnosed by clinical and histopathological examination, and the severity of disease and its impact on quality of life were evaluated by Psoriasis Area and Severity Index (PASI) and Dermatology Life Quality Index (DLQI) scores. Comparisons were made between controls and patients, but also between different clinical types of psoriasis according to disease severity. The rank of C/T alleles for rs550675 (COL9A1 gene) was higher in the patients versus the control group (p = 0.026), while the G/A alleles for rs10204525 (PDCD1 gene) had no differences between the two groups (p = 0.450). Case and control comparisons also showed statistical significance between homozygous CC/TT genotypes (p = 0.039). After subdividing the three types of psoriasis (plaque psoriasis, arthropathic psoriasis and palmoplantar psoriasis) according to disease severity, there were differences between CC/CT genotype (p = 0.0246) and CC/TT (p = 0.007) genotype in patients with plaque psoriasis in favor of severe disease. At the same time, the GA/GG versus AA pattern was significantly higher in patients with plaque psoriasis.
1. Introduction
Psoriasis is a multifactorial condition, in which the immune system is a topic of great interest for research in these times. In addition to the various cytokines involved in the immunopathology of the disease, genetic polymorphisms could be the key to the administration of targeted therapies []. Psoriasis is known to be a disease with the involvement of many genes in which susceptible alleles, along with triggers such as infections, medications, psychological factors, produce various clinical manifestations []. The Online Mendelian Inheritance in Man (IMMO) database currently lists 15 loci that increase psoriasis susceptibility: PSORS1 (6p21.33), PSORS2 (17q25), PSORS3 (4q), PSORS4 (1q21), PSORS5 (3q21), PSORS6 (19p), PSORS7 (1p), PSORS8 (16q), PSORS9 (4q31), PSORS10 (18p11), PSORS11 (5q31-q33), PSORS12 (20q13), PSORS13 (by variations in the TRAF3IP2 gene on 6q21), PSORS 14 (2q14), PSORS15 (by variation in the AP1S3 gene on 2q36) []. PSORS1 is the most studied locus that increases psoriasis susceptibility by 35–50%. It encodes an important histocompatibility complex class I receptor (MHC I) that is involved in the antigen–presentation immune response to CD8+ T lymphocytes, which are found in abundance in psoriatic skin []. Genome-wide association studies (GWAS) have discovered new information about many chronic autoimmune conditions, including psoriasis, by being able to analyze millions of markers in the genome and finding a large number of susceptible or causative loci of psoriasis []. Studies conducted on single nucleotide polymorphism (SNP) to discover genetic differences between cutaneous-only psoriasis and arthropathic psoriasis demonstrated that the presence of glutamic acid at position 45 of the human leukocyte antigen (HLA)-B (carried on HLA-B*27, HLA-B*38 and HLA-B*39) and asparagine or serine at position 97 (carried on the HLA-B*27 and HLA-B*27:05) were associated with arthropathic psoriasis. The presence of asparagine at position 97 within the HLA-B gene, carried by both HLA-B27 and HLA-B27:05 alleles, significantly increases the risk of arthropathic psoriasis. This association remains significant even after performing conditional analysis using HLA-B*27 as a covariate []. Six distinct genes have been associated with pustular psoriasis and related clinical syndromes, some of which have limited manifestations of pustular psoriasis. These genes are responsible for encoding various proteins, including the interleukin (IL)-36 receptor antagonist (IL36RN), caspase recruitment domain-containing protein 14 (CARD14), adapter protein complex 1 subunit sigma 3 (AP1S3), TNFAIP3-interacting protein 1 (TNIP1), serine protease inhibitor gene serpin family A member 3 (SERPINA3), and the interleukin-1 receptor antagonist (IL-1RA) gene (IL1RN) [,]. Genetic studies in psoriasis have provided useful information not only about the risk of developing the disease or a clinical form of disease, but also about the response to treatment. Research focused on pharmacogenetics, pharmacogenomics, and the impact of genetic variations on drug efficacy and adverse reactions could provide valuable insights in this context. Although studies are variable, it appears that genetic variations in the Vitamin D Receptor (VDR) gene might have a link to treatment response involving calcipotriol or other vitamin D analogs among people with psoriasis []. Associations between increased minor allele frequency distribution with better apremilast therapy outcomes were found for SNPs located at IL1β, IL4, IL23R, Tumour Necrosis Factor alpha (TNFα) []. A study in the Greek population revealed that the presence of rs10484554 in the HLA-C gene demonstrated an affiliation with favorable responses to anti-TNF-α agents, but this association was not observed in relation to treatment with Ustekinumab. However, rs151823 and rs26653 in the Endoplasmic Reticulum Aminopeptidase 1 (ERAP1) gene showed connections with positive responses to anti-IL-12/23 therapy []. Another study concluded that people who have at least one SNP rs35569429 removal allele showed an unfavorable response to treatment with Ustekinumab []. This accumulation of information arouses the interest of researchers in deepening genetic studies of various diseases. The homozygous AG genotype and the G allele of rs10204525 increased the risk of developing psoriasis in a study carried out on the PDCD1 gene in the Chinese population and we were, therefore, interested in testing the link of this gene in the European population as well []. The association of the COL9A1 gene with joint diseases sparked our interest to seek a connection of its expression in psoriatic arthritis [,]. In this study, we evaluated the rs10204525 genotypes for the PDCD1 gene and rs550675 for the COL9A1 gene, comparing them between a group of psoriasis patients and a control group. PDCD1 is an activated T cell receptor that inhibits immunity, regulates T-activated cell function, differentiates CD4+ T cells into regulatory T cells; thus, it is involved in autoimmune diseases, antitumor and antimicrobial defense []. COL9A1 gene encodes one of the chains of collagen IX found in skin, bones, cartilage, tendons, and ligaments []. Here, we present a study with a group of 45 patients diagnosed with psoriasis and 43 healthy patients of the same sex and similar age, with a BMI (Body Mass Index) mean of 26.55 for the control group (min = 20.49, max = 34.19) and 29.29 for psoriatic patients (min = 19.96, max = 50) in which polymorphisms of PDCD1 and COL9A1 genes were analyzed.
2. Materials and Methods
In this study, we evaluated forty-five subjects with mild to severe psoriasis who were examined at the University Clinic of Dermatology Timisoara from April 2022 to April 2023. Forty-five subjects with psoriasis vulgaris diagnosed by clinical examination and histopathology results were enrolled in the study. Patients with other forms of psoriasis (pustular or erythrodermic psoriasis), known subjects with malignant tumors or hematopoietic malignancies, patients with other dermatological or nondermatological autoimmune diseases, and other forms of arthropathy except arthropathic psoriasis were excluded. We recruited 26 women with a mean age of 58.84 years and 19 men with a mean age of 56 years. From both of these groups, 28 had clinical forms of plaque psoriasis, 10 also associated palmoplantar elements, and 7 combined the arthropathic form. The severity of the disease and its impact on the patient’s quality of life were assessed by PASI and DLQI scores. The control group consisted of 43 subjects of the same sex and age close to psoriatic patients who had no recent history or clinical signs of dermatological diseases. The study was carried out in accordance with the Helsinki Declaration and approved by the Ethics Committee of the University of Medicine and Pharmacy “Victor Babes” Timisoara (No. 23/28.03.2022) for human studies. Information consent was obtained from all participants in the study.
2.1. DNA Isolation
2 mL of peripheral blood was collected in vacutainers containing ethylenediaminetetraacetic acid (EDTA) from all subjects enrolled in the study. Genomic DNA was extracted from blood samples by using the MagCore® Extractor System and MagCore® Genomic DNA Whole Blood Kit (RBC Bioscience, New Taipei City, Taiwan), following the manufacturer’s protocol. DNA samples were stored at −20 °C.
2.2. Genotyping
Allelic discrimination was done by using the following TaqMan genotyping assays for allelic discrimination: C___2989110_1_ (rs550675) and C____172862_10 (rs10204525). The experiments were run on a QuantStudio 7 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturer protocols. The genotype was determined by measuring the fluorescence of specific alleles using software for allele discrimination (Applied Biosystems, Foster City, CA, USA). For quality control, we repeated the analysis for approximately 5% of the samples, randomly selected. The results showed no discrepancies.
2.3. Statistical Analysis
All data has been collected in Microsoft Excel version 2309 and processed in MedCalc® Statistical Software version 20.216 (MedCalc Software Ltd., Ostend, Belgium; https://www.medcalc.org, accessed on; 20 October 2023). The comparisons between two series of scores were made with non-parametric Mann–Whitney test and for more than two series of scores, with non-parametric Kruskal–Wallis test. The frequency of genotypes and alleles was compared with Chi-squared test. A p-value less than 0.05 indicates signification.
3. Results
rs10204525 for PDCD1 gene and rs550675 for COL9A1 gene were genotyped in 45 psoriasis patients and 43 controls. Two of the patient group blood samples could not be used to determine rs10204525 due to sample quality, being excluded from statistical analysis of this genotype. Based on the inclusion criteria, 26 women and 19 men with psoriasis were recruited. Average PASI score was 14.42 (minimum = 2, maximum = 30) and mean DLQI was 11.62 (minimum = 0, maximum = 30). The minimum PASI score and DLQI belonged to patients on biologic therapy for more than six months. The biologic therapies used to treat patients were as follows: anti TNF-α (Adalimumab for 2 patients with plaque psoriasis, Etanercept for 1 patient with plaque psoriasis), anti IL-12/23 (Ustekinumab for 2 patients with arthropathic psoriasis), anti IL-23 (Guselkumab for 1 patient with plaque psoriasis, Risankizumab for 1 patient with palmoplantar psoriasis) and anti IL-17 (Secukinumab for 1 patient with arthropathic psoriasis). Patients were divided according to psoriatic disease severity, clinical forms of disease, special areas affected, ongoing treatment and associated comorbidities. The patient profile can be found in Table 1.

Table 1.
Characteristics of patients included in the study.
The distribution of the two genes and alleles was according to the Hardy–Weinberg Equilibrium (HWE). For COL9A1 gene variant rs550675, the C/T alleles were evaluated by comparison between the control group and the patient group, but also between the patients, taking into account different parameters. Comparisons between allele frequency and the genotypes between the control group and patient group were performed by Chi-squared test, and the statistical significance is presented by p-value and shown in Table 2.

Table 2.
The two variants investigated in the psoriasis group and control group.
The rank of C/T alleles for rs550675 (COL9A1 gene) has statistically significant values in the psoriatic patient group (p = 0.026), while the G/A alleles for rs10204525 (PDCD1 gene) show no changes between the two groups (p = 0.450). The analysis of rs550675 and rs10204525 is shown in Table 3.

Table 3.
Non-parametric Mann–Whitney test for investigated SNPs.
Statistical analyses were performed between mild and severe psoriasis according to PASI score (profile shown in Table 4), independent of clinical forms of psoriasis, but no statistically significant data were obtained. The Mann–Whitney and Kruskal–Wallis tests were used to compare the scores, and the frequency of alleles and genotypes was compared by applying the Chi-squared test. Frequencies results are presented in Table 5.

Table 4.
Descriptive statistics for PASI and DLQI scores.

Table 5.
PDCD1 and COL9A1 polymorphism in patients with mild or severe psoriasis.
The genotype frequency for rs550675 in COL9A1 gene was 40.0% (CC), 53.3% (CT) and 6.66% (TT) in psoriasis cases and 23.3% (CC), 58.1% (CT), 18.6% (TT) for controls. When comparing CC/TT genotypes between the psoriatic and healthy group, the TT genotype is associated with psoriasis (p = 0.039). The frequency of genotypes for rs10204525 in PDCD1 gene was 77.8% (GG), 15.6% (GA), 2.2% (AA), and 4.4% could not be determined in the patient group; and in the control group, the frequency was 74.4% (GG), 23.3% (GA), 2.3% (AA). Statistical analysis of these genotypes shows that rs10204525 was not associated with psoriasis susceptibility. These data are summarized in Table 6.

Table 6.
Analysis of rs550675 and rs10204525 variants in the patient group and the control group.
Figure 1a,b show genotype percentage distribution of the two genes studied according to disease severity for each type of psoriasis.
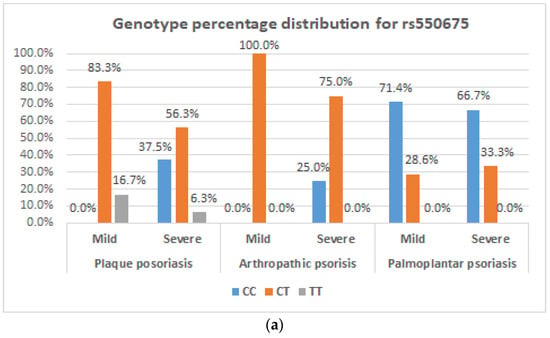
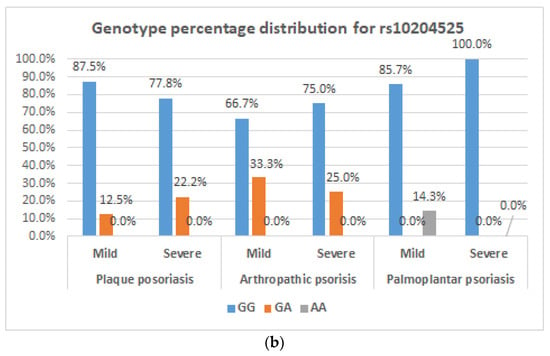
Figure 1.
Genotype distribution for the COL9A1 gene and PDCD1 gene. (a) Frequency of C/T genotypes of the COL9A1 gene in plaque psoriasis, arthropathic psoriasis, palmoplantar psoriasis depending on disease severity; (b) frequency of G/A genotypes of the PDCD1 gene in plaque psoriasis, arthropathic psoriasis, palmoplantar psoriasis depending on disease severity.
Statistical tests (Chi-squared, Kruskal–Wallis) applied to the frequency of genotypes and alleles according to the three forms of psoriasis had no statistical significance. We approached associations not only by frequencies but also by scores (between the three diagnoses), but no differences were observed. Table 7 shows the frequency of genotypes and alleles in different clinical forms of disease.

Table 7.
Genotype and allele frequency for rs10204525 for PDCD1 gene and rs550675 for COL9A1 gene depending on clinical psoriasis form.
Comparative tests were performed between genotypes and alleles of COL9A1 and PDCD1 gene polymorphisms for each type of psoriasis depending on disease severity. The Chi-squared test generated statistically significant values only when comparing CC/CT (p = 0.0245) and CC/TT (p = 0.007) genotypes and C/T allele frequency (p = 0.0154) in plaque psoriasis. The data are summarized in Table 8.

Table 8.
Comparative tests for each type of psoriasis depending on the severity of the disease.
Comparisons based on inheritance models showed that the GA/GG genotypes are more associated with plaque psoriasis than arthropathic psoriasis, and the CT/TT genotypes are more common in arthropathic psoriasis than palmoplantar psoriasis (Table 9).

Table 9.
Statistical analyses according to models of inheritance.
4. Discussion
Psoriasis is known as a heterogamous condition with no genetic homogeneity, which comprises multiple distinct disease phenotypes that are linked to various genetic variations. The prevalence of psoriasis differs, being reported between 0.27% and 11.4% depending on sex, age, geographical area, ethnicity, genetic and environmental factors. The lower prevalence of disease is found in East Asia (0.04% to 0.30%), and it is more common in Australia (0.50% to 5.73%) and Western Europe (0.87% to 2.74%) []. It was observed that the risk of psoriasis is 2–3 times higher in monozygotic twins compared to dizygotic twins, concluding that discordance between familial disease aggregations and lack of disease is based on the multifactorial pathogenesis of psoriasis []. Studies on family aggregations have revealed a high prevalence of the disease in relatives, the risk of relapse being between 4 and 18.75% in first-degree relatives affected by psoriasis []. A study published in 2014 on family aggregation of moderate-to-severe psoriasis showed a family history of disease in 59.37%, with 27.18% having at least one affected parent, 16.56% second-degree relatives and 5.31% third-degree relatives []. Psoriasis is not a disease with 100% genetic determinism, so various favorable factors lead to the onset of the disease or its aggravation. Psoriasis plaques are characterized by chronic inflammation, accelerated keratinocyte proliferation, and exaggerated inflammatory responses generated by a multicellular influx that produces a complex cytokine cascade. Injuries compromise epithelial barriers, leaving them vulnerable to external threats. Thus, the survival of the body depends on the rapid restoration of the barrier after damage. Immunocompromised individuals and immunodeficient animals have profound defects in epithelial repair. During the wound healing process, stem cell offspring are not limited to their niche, but rather migrate to the affected area when tissue damage occurs. Throughout the wound repair process, groups of different cells participate. Given the amplified inflammatory reactions in psoriatic skin, healing of the skin after injury is considered a persistent and aberrant reaction [,,,]. Also, studies have shown genetic links between psoriasis and various comorbidities that were based on immunogenic changes in different genes, alleles and loci []. Recent strides in technology and comprehensive GWAS studies have provided substantial proof of connections between psoriasis and numerous genes situated both within and outside the MHC []. Psoriasis is influenced by a complex interaction between genetic loci and environmental factors. Over 40 SNPs related to psoriasis have been identified through GWAS and targeted approaches, many of which are close to genes linked to adaptive and innate immunity pathways. The MHC class I, especially the HLA-C*06:02 allele, is the main genetic factor contributing to susceptibility to psoriasis. Various other genes associated with innate immunity and signaling pathways were also involved []. SNPs that affect processes such as skin barrier functions (Late Cornified Envelope 3E (LCE3E) and Late Cornified Envelope 3C (LCE3C)), IL-23 signaling (IL-23A, IL-23R, and IL-23) nuclear factor-kB and interferon signaling (Nuclear Factor Of Kappa Light Polypeptide Gene Enhancer In B-Cells Inhibitor, Alpha (NFKBIA), Long Intergenic Non-Protein Coding RNA 1185 (LINC01185), Tyrosine Kinase 2 (TYK2), Potassium Voltage-Gated Channel Subfamily H Member 7 (KCNH7), IL-28RA, Tumor Necrosis Factor Alpha-Induced Protein 3 (TNFAIP3), Annexin A6 (ANXA6)) and IL-17 cell response (REV3 Like, DNA Directed Polymerase Zeta Catalytic Subunit ((REV3L), TYK2, IL-23R) were correlated with psoriasis risk [,].
In our study, we analyzed the link between PDCD1 (rs10204525), COL91A (rs550675) gene polymorphisms and psoriasis from an Eastern European population to bring new information into the complex genetic picture of this disease.
The PDCD1 gene is mapped to 2q.37.3 and encodes a transmembrane glycoprotein (PD-1) which is a receptor with immune-inhibitory properties that gets expressed in activated T cells and participates in the control of T-cell functions, particularly those of effector CD8+ T cells. Additionally, it can facilitate the transformation of CD4+ T cells into T regulatory cells. PDCD1’s presence extends to numerous tumor types, including melanomas, and it has been established to exert an influence on antitumor immune responses. Furthermore, this protein has demonstrated its involvement in preventing autoimmune reactions []. The effect on the immune response of PD-1 is achieved by its ligands (PDL-1 and PDL-2), the former being expressed on various hematopoietic cells, non-hematopoietic cells but also non-immune cells such as endothelial cells, stromal cells, epithelial cells and keratinocytes []. A study of 10 psoriatic patients who were biopsied from skin lesions and immunohistochemically analyzed with specific anti-PD-1 antibodies was performed. The results showed that PD-1 levels increased with inflammation in psoriatic plaques. Also, in this study, PD-1 was determined in experimental mice induced by applying imiquimod 62.5 topically in 5%. Thus, elevated levels of PD-1 compared to the healthy sample were detected in samples collected from regional lymph nodes draining psoriatic skin, and when monoclonal antibody neutralizing against PD-1 was administered, mice with psoriasis showed exacerbation of lesions []. An article published by Dimitra Vodouri et al. in 2017 presented 5 patients with malignant tumor pathologies, 4 of whom had skin psoriasis lesions and one had a close family history of psoriasis. All 5 patients received anti-PD1/PDL1 immunotherapy for antitumor therapeutic purposes and experienced exacerbation of psoriatic skin lesions or their onset []. Six polymorphisms of the PDCD1 gene coding the PD-1 protein were studied in a Chinese population of psoriasis patients, including 128 subjects. Genomic DNA was extracted from peripheral blood and PD1.6 (rs10204525) G allele was observed to increase the risk of psoriasis []. Another study determined Messenger Ribonucleic Acid (mRNA) for the PDCD1 gene in the peripheral blood of 72 psoriasis patients and 35 healthy subjects, showing that PDCD1 expression is significantly lower in psoriatic patients []. Many studies have demonstrated the protective role of PD-1 and its ligands in psoriatic patients, but polymorphisms of the gene encoding this glycoprotein have not yet been extensively studied. In our study, we determined the PD1.6 polymorphism (rs10204525) for the PDCD1 gene and observed a higher frequency of the GG genotype in patients compared to the control group but without statistical significance. Different genetic results could be due to population differences, but also the small batches on which studies have been done for this gene, requiring a larger number of subjects. COL9A1 gene is located on chromosome 6q13 and encodes one of the three proteins of the type IX collagen chain, which is a component of the extracellular matrix []. This protein is predominantly found in hyaline cartilage and the vitreous body of the eye. The gene that encodes it is associated with osteoarthritis, epiphyseal dysplasia, and Stickler syndrome []. Collagen is the main component of the articular cartilage matrix, the collagen network having an important role in joint biomechanics []. Type II collagen is the main component of articular cartilage, but other types of collagens III, VI, IX, X, XI, XII and XIV also contribute to the formation of the mature matrix. The exact roles fulfilled by collagens IX and XI within this complex arrangement are not yet entirely elucidated. However, their significance is unmistakable, given that mutations found within the COL9 and COL11 genes result in chondrodysplasia phenotypes characterized by the premature onset of osteoarthritis []. In our study, we found a significant association of rs550675 (C/T) variant located in COL9A1 with psoriasis, but without being specific to psoriatic arthritis, as we would have expected. However, this could be investigated in the future by following patients over a long period of time and correlated with the development of psoriatic arthritis throughout the disease. The role of gene coding for different types of collagens in psoriasis and psoriatic arthritis has been studied. In a review published in 2020, collagen genes with potential involvement in the pathogenesis of psoriasis and psoriatic arthritis were synthesized, listing gene coding for collagen types I, II, III, V, VI, VII, VIII, IX, X, XI and XVII []. rs12488457 (A/C, COL6A5), rs13081855 (G/T, COL8A1) were associated with a higher risk of psoriasis and arthropathic psoriasis, rs3812111 (COL10A1) was associated with psoriatic arthritis, and the G allele of rs2910164 (C/G, MIR146A) showed high susceptibility to both diseases in a study published by Caputo V. et al. []. The study of epigenetic modification represents a revolutionary perspective for the targeted treatment. Epigenetic changes in COL9A1 have been studied in osteoarthritis, and Lysine-Specific Demethylase-1 (LSD1) is responsible for the negative regulation of the COL9A1 gene by specific chromatin marks in each promoter. The treatment with 5-azadeoxycytidine prevented the hypermethylation of the COL9A1 promoter associated with negative gene regulation [,,]. Studies have been conducted on the regulation of PD-1 expression after various immune stimulation, but epigenetic changes in the coding gene under the influence of drugs have not been widely studied. The epigenetic mechanisms of PDCD1 consist mainly of dynamic changes by DNA methylation after the activation and depletion of T cells and histone changes in gene-regulatory regions during the activation of T cells and induced expression of PD-1, which are related to active gene expression. The changes in PD-1 expression were demonstrated by immune stimulation in viral infections, antitumor therapies (nivolumab/pembrolizumab) or peptide immunotherapy and represent future indications for the development of targeted therapies for autoimmune, inflammatory or antitumor diseases [,,].
5. Limitations
The limitations of the presented paper are the diversity of clinical forms of psoriasis. The participants are of Eastern European origin; therefore, studies on subjects from different populations are needed to establish if the results are found in other populations. Although the control group was chosen with similar characteristics and no dermatological conditions, they may have unknown comorbidities and dermatological diseases that we could not determine at the investigation time so genetic models could be influenced.
6. Conclusions
Understanding the pathogenesis of psoriasis is the path to targeted, effective and safe treatment for the patient. Being a multifactorial disease, new information about the immune response, cytokines involved in the disease, and various genetic factors related to psoriasis help researchers and health practitioners in addressing the disease and patient-focused therapy. Here, we presented the results obtained for genotype rs10204525 of the PDCD gene; in the dominant GA/GG versus AA model, it is positively correlated with plaque psoriasis compared to arthropathic psoriasis in the sample we have studied. In fact, there are significant results in the literature for this gene and the protein encoded by it, as mentioned earlier. The second SNP we studied, encoding the alpha chain of collagen type IX, was statistically significantly correlated with psoriasis, and has induced interest in a further study of the genes that encode components of the extracellular matrix. Thus, we believe that further studies on large and ethnically diverse populations are needed to document this information and track disease progression over a longer period.
Author Contributions
Conceptualization, D.-S.L.-C. and N.I.A.; writing—original draft preparation, D.-S.L.-C. and L.C.P.; data analysis, A.T.; investigation, D.-S.L.-C. and L.C.P.; writing—review and editing, N.I.A. and S.S.F.; supervision, N.I.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Ethics Committee of “Victor Babes” University of Medicine and Pharmacy, Timisoara (Nr. 23/28.03.2022) for studies involving humans [].
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy and ethical reasons.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Prieto-Pérez, R.; Cabaleiro, T.; Daudén, E.; Ochoa, D.; Roman, M.; Abad-Santos, F. Genetics of Psoriasis and Pharmacogenetics of Biological Drugs. Autoimmune Dis. 2013, 2013, 613086. [Google Scholar] [CrossRef]
- Capon, F. The Genetic Basis of Psoriasis. Int. J. Mol. Sci. 2017, 18, 2526. [Google Scholar] [CrossRef]
- The Online Mendelian Inheritance in Man. Available online: https://www.omim.org/entry/177900 (accessed on 17 July 2023).
- Kocaaga, A.; Kocaaga, M. Psoriasis: An Immunogenetic Perspective. Glob. Med. Genet. 2022, 09, 082–089. [Google Scholar] [CrossRef]
- Dand, N.; Mahil, S.K.; Capon, F.; Smith, C.H.; Simpson, M.A.; Barker, J.N. Psoriasis and Genetics. Acta Derm. Venereol. 2020, 100, 55–65. [Google Scholar] [CrossRef]
- Soomro, M.; Hum, R.; Barton, A.; Bowes, J. Genetic Studies Investigating Susceptibility to Psoriatic Arthritis: A Narrative Review. Clin. Ther. 2023, 45, 810–815. [Google Scholar] [CrossRef] [PubMed]
- Uppala, R.; Tsoi, L.C.; Harms, P.W.; Wang, B.; Billi, A.C.; Maverakis, E.; Michelle Kahlenberg, J.; Ward, N.L.; Gudjonsson, J.E. “Autoinflammatory psoriasis”-genetics and biology of pustular psoriasis. Cell Mol. Immunol. 2021, 18, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Aksentijevich, I.; Masters, S.L.; Ferguson, P.J.; Dancey, P.; Frenkel, J.; Van Royen-Kerkhoff, A.; Laxer, R.; Tedgård, U.; Cowen, E.W.; Pham, T.-H.; et al. An autoinflammatory disease with deficiency of the interleukin-1–receptor antagonist. N. Engl. J. Med. 2009, 360, 2426–2437. [Google Scholar] [CrossRef] [PubMed]
- Berna-Rico, E.; Perez-Bootello, J.; de Aragon, C.A.-J.; Gonzalez-Cantero, A. Genetic Influence on Treatment Response in Psoriasis: New Insights into Personalized Medicine. Int. J. Mol. Sci. 2023, 24, 9850. [Google Scholar] [CrossRef]
- Verbenko, D.A.; Karamova, A.E.; Artamonova, O.G.; Deryabin, D.G.; Rakitko, A.; Chernitsov, A.; Krasnenko, A.; Elmuratov, A.; Solomka, V.S.; Kubanov, A.A. Apremilast Pharmacogenomics in Russian Patients with Moderate-to-Severe and Severe Psoriasis. J. Pers. Med. 2020, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Masouri, S.; Stefanaki, I.; Ntritsos, G.; Kypreou, K.P.; Drakaki, E.; Evangelou, E.; Nicolaidou, E.; Stratigos, A.J.; Antoniou, C. A Pharmacogenetic Study of Psoriasis Risk Variants in a Greek Population and Prediction of Responses to Anti-TNF-α and Anti-IL-12/23 Agents. Mol. Diagn. Ther. 2016, 20, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Hua, S.; Fan, B.; Mao, W.; Xu, R.; Wang, Y.; Kuai, L.; Ding, X.; Li, B.; Chen, J.; Miao, X. Association between PDCD1 gene polymorphisms and psoriasis susceptibility in the Chinese population. Int. J. Dermatol. 2021, 60, 1411–1417. [Google Scholar] [CrossRef] [PubMed]
- Spector, T.D.; MacGregor, A.J. Risk factors for osteoarthritis: Genetics. Osteoarthr. Cartil. 2004, 12, 39–44. [Google Scholar] [CrossRef]
- Mustafa, Z.; Chapman, K.; Irven, C.; Carr, A.J.; Clipsham, K.; Chitnavis, J.; Sinsheimer, J.S.; Bloomfield, V.A.; McCartney, M.; Cox, O.; et al. Linkage analysis of candidate genes as susceptibility loci for osteoarthritis--suggestive linkage of COL9A1 to female hip osteoarthritis. Rheumatology 2000, 39, 299–306. [Google Scholar] [CrossRef]
- National Library of Medicine, National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/gene/5133 (accessed on 18 July 2023).
- GeneCards The Humane Gene Database. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=COL9A1#expression (accessed on 18 July 2023).
- Parisi, R.; Iskandar, I.Y.K.; Kontopantelis, E.; Augustin, M.; Griffiths, C.E.M.; Ashcroft, D.M. National, regional, and worldwide epidemiology of psoriasis: Systematic analysis and modelling study. BMJ 2020, 369, m1590. [Google Scholar] [CrossRef] [PubMed]
- Nedoszytko, B.; Szczerkowska-Dobosz, A.; Stawczyk-Macieja, M.; Owczarczyk-Saczonek, A.; Reich, A.; Bartosińska, J.; Batycka-Baran, A.; Czajkowski, R.; Dobrucki, I.; Dobrucki, L.; et al. Pathogenesis of psoriasis in the “omic” era. Part II. Genetic, genomic and epigenetic changes in psoriasis. Postep. Dermatol. Alergol. 2020, 37, 283–298. [Google Scholar] [CrossRef]
- Huang, Y.-H.; Kuo, C.-F.; Huang, L.-H.; Hsieh, M.-Y. Familial Aggregation of Psoriasis and Co-Aggregation of Autoimmune Diseases in Affected Families. J. Clin. Med. 2019, 8, 115. [Google Scholar] [CrossRef]
- Di Lernia, V.; Ficarelli, E.; Lallas, A.; Ricci, C. Familial aggregation of moderate to severe plaque psoriasis. Clin. Exp. Dermatol. 2014, 39, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Nickoloff, B.J.; Bonish, B.K.; Marble, D.J.; Schriedel, K.A.; DiPietro, L.A.; Gordon, K.B.; Lingen, M.W. Lessons Learned from Psoriatic Plaques Concerning Mechanisms of Tissue Repair, Remodeling, and Inflammation. J. Investig. Dermatol. Symp. Proc. 2006, 11, 16–29. [Google Scholar] [CrossRef]
- Han, J.; Lin, K.; Choo, H.; Chen, Y.; Zhang, X.; Xu, R.-H.; Wang, X.; Wu, Y. Distinct bulge stem cell populations maintain the pilosebaceous unit in a β-catenin-dependent manner. iScience 2023, 26, 105805. [Google Scholar] [CrossRef]
- Chen, H.; Wang, X.; Chen, Y.; Han, J.; Kong, D.; Zhu, M.; Fu, X.; Wu, Y. Pten loss in Lgr5+ hair follicle stem cells promotes SCC development. Theranostics. 2019, 9, 8321–8331. [Google Scholar] [CrossRef]
- Chen, H.; Wang, X.; Han, J.; Fan, Z.; Sadia, S.; Zhang, R.; Guo, Y.; Jiang, Y.; Wu, Y. AKT and its related molecular feature in aged mice skin. PLoS ONE 2017, 12, e0178969. [Google Scholar] [CrossRef] [PubMed]
- Merve, H.M.; Sevilay, K.; Sibel, O.; Başak, B.; Ceren, C.G.; Demirci, T.; Cüneyt, A. Psoriasis and Genetics. In An Interdisciplinary Approach to Psoriasis; InTech: London, UK, 2017. [Google Scholar]
- Puig, L.; Julià, A.; Marsal, S. The Pathogenesis and Genetics of Psoriasis. Actas Dermo-Sifiliográficas (Engl. Ed.) 2014, 105, 535–545. [Google Scholar] [CrossRef]
- Ray-Jones, H.; Eyre, S.; Barton, A.; Warren, R.B. One SNP at a Time: Moving beyond GWAS in Psoriasis. J. Investig. Dermatol. 2016, 136, 567–573. [Google Scholar] [CrossRef] [PubMed]
- Villarreal-Martínez, A.; Gallardo-Blanco, H.; Cerda-Flores, R.; Torres-Muñoz, I.; Gómez-Flores, M.; Salas-Alanís, J.; Ocampo-Candiani, J.; Martínez-Garza, L. Candidate gene polymorphisms and risk of psoriasis: A pilot study. Exp. Ther. Med. 2016, 11, 1217–1222. [Google Scholar] [CrossRef][Green Version]
- Ran, D.; Cai, M.; Zhang, X. Genetics of psoriasis: A basis for precision medicine. Precis. Clin. Med. 2019, 2, 120–130. [Google Scholar] [CrossRef]
- Adamczyk, M.; Krasowska, D. PD1/PD-L1 pathway in psoriasis and psoriatic arthritis: A review. Postep. Dermatol. Alergol. 2021, 38, 925–930. [Google Scholar] [CrossRef]
- Peng, S.; Cao, M.; Sun, X.; Zhou, Y.; Chen, C.; Ma, T.; Li, H.; Li, B.; Zhu, B.; Li, X. Recombinant programmed cell death 1 inhibits psoriatic inflammation in imiquimod-treated mice. Int. J. Mol. Med. 2020, 46, 869–879. [Google Scholar] [CrossRef] [PubMed]
- Voudouri, D.; Nikolaou, V.; Laschos, K.; Charpidou, A.; Soupos, N.; Triantafyllopoulou, I.; Panoutsopoulou, I.; Aravantinos, G.; Syrigos, K.; Stratigos, A. Anti-PD1/PDL1 induced psoriasis. Curr. Probl. Cancer 2017, 41, 407–412. [Google Scholar] [CrossRef]
- Bartosińska, J.; Zakrzewska, E.; Raczkiewicz, D.; Purkot, J.; Michalak-Stoma, A.; Kowal, M.; Krasowska, D.; Chodorowska, G.; Giannopoulos, K. Suppressed Programmed Death 1 Expression on CD4+ and CD8+ T Cells in Psoriatic Patients. Mediat. Inflamm. 2017, 2017, 5385102. [Google Scholar] [CrossRef]
- GeneCards The Humane Gene Database. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=COL9A1#aliases_descriptions (accessed on 7 August 2023).
- National Library of Medicine, National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/gene/1297 (accessed on 7 August 2023).
- Responte, D.J.; Natoli, R.M.; Athanasiou, K.A. Collagens of Articular Cartilage: Structure, Function, and Importance in Tissue Engineering. Crit. Rev. Biomed. Eng. 2007, 35, 363–411. [Google Scholar] [CrossRef]
- Eyre, D. Articular cartilage and changes in Arthritis: Collagen of articular cartilage. Arthritis Res. Ther. 2001, 4, 30–35. [Google Scholar] [CrossRef][Green Version]
- Caputo, V.; Strafella, C.; Termine, A.; Dattola, A.; Mazzilli, S.; Lanna, C.; Cosio, T.; Campione, E.; Novelli, G.; Giardina, E.; et al. Overview of the molecular determinants contributing to the expression of Psoriasis and Psoriatic Arthritis phenotypes. J. Cell. Mol. Med. 2020, 24, 13554–13563. [Google Scholar] [CrossRef]
- Caputo, V.; Strafella, C.; Termine, A.; Campione, E.; Bianchi, L.; Novelli, G.; Giardina, E.; Cascella, R. RNAseq-Based Prioritization Revealed COL6A5, COL8A1, COL10A1 and MIR146A as Common and Differential Susceptibility Biomarkers for Psoriasis and Psoriatic Arthritis: Confirmation from Genotyping Analysis of 1417 Italian Subjects. Int J Mol Sci. 2020, 21, 2740. [Google Scholar] [CrossRef]
- Durand, A.L.; Dufour, A.; Aubert-Foucher, E.; Oger-Desfeux, C.; Pasdeloup, M.; Lustig, S.; Servien, E.; Vaz, G.; Perrier-Groult, E.; Mallein-Gerin, F.; et al. The Lysine Specific Demethylase-1 Negatively Regulates the COL9A1 Gene in Human Articular Chondrocytes. Int. J. Mol. Sci. 2020, 21, 6322. [Google Scholar] [CrossRef]
- Barter, M.J.; Bui, C.M.; Young, D.A. Epigenetic mechanisms in cartilage and osteoarthritis: DNA methylation, histone modifications and microRNAs. Osteoarthr. Cartil. 2012, 20, 339–349. [Google Scholar] [CrossRef]
- Imagawa, K.; de Andrés, M.C.; Hashimoto, K.; Itoi, E.; Otero, M.; Roach, H.I.; Goldring, M.B.; Oreffo, R.O.C. Association of Reduced Type IX Collagen Gene Expression in Human Osteoarthritic Chondrocytes With Epigenetic Silencing by DNA Hypermethylation. Arthritis Rheumatol. 2014, 66, 3040–3051. [Google Scholar] [CrossRef]
- Bally, A.P.; Austin, J.W.; Boss, J.M. Genetic and Epigenetic Regulation of PD-1 Expression. J. Immunol. 2016, 196, 2431–2437. [Google Scholar] [CrossRef]
- McPherson, R.C.; Konkel, J.E.; Prendergast, C.T.; Thomson, J.P.; Ottaviano, R.; Leech, M.D.; Kay, O.; Zandee, S.E.; Sweenie, C.H.; Wraith, D.C.; et al. Epigenetic modification of the PD-1 (Pdcd1) promoter in effector CD4+ T cells tolerized by peptide immunotherapy. eLife 2014, 3, e03416. [Google Scholar] [CrossRef]
- Liang, Y.; Turcan, S. Epigenetic Drugs and Their Immune Modulating Potential in Cancers. Biomedicines 2022, 10, 211. [Google Scholar] [CrossRef]
- Lupea-Chilom, D.-S.; Solovan, C.S.; Farcas, S.S.; Gogulescu, A.; Andreescu, N.I. Latent Tuberculosis in Psoriasis Patients on Biologic Therapies: Real-World Data from a Care Center in Romania. Medicina 2023, 59, 1015. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).