Abstract
The term artificial intelligence (AI) was coined in the 1950s and it has successfully made its way into different fields of medicine. Forensic sciences and AI are increasingly intersecting fields that hold tremendous potential for solving complex criminal investigations. Considering the great evolution in the technologies applied to forensic genetics, this literature review aims to explore the existing body of research that investigates the application of AI in the field of forensic genetics. Scopus and Web of Science were searched: after an accurate evaluation, 12 articles were included in the present systematic review. The application of AI in the field of forensic genetics has predominantly focused on two aspects. Firstly, several studies have investigated the use of AI in haplogroup analysis to enhance and expedite the classification process of DNA samples. Secondly, other research groups have utilized AI to analyze short tandem repeat (STR) profiles, thereby minimizing the risk of misinterpretation. While AI has proven to be highly useful in forensic genetics, further improvements are needed before using these applications in real cases. The main challenge lies in the communication gap between forensic experts: as AI continues to advance, the collaboration between forensic sciences and AI presents immense potential for transforming investigative practices, enabling quicker and more precise case resolutions.
1. Introduction
Although the term artificial intelligence (AI) was coined in the 1950s by McCarthy et al. [1], other important concepts, such as neural networks, were defined in the 1940s [2]. Based on the definition of McCarthy, AI represents the science and engineering of making intelligent machines, especially intelligent computer programs [2]. The most significant developments in the last sixty years have been in search and machine learning (ML) algorithms, as well as in the integration of statistical analysis, and applying this knowledge in covering different important aspects of healthcare [3]. Forensic sciences and AI are increasingly intersecting fields that hold tremendous potential in solving complex criminal investigations [4,5,6]. AI technologies, such as ML and computer vision, have improved how forensic evidence is analyzed and interpreted. AI is the study of creating computer systems that are capable of activities like pattern recognition, problem-solving, and decision-making that normally require human intellect [7]. In the last decade, there has been a surge of interest surrounding the domain of AI due to the availability of various open-source module packages such as TensorFlow, Keras, and PyTorch, as well as commercial software such as PLS_Toolbox and Solo 9.0 [8].
Most of the research investigating the role of AI in forensic sciences has focused on areas such as sex and age estimation [9,10,11,12,13,14,15,16,17,18], physical attributes, and in forensic odontology and anthropology for human identification purposes [12,19,20,21,22,23,24,25,26]. Furthermore, AI could be very helpful in the evaluation of DNA methylation-based age prediction. Specifically, Thong et al. showed that an artificial neural network (ANN) had a higher accuracy in age prediction than a regression model, even though regression models are commonly used to estimate age [27]. Other experimental papers have explored the challenging field of definition of the cause of death: in rare cases, it can be difficult to define the manner of death and AI could be helpful. For example, in cases of drowning, several research papers have focused on the application of AI in automating diatom testing, with the aim of achieving higher levels of efficiency and accuracy in the definition of the exact cause of death [11,28,29,30,31,32,33]. Other authors have tried to apply AI in the evaluation of firearm lesions, demonstrating that digitizing and analyzing the fired projectile specimens could be used for firearm identification [34]. At the same time, AI systems could be applied in order to identify gun-shot entrance/exit wounds [35]. Several applications have been used to perform biomechanical studies on patterns of cranial bone fracture [36]. Furthermore, other papers have focused on the application of AI for the definition of the post-mortem interval (PMI) [37]. Finally, in medico-legal sciences, AI could play a pivotal role in the management of medical liability [38,39].
The integration of AI algorithms and forensic genetics has the potential to enhance the accuracy, efficiency, and reliability of forensic investigations, ultimately leading to better outcomes in solving crimes and identifying perpetrators [40]. The seminal paper that focused on the application of AI to forensic genetics was published in 2008 [41]. In that article, the authors discussed the application of multiple independent ML methods to develop formal classification functions, resulting in an integrated high-throughput analysis system for cost-effective and accurate classification of large numbers of samples into haplogroups. Moreover, AI tools are applied to a wide range of genetic and genomic analyses, such as the creation of genomic annotations, the identification of functional genomic components, and the comprehension of gene expression mechanisms [42].
Forensic science is defined as multidisciplinary, considering the wide range of disciplines integrated into this field. For this reason, to solve a crime, it could be necessary to analyze different biological data. In this context, the intricacy and richness of biological data (DNA, RNA, protein, and epigenetic markers), as well as the differentiation between human and non-human evidence, make this endeavor difficult [43]. Complex analyses are needed for the processing of generated data, and these tasks cannot be completed without computational support [44]. Current applications of ML in forensic biology can be divided into three categories: applications related to human identification, applications related to forensic intelligence, and applications related to increasing the evidential value of DNA evidence [40].
Forensic anthropology, forensic odontology, forensic pathology, forensic genetics, and other forensic fields are among the five subsections into which one group of authors separated their findings [4]. The keywords they used to analyze the implications between AI and forensic genetics were, however, too limited, as we discovered after closely examining their use of the following terms: “artificial intelligence”, “deep learning”, “forensic”, “medicolegal”, “forensic anthropology”, “forensic odontology”, “forensic pathology”, “forensic genetics”, “forensic radiology”, “forensic medicine”, “forensic entomology”, “ballistics”, “traffic medicine”, “death”, “postmortem interval”, “DNA”, and “ethics”.
Moreover, considering the great evolution in the technologies applied to forensic genetics, this literature review aims to explore the existing body of research that investigates the application of AI in the field of forensic genetics. Analyzing the published articles, the goal of this review is to provide a comprehensive overview and potential future directions in this interdisciplinary domain.
2. Materials and Methods
A systematic review was conducted according to the PRISMA guidelines [45].
Scopus and WOS databases were used as the search engines from 1 January 1980 to 3 December 2023. The following keywords were used: (artificial intelligence) AND (Forensic genetics)—21; (artificial intelligence) AND (DNA mixture profiling)—10; (artificial intelligence) AND (forensic biological evidence)—7; (artificial intelligence) AND (forensic human identification)—98; (artificial intelligence) AND (NGS forensic analysis)—1; (artificial intelligence) AND (forensic genetics laboratory)—4; (artificial intelligence) AND (massively parallel sequencing)—11; (artificial intelligence) AND (forensic DNA typing)—2; (artificial intelligence) AND (Y AND X chromosome STR)—1; (artificial intelligence) AND (STR typing)—1; (artificial intelligence) AND (STRs)—5.
2.1. Inclusion and Exclusion Criteria
The following exclusion criteria were used: articles not in English (12), conference papers (25), reviews (20), book chapters (3), books (2), conference reviews (2), short surveys (2), editorials (1), and notes (1). The inclusion criteria were as follows: original articles, articles in English.
2.2. Quality Assessment and Data Extraction
F.S first assessed all the articles by evaluating their titles, abstracts, and the entire text. Subsequently, M.S. conducted an independent reanalysis of the selected articles. If there were any conflicting opinions among the articles, they were referred to C.P. for further evaluation.
2.3. Characteristics of Eligible Studies
Out of a pool of 161 articles, 16 duplicates were eliminated, and 68 studies were excluded because of specific criteria. The remaining 77 articles (reported in the Supplementary Materials Table) were first evaluated based on the abstract content: the 36 articles marked in red in the Supplementary Materials Table were removed after abstract evaluation. Forty-one full-text papers were further analyzed, excluding 29 articles (marked in yellow in the Supplementary Materials Table) and including 12 articles (marked in green in the Supplementary Materials Table). Finally, the systematic review included a total of 12 articles (Figure 1).
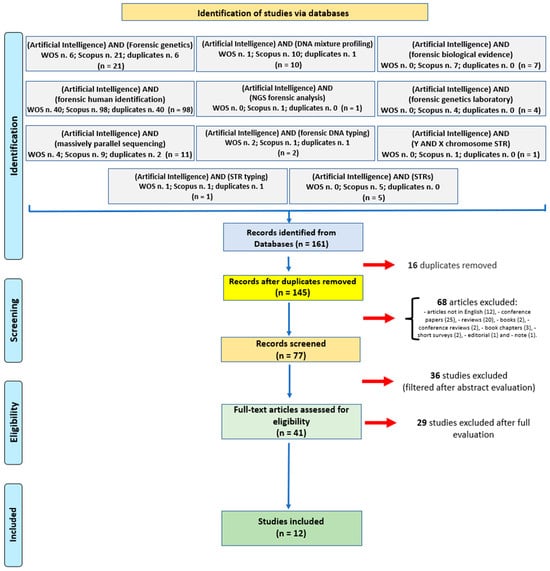
Figure 1.
Flow diagram illustrating included and excluded studies in this systematic review.
3. Results
According to the assessment of the first author, the selected articles were contributed by research groups from various countries including the United States (3), Australia (2), China (2), India (1), Netherlands (1), Poland (1), Portugal (1), and the United Kingdom (1). When examining the distribution of articles based on the year of publication, the earliest relevant paper to our research topic was published in 2008. Notably, a significant number of studies were conducted in the last three years. This review included publications in 2008 (1), 2011 (1), 2014 (1), 2016 (1), 2018 (1), 2019 (1), 2020 (1), 2021 (2), 2022 (1), and 2023 (2).
The application of AI in the field of forensic genetics has predominantly focused on two aspects. Firstly, several studies have investigated the use of AI in haplogroup analysis to enhance and expedite the classification process of DNA samples. Secondly, other research groups have used AI to analyze STR profiles, thereby minimizing the risk of misinterpretation. Information about each publication (first author, year, and nationality), the AI application used in the study, and the main findings are reported in Table 1.

Table 1.
The AI applications used in each included study, and the main findings.
In the field of AI, researchers have harnessed its power to improve various aspects of forensic genetics, including haplogroup classification. Haplogroups are groups of individuals who share a common ancestor through their maternal or paternal lineage. These groups can be distinguished by specific variations in their mitochondrial DNA (mtDNA) or on the Y chromosome in males. Forensic geneticists often use haplogroup classification to determine the ancestral origins of unidentified remains or individuals in criminal investigations. By understanding a person’s haplogroup, it becomes easier to narrow down potential geographic origins and genetic relationships. AI can significantly enhance the efficiency and accuracy of haplogroup classification in forensic genetics: using the traditional approach, haplogroup classification required experts to manually analyze genetic data, which was a time-consuming and labor-intensive process. AI algorithms can automate this classification process, greatly reducing the time and effort required. Furthermore, AI systems can learn from large datasets and adapt to new information [41,46,51,55,57].
Analyzing the application of AI in the interpretation of STRs, these tools could be very useful in the creation of automated DNA profile interpretation systems. These systems use ML algorithms to analyze DNA profiles generated from STR typing, analyzing a vast amount of data and identifying patterns that may not be easily recognizable by human examiners. In this way, it is possible to reduce the risk of human error, enhancing the speed and accuracy of DNA matching in forensic genetics. By comparing an individual’s DNA profile to a massive DNA database, AI algorithms can quickly identify potential matches. This process, known as DNA fingerprinting, has been instrumental in solving cold cases and exonerating wrongfully convicted individuals [47,48,50,56].
Although AI has brought several benefits to STR interpretation in forensic genetics, challenges still exist. Ensuring the transparency and interpretability of AI models is crucial to gaining the trust of forensic examiners and legal professionals. Additionally, the ethical implications of AI in forensic genetics, such as privacy concerns and potential misuse of technology, must be carefully addressed. The ethical issues and challenges of using AI in forensic science focused on criminal cases: there is a need for a comprehensive definition of AI systems, accountability, and legal procedures, emphasizing the application of these tools under human control [58].
4. Discussion
AI techniques, such as ML, data mining, and pattern recognition, can be effectively employed to analyze genetic data and facilitate the identification of suspects. These technologies enable forensic experts to process and obtain valuable results from vast amounts of data, including fingerprints, DNA samples, and surveillance footage, facilitating faster and more accurate identification of suspects [40]. Moreover, AI-driven facial recognition systems assist in identifying criminals from images or videos, assisting law enforcement agencies in solving crimes and bringing justice to victims [40]. There are limitations and potential biases that may arise when utilizing AI algorithms in forensic genetics, and, as such, emphasis must be placed on the importance of transparency, interpretability, and fairness in algorithmic decision making [59]. To reduce AI bias, several strategies could be applied, such as robust data augmentation for each algorithm used, the application of counterfactual fairness, and the imperative for diverse, representative datasets alongside unbiased data collection methods [60]. Moreover, each tool must report its known limitations. As previously described, an important step is represented by the definition of ML algorithms. In this sense, ANNs are networks that, by employing a condensed set of ideas from biological neural systems, mimic a biological neural network. ANNs simulate the electrical activity of the brain and nervous system: there are neurodes arranged as a layer or vector: each output represents an input for the following layers. To date, in the forensic context, ANN applications have been useful in criminal detection through the comparison of faces [61]. Another algorithm is the Random Forest which operates by constructing a multitude of decision trees based on samples, their variables, and their class. An example of its application is the age estimation of bloodstains based on temporal colorimetric analysis. The SVM is an ML algorithm that determines boundaries between data points based on predefined classes, labels, or outputs [62]. It uses supervised learning models to solve complex problems related to classification, regression, and outlier detection. In forensic sciences, it is frequently used in anthropology studies [63]. Another algorithm that could be used for ML is XGBoost; it belongs to the ensemble learning category, specifically the gradient boosting framework, using decision trees as base learners and employing regularization techniques to enhance model generalization. This algorithm could be used in a forensic context to evaluate biogeographic ancestry [56].
A fundamental step is the evaluation of the algorithm used for the ML. For example, the classification accuracy represents the ratio of the total number of input samples to the number of accurate predictions; it is used to evaluate the accuracy. Another method used for algorithm evaluation is the Area Under Curve (AUC). It is used for binary classification problems: the likelihood that a randomly selected positive example will be ranked higher than a randomly selected negative example is known as the AUC of a classifier. The F1 Score represents the harmonic mean of memory and accuracy. The F1 Score is used to provide two key performance indicators and it has a range of (0, 1). The indicators are the number of examples that the classifier properly classifies (its precision) and the number of instances that it does not miss (its robustness) [64,65].
Different genomics and genetics issues have been addressed by ML techniques, such as functional gene annotations, pattern recognition in DNA sequences, and annotation of genomic sequence elements. Histone modification, transcription factor (TF) binding ChIP-seq data, chromatin accessibility assays, microarray or RNA-seq expression data, and other genomic assay data can all be fed into ML. Gene expression data can be utilized to find potentially useful illness biomarkers and differentiate between various disease characteristics. It may be possible to identify new types of functional elements by using chromatin data to annotate the genome in an “unsupervised” manner [40].
In order to comprehend the mechanics underlying gene expression, ML techniques have also been applied. While some methods take into account TF binding profiles at the gene promoter region or ChIP-seq histone modification, others predict gene expression only based on the DNA sequence. By developing a network model, more advanced techniques try to jointly model the expression of every gene in a cell [66].
ML researchers have a history of concentrating on a subset of statistical problems and placing a strong emphasis on the analysis of big, diverse data sets. The ML literature generally lacks basic statistical notions like power calculations, statistical confidence estimation, and likelihood estimate calibration [67].
Due to their complexity and diversity, the detection of genetic markers employed for forensic DNA typing necessitates a substantial amount of data, which may make manual implementation challenging [68]. In addition, handling such data, interpreting it, and comparing it by hand is difficult, labor-intensive, and prone to inaccuracy, despite it occurring rarely [69]. Therefore, analyzing and interpreting such a large amount of data may be made easier by artificial intelligence, which has access to bioinformatic techniques, mathematical algorithms, and statistical computations. Based on the data obtained through this literature review, to date the application of AI in the field of the forensic genetics is limited to two main fields: haplogroup classification and STR electropherogram interpretation.
From a practical standpoint, AI can expedite the forensic investigation process, helping forensic geneticists and law enforcement agencies identify human remains or unknown individuals more efficiently. This is particularly relevant in cases involving missing persons or mass disasters, where the identification process can be challenging, considering the great amount of data that could be handled. The application of AI algorithms in haplogroup classification can streamline and improve the accuracy of this process, benefiting forensic investigations and scientific research alike. With further advancements and continued collaboration between these fields, AI can revolutionize the way forensic genetics and haplogroup classification are conducted, enabling faster and more accurate analyses of genetic data [5].
In the same manner, AI has transformed the field of forensic genetics, especially in the interpretation of STRs. The interpretation of DNA profiling is related to the peak height intensities expressed in relative fluorescence units (RFU), which are shown on an electropherogram (EPG), a graph of fluorescence vs. time. Allelic peak heights for a perfect PCR process are correlated with the amounts that come from the contributing individuals. These peak heights, however, are random and will differ between PCR procedures, particularly in cases where there is little DNA or when there is degradation or allelic drop-out—a result of low template and/or damaged DNA that leads to incomplete DNA profiles, stutter, or artifacts [70,71]. Due to these factors, forensic genetic analyses may complicate the manual interpretation of mixed DNA profiles, and STRs in particular may be amplified and then sequenced [72]. These restrictions are removed and the full potential of the DNA typing data is realized by statistical software packages that integrate probabilistic interpretation models [73,74]. Fully continuous DNA mixture interpretation software, such as STRmixTM, has been extensively tested and used in forensic DNA labs to analyze STR data [75]. STRmixTM determines the probability of the profile given all potential genotype combinations by utilizing quantitative data from an EPG, such as peak heights. We give the normalized probability density a number, or weight. The likelihood of the EPG given each potential genotype combination at a locus is given a relative weight by STRmixTM [70]. Therefore, using probabilistic genotyping software facilitates the control of imported trace samples, particularly those with damaged DNA [75,76]. It also enables the comparison of the presence or absence of alleles in the reference profile and the trace sample directly. Additionally, the availability of software tools reduces analysis time and human weariness [77]. Prior to interpretation, the electropherogram must be inspected to confirm if the profile is indicative of a DNA component or an artifact. There are two ways to go about this phase. When done by hand, it takes a while and can produce different results depending on the analyst reading the layout. To get around this restriction, an analytical threshold—below which the data are discarded—or a double reading method by two separate analysts is used. However, these methods may squander a significant amount of genetic material and require an unnecessary amount of analytical time. However, sophisticated EPG scanning systems like GeneMarker, OSIRIS, and Genemapper may be able to automatically eliminate artifacts. Even with this capability, a lot of artifacts still need to be manually removed before the EPG can be utilized for criminal investigations, which prolongs the reading process. These days, ANNs could be used to create an expert EPG reading system that can discriminate between alleles and artifact, reducing the amount of manual control that is required [48].
Moreover, the application of the latest techniques in forensic DNA profiling that have been established in the last few years employing MPS, also known as NGS, is another crucial question regarding the mixed profile [78]. These techniques can simultaneously identify SNPs, STRs, and in/del (insertions and deletions). Using spatially separated and clonally amplified DNA templates, MPS is related to a range of high-speed sequencing platforms (sometimes referred to as “second generation” or “next generation”) that share a common technical approach to sequence many fragments in parallel [79]. These methods include multiple steps, including library construction, enrichment, and bridge amplification or emulsion PCR. STRs may be amplified and then sequenced; this method allows nucleotide variations found in the flanking regions and repeat motifs of interest to be analyzed. The data produced by these new methods are compatible with previously archived data [80]. Moving forward with MPS will allow the forensic community to continue using millions of DNA profiles that are currently stored in DNA databases, while also expanding its use of forensic DNA typing with increased genetic diversity in STR and SNP. Continued advances in AI are likely to play a crucial role in solving crimes and promoting justice in the future [40,81].
Great progress has been made in other fields of forensic sciences such as forensic odontology. One of the first applications was reported by Chen et al. [19], who described a dental biometric system for human identification using dental radiographs. The described system is based on two main stages: feature extraction and matching. Tooth correspondences between postmortem and antemortem radiographs are established, and a distance based on corresponding teeth measures similarity. The potential applications of artificial intelligence technology in forensic odontology can be categorized into four areas: analysis of human bite marks, identification of sex determination, age estimation, and dental comparison [82]. For example, considering the international migration scenario that is considered a present concern worldwide, determining a minor’s age represents an important goal for upholding the rights of all minors [83]. Methods should be replicable, accurate, and objective: in this regard, the use of new AI tools is necessary to guarantee a trustworthy procedure.
Patil and Ingle [84] analyzed the possibility of applying an algorithm to fingerprint patterns, which are determined by a combination of genetic and environmental factors, to determine gender and age group, and in predicting blood group and classifying common clinical diseases that have a genetic basis, including hypertension, type-2 diabetes, and arthritis. After all, electronic fingerprint recognition has been commercially applied with identification purposes and could be translated to the forensic field [85]. Indeed, to date, the use of fingerprints for identification purposes in the forensic field is one of the more affordable methods, even though recognizing fragmented fingerprints is a difficulty for forensic experts; in this way, the development of AI represents an important tool [86]. Moreover, in a recent study the main goal of the authors was to calculate the sexual dimorphism in unfused or disarticulated hyoids, using ML methods [87].
AI and forensic genetics have emerged as two rapidly evolving fields that are revolutionizing the way investigations are conducted in various domains, especially in the field of criminal justice. Considering the paucity of research, to improve the AI application in forensic genetics, there is a need for interdisciplinary collaboration between AI researchers, forensic scientists, and legal experts to ensure the responsible and ethical implementation of these technologies in the criminal justice system. AI may improve the accuracy of DNA profiling, predicting physical traits and ancestry from DNA samples, and identifying potential familial relationships among individuals [88]. Interestingly, there is a possibility of exploring the use of AI in data interpretation in the field of DNA transfer. DNA transfer in forensic investigation is a crucial process: the sensitivity and specificity of DNA analysis techniques have greatly improved over the years, allowing investigators to detect and analyze even the smallest traces of DNA that could be transferred in a direct/indirect manner [89,90,91,92,93]. The ability to transfer DNA from one surface to another unintentionally, referred to as secondary or indirect transfer, can occur through a variety of mechanisms, such as direct contact or environmental factors [94,95]. Forensic investigators must carefully consider the context of DNA findings, evaluating the likelihood of direct or indirect transfer, and cross-checking various pieces of evidence to ensure accuracy in their conclusions [96]. Of particular interest is indirect transfer that occurs when DNA is transferred between individuals or objects indirectly, often through third-party or environmental factors. One common example is the transfer of DNA from an object to a person and then onto another object [97,98,99]. Factors such as the duration of contact, intensity of contact, and the nature of the surface can affect the presence and persistence of DNA [100,101]. For instance, if an individual handles a weapon that is later used in a crime, their DNA may be transferred from the weapon onto the perpetrator, providing a link between the suspect and the crime scene. DNA can be carried through the air by particles such as dust, dander, or skin cells, settling onto surfaces and ultimately being transferred to other objects or individuals [102]. It is crucial for forensic investigators to consider these mechanisms and recognize potential sources of indirect transfer when analyzing DNA evidence. Understanding the principles of DNA transfer (i.e., transfer, persistence, prevalence and recovery of DNA) is essential for forensic investigators to accurately interpret DNA evidence and present reliable conclusions in a court of law [100,103]. The Bayesian model has become widely used in evaluating indirect DNA transfer in the forensic field, providing a statistical framework for assessing the likelihood of such transfers. One key aspect of the Bayesian model is the use of prior probabilities: these represent the initial beliefs about the likelihood of different scenarios. In this context, the application of AI tools for the analysis of the activity level could help the investigator to evaluate the weight of evidence in the worked case. However, it is important to note that the Bayesian model is dependent on the input data and assumptions made. Biases or inaccuracies in the data or incorrect assumptions can impact the validity of the results [104,105,106].
AI could be helpful in the prediction of physical traits and ancestry from DNA samples. By training machine learning models on extensive genetic databases, researchers have developed algorithms that can accurately predict a person’s physical characteristics, such as eye or hair color, improving so-called forensic DNA phenotyping. Additionally, AI can provide insights into a person’s ancestry or geographical origin by analyzing STRs and other genetic markers, such as SNPs; to date, it is possible to use a set of ancestry informative markers to determine the ancestry of an individual [88,107]. While the simultaneous analysis of hundreds of DNA predictors with targeted MPS can be valuable in criminal investigations, particularly in the identification of the suspect or the identity of unknown human remains, it should be desirable to conduct further studies to achieve the forensic validation of these tools [108].
5. Conclusions
AI algorithms are playing a crucial role in revolutionizing forensic investigations on a global scale. The fields of AI and forensic genetics are rapidly evolving, particularly within the criminal justice domain. While AI has proven to be highly useful in interpreting STR profiles and conducting haplotype analysis, further improvements are needed before deploying these applications in real cases. The main challenge lies in the communication gap between forensic experts, crime investigators, and lawyers, causing statistical evidence to be misunderstood and misinterpreted in court, leading to erroneous decisions. Thus, there is a pressing need to develop methods that facilitate effective communication and act as a bridge to address these issues. As AI continues to advance, the collaboration between forensic sciences and AI presents immense potential for transforming investigative practices, enabling quicker and more precise case resolutions. However, it is important to acknowledge that although AI has benefited forensic genetics, considering the variability of forensic samples, it is difficult to expect AI tools to effectively understand, analyze, and interpret genetic data as efficiently as human forensic experts, scientists, and investigators. AI can be seen as a valuable aid, but it should never replace the essential role of human expertise.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app14052113/s1.
Author Contributions
Conceptualization, F.S. and M.S.; methodology, F.S., M.E., G.C., S.S., M.C., M.A.A.K., D.G.A. and M.S.; formal analysis, F.S., M.E. and M.S.; data curation, F.S., M.E., G.C., S.S., M.C., M.A.A.K., D.G.A. and M.S.; writing—original draft preparation, F.S.; writing—review and editing, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We wish to thank the Scientific Bureau of the University of Catania for language support.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Smith, C.; McGuire, B.; Huang, T.; Yang, G. The History of Artificial Intelligence; University of Washington: Washington, DC, USA, 2006. [Google Scholar]
- Mccarthy, J. From here to human-level AI. Art. Int. 2007, 171, 1174–1182. [Google Scholar] [CrossRef]
- Oliva, A.; Grassi, S.; Vetrugno, G.; Rossi, R.; Della Morte, G.; Pinchi, V.; Caputo, M. Management of Medico-Legal Risks in Digital Health Era: A Scoping Review. Front. Med. 2022, 8, 821756. [Google Scholar] [CrossRef] [PubMed]
- Galante, N.; Cotroneo, R.; Furci, D.; Lodetti, G.; Casali, M.B. Applications of Artificial Intelligence in Forensic Sciences: Current Potential Benefits, Limitations and Perspectives. Int. J. Legal Med. 2022, 137, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.M. Recent Advances in Forensic Biology and Forensic DNA Typing: INTERPOL Review 2019–2022. Forensic Sci. Int. 2023, 6, 100311. [Google Scholar] [CrossRef] [PubMed]
- Esposito, M.; Sessa, F.; Cocimano, G.; Zuccarello, P.; Roccuzzo, S.; Salerno, M. Advances in Technologies in Crime Scene Investigation. Diagnostics 2023, 13, 3169. [Google Scholar] [CrossRef]
- Aldoseri, A.; Al-Khalifa, K.N.; Hamouda, A.M. Re-Thinking Data Strategy and Integration for Artificial Intel-ligence: Concepts, Opportunities, and Challenges. Appl. Sci. 2023, 13, 7082. [Google Scholar] [CrossRef]
- Collins, C.; Dennehy, D.; Conboy, K.; Mikalef, P. Artificial Intelligence in Information Systems Research: A Systematic Literature Review and Research Agenda. Int. J. Inf. Manag. 2021, 60, 102383. [Google Scholar] [CrossRef]
- der Mauer, M.A.; Well, E.J.-V.; Herrmann, J.; Groth, M.; Morlock, M.M.; Maas, R.; Säring, D. Automated Age Estimation of Young Individuals Based on 3D Knee MRI Using Deep Learning. Int. J. Leg. Med. 2020, 135, 649–663. [Google Scholar] [CrossRef]
- Cao, Y.; Ma, Y.; Vieira, D.N.; Guo, Y.; Wang, Y.; Deng, K.; Chen, Y.; Zhang, J.; Qin, Z.; Chen, F.; et al. A Potential Method for Sex Estimation of Human Skeletons Using Deep Learning and Three-Dimensional Sur-face Scanning. Int. J. Leg. Med. 2021, 135, 2409–2421. [Google Scholar] [CrossRef]
- Bewes, J.; Low, A.; Morphett, A.; Pate, F.D.; Henneberg, M. Artificial Intelligence for Sex Determination of Skeletal Remains: Application of a Deep Learning Artificial Neural Network to Human Skulls. J. Forensic Leg. Med. 2019, 62, 40–43. [Google Scholar] [CrossRef]
- Mohtarami, S.A.; Hedjazi, A.; Manouchehri, R.H. Determine the Age Range Based on Machine-Learning Methods from Skeletal Angles of the Face (Glabella and Maxilla Angle and Length and Width of Piriformis) in a CT Scan. Int. J. Med. Toxicol. Forensic Med. 2022, 12, 38605. [Google Scholar] [CrossRef]
- Kim, Y.H.; Ha, E.-G.; Jeon, K.J.; Lee, C.; Han, S.-S. A Fully Automated Method of Human Identification Based on Dental Panoramic Radiographs Using a Con-volutional Neural Network. Dentomaxillofacial Radiol. 2021, 51, 20210383. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.; Porto, L.; Heng, D.; Murray, J.; Lygate, A.; Franco, R.; Bueno, J.; Sobania, M.; Costa, M.M.; Paranhos, L.R.; et al. Diagnostic Performance of Convolutional Neural Networks for Dental Sexual Dimorphism. Sci. Rep. 2022, 12, 17279. [Google Scholar] [CrossRef]
- Gómez, Ó.; Mesejo, P.; Ibáñez, Ó.; Valsecchi, A.; Bermejo, E.; Cerezo, A.; Pérez, J.; Alemán, I.; Kahana, T.; Damas, S.; et al. Evaluating Artificial Intelligence for Comparative Radiography. Int. J. Leg. Med. 2023, 138, 307–327. [Google Scholar] [CrossRef] [PubMed]
- Kondou, H.; Morohashi, R.; Ichioka, H.; Bandou, R.; Matsunari, R.; Kawamoto, M.; Idota, N.; Ting, D.; Kimura, S.; Ikegaya, H. Deep Neural Networks-Based Age Estimation of Cadavers Using CT Imaging of Vertebrae. Int. J. Environ. Res. Public Health 2023, 20, 4806. [Google Scholar] [CrossRef] [PubMed]
- Baydogan, M.P.; Baybars, S.C.; Tuncer, S.A. Age-Net: An Advanced Hybrid Deep Learning Model for Age Estimation Using Orthopantomograph Images. Trait. Signal 2023, 40, 1553–1563. [Google Scholar] [CrossRef]
- Bu, W.-Q.; Zhang, D.; Du, S.-Y.; Han, M.-Q.; Wu, Z.-X.; Tang, Y.; Chen, T.; Guo, Y.-C.; Meng, H.-T. Automatic Sex Estimation Using Deep Convolutional Neural Network Based on Orthopantomogram Images. Forensic Sci. Int. 2023, 348, 111704. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Jain, A. Dental Biometrics: Alignment and Matching of Dental Radiographs. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1319–1326. [Google Scholar] [CrossRef]
- Campomanes-Alvarez, C.; Ibáñez, O.; Cordón, O.; Wilkinson, C. Hierarchical Information Fusion for Decision Making in Craniofacial Superimposition. Inf. Fusion 2018, 39, 25–40. [Google Scholar] [CrossRef]
- Heinrich, A.; Güttler, F.; Wendt, S.; Schenkl, S.; Hubig, M.; Wagner, R.; Mall, G.; Teichgräber, U. Forensic Odontology: Automatic Identification of Persons Comparing Antemortem and Postmortem Panoramic Radiographs Using Computer Vision. RoFo Fortschritte auf dem Gebiet der Rontgenstrahlen und der Bildgebenden Verfahren 2018, 190, 1152–1158. [Google Scholar] [CrossRef]
- Thurzo, A.; Jančovičová, V.; Hain, M.; Thurzo, M.; Novák, B.; Kosnáčová, H.; Lehotská, V.; Varga, I.; Kováč, P.; Moravanský, N. Human Remains Identification Using Micro-CT, Chemometric and AI Methods in Forensic Experimental Reconstruction of Dental Patterns after Concentrated Sulphuric Acid Significant Impact. Molecules 2022, 27, 4035. [Google Scholar] [CrossRef] [PubMed]
- Kira, K.; Chiba, F.; Makino, Y.; Torimitsu, S.; Yamaguchi, R.; Tsuneya, S.; Motomura, A.; Yoshida, M.; Saitoh, N.; Inokuchi, G.; et al. Stature Estimation by Semi-Automatic Measurements of 3D CT Images of the Femur. Int. J. Leg. Med. 2022, 137, 359–377. [Google Scholar] [CrossRef]
- Venema, J.; Peula, D.; Irurita, J.; Mesejo, P. Employing Deep Learning for Sex Estimation of Adult Individuals Using 2D Images of the Humerus. Neural Comput. Appl. 2022, 35, 5987–5998. [Google Scholar] [CrossRef]
- Murray, J.; Heng, D.; Lygate, A.; Porto, L.; Abade, A.; Manica, S.; Franco, A. Applying Artificial Intelligence to Determination of Legal Age of Majority from Radiographic Data. Morphologie 2024, 108, 100723. [Google Scholar] [CrossRef] [PubMed]
- Barrington, S.; Farid, H. A comparative Analysis of Human and AI Performance in Forensic Estimation of Physical Attributes. Sci. Rep. 2023, 13, 4784. [Google Scholar] [CrossRef] [PubMed]
- Thong, Z.; Tan, J.Y.Y.; Loo, E.S.; Phua, Y.W.; Chan, X.L.S.; Syn, C.K.-C. Artificial Neural Network, Predictor Variables and Sensitivity Threshold for DNA Methylation-Based Age Prediction Using Blood Samples. Sci. Rep. 2021, 11, 1744. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Xue, Y.; Knoops, R.; Yu, D.; Balmashnova, E.; Kang, X.; Falgari, P.; Zheng, D.; Liu, P.; Chen, H.; et al. Automated Diatom Searching in the Digital Scanning Electron Microscopy Images of Drowning Cases Using the Deep Neural Networks. Int. J. Leg. Med. 2020, 135, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhou, Y.; Vieira, D.N.; Cao, Y.; Deng, K.; Cheng, Q.; Zhu, Y.; Zhang, J.; Qin, Z.; Ma, K.; et al. An Efficient Method for Building a Database of Diatom Populations for Drowning Site Inference Using a Deep Learning Algorithm. Int. J. Leg. Med. 2021, 135, 817–827. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, J.; Huang, J.; Deng, K.; Zhang, J.; Qin, Z.; Wang, Z.; Zhang, X.; Tuo, Y.; Chen, L.; et al. Digital Whole-Slide Image Analysis for Automated Diatom Test in Forensic Cases of Drowning Using a Convolutional Neural Network Algorithm. Forensic Sci. Int. 2019, 302, 109922. [Google Scholar] [CrossRef]
- Deng, J.; Guo, W.; Zhao, Y.; Liu, J.; Lai, R.; Gu, G.; Zhang, Y.; Li, Q.; Liu, C.; Zhao, J. Identification of Diatom Taxonomy by a Combination of Region-Based Full Convolutional Network, Online Hard Example Mining, and Shape Priors of Diatoms. Int. J. Leg. Med. 2021, 135, 2519–2530. [Google Scholar] [CrossRef]
- Martos, R.; Guerra, R.; Navarro, F.; Peruch, M.; Neuwirth, K.; Valsecchi, A.; Jankauskas, R.; Ibáñez, O. Computer-Aided Craniofacial Superimposition Validation Study: The Identification of the Leaders and Participants of the Polish-Lithuanian January Uprising (1863–1864). Int. J. Leg. Med. 2022, 138, 107–121. [Google Scholar] [CrossRef]
- Zhang, J.; Vieira, D.N.; Cheng, Q.; Zhu, Y.; Deng, K.; Zhang, J.; Qin, Z.; Sun, Q.; Zhang, T.; Ma, K.; et al. DiatomNet v1.0: A Novel Approach for Automatic Diatom Testing for Drowning Diagnosis in Forensically Biomedical Application. Comput. Methods Programs Biomed. 2023, 232, 107434. [Google Scholar] [CrossRef]
- Li, D. Ballistics Projectile Image Analysis for Firearm Identification. IEEE Trans. Image Process. 2006, 15, 2857–2865. [Google Scholar] [CrossRef]
- Cheng, J.; Schmidt, C.; Wilson, A.; Wang, Z.; Hao, W.; Pantanowitz, J.; Morris, C.; Tashjian, R.; Pantanowitz, L. Artificial Intelligence for Human Gunshot Wound Classification. J. Pathol. Inform. 2024, 15, 100361. [Google Scholar] [CrossRef] [PubMed]
- Haut, R.C.; Wei, F. Biomechanical Studies on Patterns of Cranial Bone Fracture Using the Immature Porcine Model. J. Biomech. Eng. 2017, 139, 021001. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, Y.-J.; Liu, M.-F.; Li, N.; Dang, L.-H.; An, G.-S.; Lu, X.-J.; Wang, L.-L.; Du, Q.-X.; Cao, J.; et al. Multi-Omics Integration Strategy in the Post-Mortem Interval of Forensic Science. Talanta 2024, 268, 125249. [Google Scholar] [CrossRef] [PubMed]
- Tozzo, P.; Angiola, F.; Gabbin, A.; Politi, C.; Caenazzo, L. The difficult role of Artificial Intelligence in Medical Liability: To Err is Not Only Human. Clin. Terapeutica 2021, 172, 527–528. [Google Scholar] [CrossRef]
- Cestonaro, C.; Delicati, A.; Marcante, B.; Caenazzo, L.; Tozzo, P. Defining Medical Liability when Artificial Intelligence Is Applied on Diagnostic Algorithms: A Systematic Review. Front. Med. 2023, 10, 1305756. [Google Scholar] [CrossRef]
- Barash, M.; McNevin, D.; Fedorenko, V.; Giverts, P. Machine Learning Applications in Forensic DNA Profiling: A Critical Review. Forensic Sci. Int. Genet. 2024, 69, 102994. [Google Scholar] [CrossRef] [PubMed]
- Schlecht, J.; Kaplan, M.E.; Barnard, K.; Karafet, T.; Hammer, M.F.; Merchant, N.C. Machine-Learning Approaches for Classifying Haplogroup from Y Chromosome STR Data. PLoS Comput. Biol. 2008, 4, e1000093. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine Learning Applications in Genetics and Genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef]
- Haddrill, P.R. Developments in Forensic DNA Analysis. Emerg. Top. Life Sci. 2021, 5, 381–393. [Google Scholar] [CrossRef]
- Paudel, R.; Ligmann-Zielinska, A. A Largely Unsupervised Domain-Independent Qualitative Data Extraction Approach for Empirical Agent-Based Model Development. Algorithms 2023, 16, 338. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews Systematic Reviews and Meta-Analyses. BMJ 2021, 372, 71. [Google Scholar] [CrossRef]
- Pereira, L.; Alshamali, F.; Andreassen, R.; Ballard, R.; Chantratita, W.; Cho, N.S.; Coudray, C.; Dugoujon, J.-M.; Espinoza, M.; González-Andrade, F.; et al. PopAffiliator: Online Calculator for Individual Affiliation to a Major Population Group Based on 17 Autosomal Short Tandem Repeat Genotype Profile. Int. J. Leg. Med. 2010, 125, 629–636. [Google Scholar] [CrossRef][Green Version]
- Mukunthan, B.; Nagaveni, N. Identification of unique repeated patterns, location of mutation in DNA Finger Printing Using Artificial Intelligence Technique. Int. J. Bioinform. Res. Appl. 2014, 10, 157–176. [Google Scholar] [CrossRef]
- Taylor, D.; Powers, D. Teaching Artificial Intelligence to Read Electropherograms. Forensic Sci. Int. Genet. 2016, 25, 10–18. [Google Scholar] [CrossRef]
- Aliferi, A.; Ballard, D.; Gallidabino, M.D.; Thurtle, H.; Barron, L.; Court, D.S. DNA Methylation-Based Age Prediction Using Massively Parallel Sequencing Data and Multiple Machine Learning Models. Forensic Sci. Int. Genet. 2018, 37, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Adelman, J.D.; Zhao, A.; Eberst, D.S.; Marciano, M.A. Automated Detection and Removal of Capillary Electrophoresis Artifacts Due to Spectral Overlap. Electrophoresis 2019, 40, 1753–1761. [Google Scholar] [CrossRef]
- Siino, V.; Sears, C. Artificially Intelligent Scoring and Classification Engine for Forensic Identification. Forensic Sci. Int. Genet. 2019, 44, 102162. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Tao, R.; Zhou, W.; Li, Y.; Meng, M.; Zhang, Y.; Yu, L.; Chen, L.; Bian, Y.; Li, C. Validation Studies of the ParaDNA® Intelligence System with Artificial Evidence Items. Forensic Sci. Res. 2019, 6, 84–91. [Google Scholar] [CrossRef]
- Volgin, L.; Taylor, D.; Bright, J.-A.; Lin, M.-H. Validation of a Neural Network Approach for STR Typing to Replace Human Reading. Forensic Sci. Int. Genet. 2021, 55, 102591. [Google Scholar] [CrossRef]
- Veldhuis, M.S.; Ariëns, S.; Ypma, R.J.; Abeel, T.; Benschop, C.C. Explainable Artificial Intelligence in Forensics: Realistic Explanations for Number of Contributor Predictions of DNA Profiles. Forensic Sci. Int. Genet. 2021, 56, 102632. [Google Scholar] [CrossRef]
- Chen, M.; Cui, W.; Bai, X.; Fang, Y.; Yao, H.; Zhang, X.; Lei, F.; Zhu, B. Comprehensive Evaluations of Individual Discrimination, Kinship Analysis, Genetic Relationship Exploration and Biogeographic Origin Prediction in Chinese Dongxiang Group by a 60-plex DIP panel. Hereditas 2023, 160, 14. [Google Scholar] [CrossRef]
- Kloska, A.; Giełczyk, A.; Grzybowski, T.; Płoski, R.; Kloska, S.M.; Marciniak, T.; Pałczyński, K.; Rogalla-Ładniak, U.; Malyarchuk, B.A.; Derenko, M.V.; et al. A Machine-Learning-Based Approach to Prediction of Biogeographic Ancestry within Europe. Int. J. Mol. Sci. 2023, 24, 15095. [Google Scholar] [CrossRef]
- Fabbri, M.; Alfieri, L.; Mazdai, L.; Frisoni, P.; Gaudio, R.M.; Neri, M. Application of Forensic DNA Phenotyping for Prediction of Eye, Hair and Skin Colour in Highly Decomposed Bodies. Healthcare 2023, 11, 647. [Google Scholar] [CrossRef] [PubMed]
- Hefetz, I. Mapping AI-ethics’ Dilemmas in Forensic Case Work: To trust AI or Not? Forensic Sci. Int. 2023, 350, 111807. [Google Scholar] [CrossRef]
- Raposo, V.L. The Use of Facial Recognition Technology by Law Enforcement in Europe: A Non-Orwellian Draft Proposal. Eur. J. Crim. Policy Res. 2023, 29, 515–533. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, E. Fairness and Bias in Artificial Intelligence: A Brief Survey of Sources, Impacts, and Mitigation Strategies. arXiv 2023, arXiv:2304.07683. [Google Scholar]
- Padma, K.R.; Don, K.R. Artificial Neural Network Applications in Analysis of Forensic Science. In Cyber Security and Digital Forensics; Wiley Online Library: New York, NY, USA, 2021. [Google Scholar]
- Seki, T.; Hsiao, Y.-Y.; Ishizawa, F.; Sugano, Y.; Takahashi, Y. Establishment of A Random Forest Regression Model to Estimate the Age of Bloodstains Based on Temporal Colorimetric Analysis. Leg. Med. 2023, 102343. [Google Scholar] [CrossRef] [PubMed]
- Nikita, E.; Nikitas, P. On the Use of Machine Learning Algorithms in Forensic Anthropology. Leg. Med. 2020, 47, 101771. [Google Scholar] [CrossRef]
- Rostamzadeh, S.; Abouhossein, A.; Saremi, M.; Taheri, F.; Ebrahimian, M.; Vosoughi, S. A Comparative Investigation of Machine Learning Algorithms for Predicting Safety Signs Comprehension Based on Socio-Demographic Factors and Cognitive Sign Features. Sci. Rep. 2023, 13, 10843. [Google Scholar] [CrossRef]
- Albahra, S.; Gorbett, T.; Robertson, S.; D’Aleo, G.; Kumar, S.V.S.; Ockunzzi, S.; Lallo, D.; Hu, B.; Rashidi, H.H. Artificial Intelligence and Machine Learning Overview in Pathology & Laboratory Medicine: A general review of data preprocessing and basic supervised concepts. Semin. Diagn. Pathol. 2023, 40, 71–87. [Google Scholar] [CrossRef]
- Li, Z.; Gao, E.; Zhou, J.; Han, W.; Xu, X.; Gao, X. Applications of Deep Learning in Understanding Gene Regulation. Cell Rep. Methods 2023, 3, 100384. [Google Scholar] [CrossRef]
- Rashidi, H.H.; Albahra, S.; Robertson, S.; Tran, N.K.; Hu, B. Common Statistical Concepts in the Supervised Machine Learning arena. Front. Oncol. 2023, 13, 1130229. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-Y.; Harbison, S. A Review of Bioinformatic Methods for Forensic DNA analyses. Forensic Sci. Int. Genet. 2018, 33, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Benschop, C.C.; Hoogenboom, J.; Hovers, P.; Slagter, M.; Kruise, D.; Parag, R.; Steensma, K.; Slooten, K.; Nagel, J.H.; Dieltjes, P.; et al. DNAxs/DNAStatistX: Development and Validation of a Software Suite for the Data Management and Probabilistic Interpretation of DNA Profiles. Forensic Sci. Int. Genet. 2019, 42, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Bright, J.-A.; Taylor, D.; McGovern, C.; Cooper, S.; Russell, L.; Abarno, D.; Buckleton, J. Developmental Validation of STRmix™, Expert Software for the Interpretation of Forensic DNA Profiles. Forensic Sci. Int. Genet. 2016, 23, 226–239. [Google Scholar] [CrossRef]
- Manabe, S.; Fukagawa, T.; Fujii, K.; Mizuno, N.; Sekiguchi, K.; Akane, A.; Tamaki, K. Development and validation of Kongoh ver. 3.0.1: Open-Source Software for DNA Mixture Interpretation in the GlobalFiler System Based on a Quantitative Continuous Model. Leg. Med. 2021, 54, 101972. [Google Scholar] [CrossRef]
- Moretti, T.R.; Just, R.S.; Kehl, S.C.; Willis, L.E.; Buckleton, J.S.; Bright, J.-A.; Taylor, D.A.; Onorato, A.J. Internal Validation of STRmix™ for the Interpretation of Single Source and Mixed DNA Profiles. Forensic Sci. Int. Genet. 2017, 29, 126–144. [Google Scholar] [CrossRef]
- Perlin, M.W.; Legler, M.M.; Spencer, C.E.; Smith, J.L.; Allan, W.P.; Belrose, J.L.; Duceman, B.W. Validating TrueAllele® DNA Mixture Interpretation. J. Forensic Sci. 2011, 56, 1430–1447. [Google Scholar] [CrossRef]
- Balding, D.J.; Buckleton, J. Interpreting Low Template DNA Profiles. Forensic Sci. Int. Genet. 2009, 4, 1–10. [Google Scholar] [CrossRef]
- Duke, K.R.; Myers, S.P. Systematic Evaluation of STRmix™ Performance on Degraded DNA Profile Data. Forensic Sci. Int. Genet. 2019, 44, 102174. [Google Scholar] [CrossRef] [PubMed]
- Bleka, Ø.; Storvik, G.; Gill, P. EuroForMix: An Open Source Software Based on a Continuous model to evaluate STR DNA profiles from a mixture of contributors with artefacts. Forensic Sci. Int. Genet. 2016, 21, 35–44. [Google Scholar] [CrossRef]
- Holland, M.M.; Parson, W. GeneMarker® HID: A Reliable Software Tool for the Analysis of Forensic STR Data. J. Forensic Sci. 2010, 56, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Carratto, T.M.T.; Moraes, V.M.S.; Recalde, T.S.F.; de Oliveira, M.L.G.; Mendes-Junior, C.T. Applications of massively parallel sequencing in forensic genetics. Genet. Mol. Biol. 2022, 45, e20220077. [Google Scholar] [CrossRef]
- Gupta, N.; Verma, V.K. Next-Generation Sequencing and Its Application: Empowering in Public Health Beyond Reality. Microb. Technol. Welfare Soc. 2019, 17, 313–341. [Google Scholar] [CrossRef]
- Dash, H.R.; Kaitholia, K.; Kumawat, R.K.; Singh, A.K.; Shrivastava, P.; Chaubey, G.; Das, S. Sequence variations, flanking region mutations, and allele frequency at 31 autosomal STRs in the central Indian population by next generation sequencing (NGS). Sci. Rep. 2021, 11, 23238. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Yadav, S.; Yadav, S.; Verma, P.; Ojha, S. Artificial Intelligence: An Advanced Evolution in Forensics And Criminal Investigation. 2022. Available online: https://www.researchgate.net/profile/Sudhanshu-Mishra/publication/362815618_Artificial_Intelligence_An_Advanced_Evolution_In_Forensics_And_Criminal_Investigation/links/632c57ca873eca0c00a8f803/Artificial-Intelligence-An-Advanced-Evolution-In-Forensics-And-Criminal-Investigation.pdf (accessed on 27 February 2024).
- Mohammad, N.; Ahmad, R.; Kurniawan, A.; Mohd Yusof, M.Y.P. Applications of Contemporary Artificial In-telligence Technology in Forensic Odontology as Primary Forensic Identifier: A Scoping Review. Front. Artif. Intell. 2022, 5, 1049584. [Google Scholar] [CrossRef]
- Pomara, C.; Zappalà, S.A.; Salerno, M.; Sessa, F.; Esposito, M.; Cocimano, G.; Ippolito, S.; Miani, A.; Missoni, E.; Piscitelli, P. Migrants’ Human Rights and Health Protection during the COVID-19 Pandemic in the Mediterra-nean Sea: What We Have Learnt from Direct Inspections in Two Italian Hotspots. Front. Public Health 2023, 11, 1129267. [Google Scholar] [CrossRef]
- Patil, V.; Ingle, D.R. An association between Fingerprint Patterns with Blood Group and Lifestyle Based Diseases: A Review. Artif. Intell. Rev. 2020, 54, 1803–1839. [Google Scholar] [CrossRef]
- Leone, M. From Fingers to Faces: Visual Semiotics and Digital Forensics. Int. J. Semiot. Law-Revue Int. Sémiotique Juridique 2020, 34, 579–599. [Google Scholar] [CrossRef]
- Chauhan, S.; Sharma, V.; Siddhartha, I. Fingerprints analysis using AI Algorithm. Int. J. Med. Toxicol. Leg. Med. 2022, 25, 273–279. [Google Scholar] [CrossRef]
- Tyagi, A.; Tiwari, P.; Bhardwaj, P.; Chawla, H. Prognosis of sexual dimorphism with unfused hyoid bone: Artificial intelligence informed decision making with discriminant analysis. Sci. Justice 2021, 61, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Kayser, M.; Branicki, W.; Parson, W.; Phillips, C. Recent advances in Forensic DNA Phenotyping of appearance, ancestry and age. Forensic Sci. Int. Genet. 2023, 65, 102870. [Google Scholar] [CrossRef] [PubMed]
- Tozzo, P.; Mazzobel, E.; Marcante, B.; Delicati, A.; Caenazzo, L. Touch DNA Sampling Methods: Efficacy Evaluation and Systematic Review. Int. J. Mol. Sci. 2022, 23, 15541. [Google Scholar] [CrossRef] [PubMed]
- Tozzo, P.; D’angiolella, G.; Brun, P.; Castagliuolo, I.; Gino, S.; Caenazzo, L. Skin Microbiome Analysis for Forensic Human Identification: What Do We Know So Far? Microorganisms 2020, 8, 873. [Google Scholar] [CrossRef] [PubMed]
- Sessa, F.; Salerno, M.; Pomara, C. The Interpretation of Mixed DNA Samples. In Handbook of DNA Profiling; Dash, H.R., Shrivastava, P., Lorente, J.A., Eds.; Springer: Singapore, 2022; pp. 997–1017. ISBN 978-981-16-4318-7. [Google Scholar]
- Sessa, F.; Salerno, M.; Bertozzi, G.; Messina, G.; Ricci, P.; Ledda, C.; Rapisarda, V.; Cantatore, S.; Turillazzi, E.; Pomara, C. Touch DNA: Impact of Handling Time on Touch Deposit and Evaluation of Different Recovery Techniques: An Experimental Study. Sci. Rep. 2019, 9, 9542. [Google Scholar] [CrossRef]
- Buckingham, A.K.; Harvey, M.L.; van Oorschot, R.A. The Origin of Unknown Source DNA from Touched Objects. Forensic Sci. Int. Genet. 2016, 25, 26–33. [Google Scholar] [CrossRef]
- Pfeifer, C.M.; Wiegand, P. Persistence of Touch DNA on Burglary-Related Tools. Int. J. Leg. Med. 2017, 131, 941–953. [Google Scholar] [CrossRef]
- Sessa, F.; Esposito, M.; Roccuzzo, S.; Cocimano, G.; Rosi, G.L.; Sablone, S.; Salerno, M. DNA Profiling from Fired Cartridge Cases: A Literature Review. Minerva Medicolegale 2023, 143, 34–43. [Google Scholar] [CrossRef]
- Coble, M.D.; Buckleton, J.; Butler, J.; Egeland, T.; Fimmers, R.; Gill, P.; Gusmão, L.; Guttman, B.; Krawczak, M.; Morling, N.; et al. DNA Commission of the International Society for Forensic Genetics: Recommendations on the Validation of Software Programs Performing Biostatistical Calculations for Forensic Genetics Applications. Forensic Sci. Int. Genet. 2016, 25, 191–197. [Google Scholar] [CrossRef]
- Tanzhaus, K.; Reiß, M.-T.; Zaspel, T. “I’ve Never Been at the Crime Scene!”—Gloves as Carriers for Secondary DNA Transfer. Int. J. Leg. Med. 2021, 135, 1385–1393. [Google Scholar] [CrossRef] [PubMed]
- Gosch, A.; Courts, C. On DNA Transfer: The Lack and Difficulty of Systematic Research and How to do it Better. Forensic Sci. Int. Genet. 2019, 40, 24–36. [Google Scholar] [CrossRef]
- Samie, L.; Hicks, T.; Castella, V.; Taroni, F. Stabbing Simulations and DNA Transfer. Forensic Sci. Int. Genet. 2016, 22, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Sessa, F.; Pomara, C.; Esposito, M.; Grassi, P.; Cocimano, G.; Salerno, M. Indirect DNA Transfer and Forensic Implications: A Literature Review. Genes 2023, 14, 2153. [Google Scholar] [CrossRef]
- van Oorschot, R.A.; Szkuta, B.; Meakin, G.E.; Kokshoorn, B.; Goray, M. DNA Transfer in Forensic Science: A review. Forensic Sci. Int. Genet. 2018, 38, 140–166. [Google Scholar] [CrossRef] [PubMed]
- Fantinato, C.; Fonneløp, A.E.; Bleka, Ø.; Vigeland, M.D.; Gill, P. The Invisible Witness: Air and Dust as DNA Evidence of Human Occupancy in Indoor Premises. Sci. Rep. 2023, 13, 19059. [Google Scholar] [CrossRef]
- van Oorschot, R.A.H.; Meakin, G.E.; Kokshoorn, B.; Goray, M.; Szkuta, B. DNA Transfer in Forensic Science: Recent Progress towards Meeting Challenges. Genes 2021, 12, 1766. [Google Scholar] [CrossRef]
- Onofri, M.; Altomare, C.; Severini, S.; Tommolini, F.; Lancia, M.; Carlini, L.; Gambelunghe, C.; Carnevali, E. Direct and Secondary Transfer of Touch DNA on a Credit Card: Evidence Evaluation Given Activity Level Propositions and Application of Bayesian Networks. Genes 2023, 14, 996. [Google Scholar] [CrossRef]
- Barker, M.J. Connecting Applied and Theoretical Bayesian Epistemology: Data Relevance, Pragmatics, and the Legal Case of Sally Clark. J. Appl. Philos. 2016, 34, 242–262. [Google Scholar] [CrossRef]
- Taylor, D.; Samie, L.; Champod, C. Using Bayesian Networks to Track DNA Movement Through Complex Transfer Scenarios. Forensic Sci. Int. Genet. 2019, 42, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C. Forensic Genetic Analysis of Bio-Geographical Ancestry. Forensic Sci. Int. Genet. 2015, 18, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Pośpiech, E.; Teisseyre, P.; Mielniczuk, J.; Branicki, W. Predicting Physical Appearance from DNA Data—Towards Genomic Solutions. Genes 2022, 13, 121. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).