Advancing Content-Based Histopathological Image Retrieval Pre-Processing: A Comparative Analysis of the Effects of Color Normalization Techniques
Abstract
1. Introduction
2. Related Work
2.1. Content-Based Image Retrieval
2.2. Color Normalization
- Proposal of a new CBHIR framework with an unsupervised feature extractor that includes color normalization as a pre-processing step;
- Analysis of CBHIR’s performance when using normalized images in comparison with original images;
- We draw attention to the relevance of color variation and its impact on CBHIR;
- We provide a comprehensive performance assessment of the proposed method. This evaluation employs a large breast cancer database scored from five distinct laboratories. This evaluation has a more restrictive K-top accuracy assessment compared to recent state-of-the-art studies and also involves an in-depth analysis of retrieving patches with the same cancer label.
3. Methodology
3.1. Pre-Processing
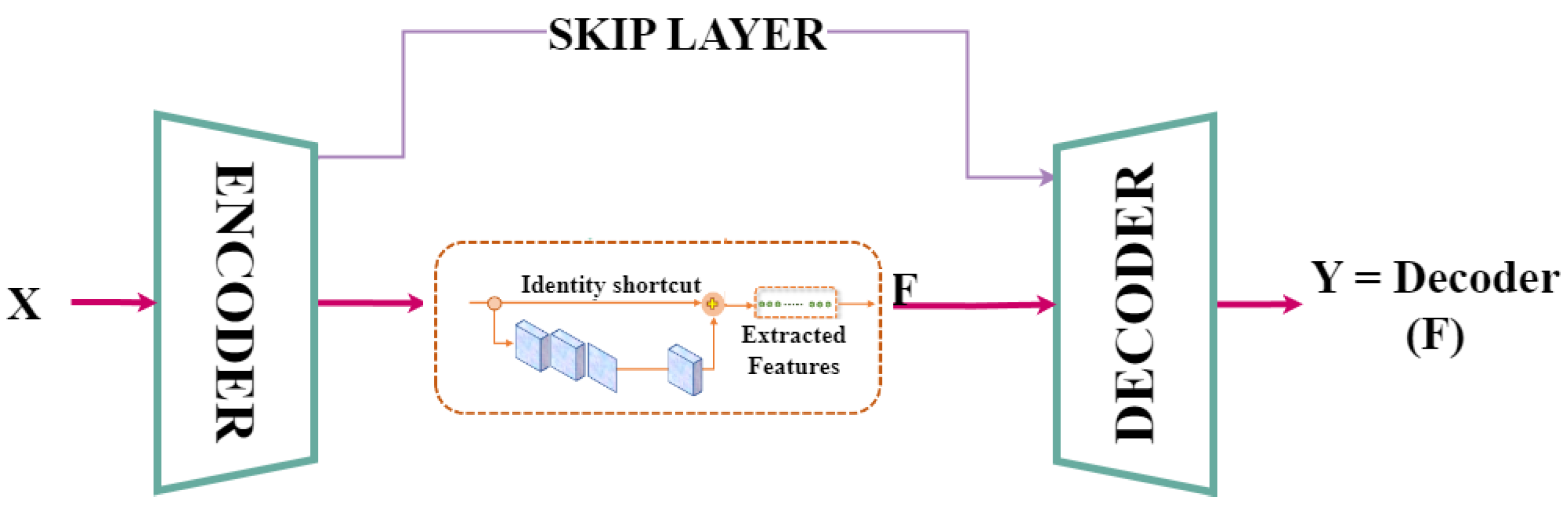
3.2. Feature Extractor
3.3. Searching
4. Material
5. Experiments and Results
5.1. Pre-Processing
5.2. CBMIR Results
5.3. Visual Evaluation
5.4. Comparing the Results of CBHIR with a Classifier
6. Conclusions
7. Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AE | Auto Encoder |
| AUC | Area Under the Curve |
| BKSVD | Bayesian K-Singular Value Decomposition |
| BOF | Bag Of Features |
| CAD | Computer-Aided Diagnosis |
| CAE | Convolutional Auto Encoder |
| CAM17 | CAMELYON17 challenge |
| CBHIR | Content-Based Histopathological Image Retrieval |
| CBMIR | Content-Based Medical Image Retrieval |
| CN | Color Normalization |
| CNN | Convolutional Neural Network |
| DCGMM | Deep Convolutional Gaussian Mixture Model |
| DL | Deep Learning |
| FE | Feature Extractor |
| GAN | Generative Adversarial Network |
| H&E | Hematoxylin and Eosin |
| MRI | Magnetic Resonance Imaging |
| Mac | Macenko |
| NMI | Normalized Median Intensity |
| NMI SD | Normalized Median Intensity Standard Deviation |
| NMI CV | Normalized Median Intensity Coefficient of Variation |
| PSNR | Peak Signal to Noise Ratio |
| ROI | Regions Of Interest |
| SVD | Singular Value Decomposition |
| UFL | Unsupervised Features Learning |
| VAE | Variational Auto Encoder |
| Vah | Vahadane |
| WSIs | Whole-Slide Images |
References
- Ibrahim, A.; Gamble, P.; Jaroensri, R.; Abdelsamea, M.M.; Mermel, C.H.; Chen, P.H.C.; Rakha, E.A. Artificial intelligence in digital breast pathology: Techniques and applications. Breast 2020, 49, 267–273. [Google Scholar] [CrossRef]
- Al-Hussaeni, K.; Karamitsos, I.; Adewumi, E.; Amawi, R.M. CNN-Based Pill Image Recognition for Retrieval Systems. Appl. Sci. 2023, 13, 5050. [Google Scholar] [CrossRef]
- Khalil, S.; Nawaz, U.; Zubariah; Mushtaq, Z.; Arif, S.; ur Rehman, M.Z.; Qureshi, M.F.; Malik, A.; Aleid, A.; Alhussaini, K. Enhancing Ductal Carcinoma Classification Using Transfer Learning with 3D U-Net Models in Breast Cancer Imaging. Appl. Sci. 2023, 13, 4255. [Google Scholar] [CrossRef]
- Fuster, S.; Khoraminia, F.; Eftestøl, T.; Zuiverloon, T.C.; Engan, K. Active Learning Based Domain Adaptation for Tissue Segmentation of Histopathological Images. In Proceedings of the 2023 31st European Signal Processing Conference (EUSIPCO), Helsinki, Finland, 4–8 September 2023; pp. 1045–1049. [Google Scholar] [CrossRef]
- Shahdoosti, H.R.; Mehrabi, A. Multimodal image fusion using sparse representation classification in tetrolet domain. Digit. Signal Process. 2018, 79, 9–22. [Google Scholar] [CrossRef]
- Alrowais, F.; Alotaibi, F.A.; Hassan, A.Q.; Marzouk, R.; Alnfiai, M.M.; Sayed, A. Enhanced Pelican Optimization Algorithm with Deep Learning-Driven Mitotic Nuclei Classification on Breast Histopathology Images. Biomimetics 2023, 8, 538. [Google Scholar] [CrossRef]
- Hegde, N.; Hipp, J.D.; Liu, Y.; Emmert-Buck, M.; Reif, E.; Smilkov, D.; Terry, M.; Cai, C.J.; Amin, M.B.; Mermel, C.H.; et al. Similar image search for histopathology: SMILY. NPJ Digit. Med. 2019, 2, 56. [Google Scholar] [CrossRef] [PubMed]
- Strittmatter, A.; Caroli, A.; Zöllner, F.G. A Multistage Rigid-Affine-Deformable Network for Three-Dimensional Multimodal Medical Image Registration. Appl. Sci. 2023, 13, 13298. [Google Scholar] [CrossRef]
- Kanwal, N.; Fuster, S.; Khoraminia, F.; Zuiverloon, T.C.; Rong, C.; Engan, K. Quantifying the effect of color processing on blood and damaged tissue detection in whole slide images. In Proceedings of the 2022 IEEE 14th Image, Video, and Multidimensional Signal Processing Workshop (IVMSP), Nafplio, Greece, 26–29 June 2022; pp. 1–5. [Google Scholar]
- Shahdoosti, H.R.; Tabatabaei, Z. MRI and PET/SPECT image fusion at feature level using ant colony based segmentation. Biomed. Signal Process. Control 2019, 47, 63–74. [Google Scholar] [CrossRef]
- Long, F.; Zhang, H.; Feng, D.D. Fundamentals of content-based image retrieval. In Multimedia Information Retrieval and Management: Technological Fundamentals and Applications; Springer: Berlin/Heidelberg, Germany, 2003; pp. 1–26. [Google Scholar]
- Tabatabaei, Z.; Colomer, A.; Moll, J.O.; Naranjo, V. Siamese Content-based Search Engine for a More Transparent Skin and Breast Cancer Diagnosis through Histological Imaging. arXiv 2024, arXiv:2401.08272. [Google Scholar]
- Smeulders, A.W.; Worring, M.; Santini, S.; Gupta, A.; Jain, R. Content-based image retrieval at the end of the early years. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 1349–1380. [Google Scholar] [CrossRef]
- Qi, X.; Wang, D.; Rodero, I.; Diaz-Montes, J.; Gensure, R.H.; Xing, F.; Zhong, H.; Goodell, L.; Parashar, M.; Foran, D.J.; et al. Content-based histopathology image retrieval using CometCloud. BMC Bioinform. 2014, 15, 287. [Google Scholar] [CrossRef]
- Tabatabaei, Z.; Colomer, A.; Engan, K.; Oliver, J.; Naranjo, V. Residual block convolutional auto encoder in content-based medical image retrieval. In Proceedings of the 2022 IEEE 14th Image, Video, and Multidimensional Signal Processing Workshop (IVMSP), Nafplio, Greece, 26–29 June 2022; pp. 1–5. [Google Scholar]
- Bianconi, F.; Kather, J.N.; Reyes-Aldasoro, C.C. Experimental assessment of color deconvolution and color normalization for automated classification of histology images stained with hematoxylin and eosin. Cancers 2020, 12, 3337. [Google Scholar] [CrossRef]
- Tabatabaei, Z.; Wang, Y.; Colomer, A.; Oliver Moll, J.; Zhao, Z.; Naranjo, V. WWFedCBMIR: World-Wide Federated Content-Based Medical Image Retrieval. Bioengineering 2023, 10, 1144. [Google Scholar] [CrossRef]
- Fuster, S.; Khoraminia, F.; Kiraz, U.; Kanwal, N.; Kvikstad, V.; Eftestøl, T.; Zuiverloon, T.C.; Janssen, E.A.; Engan, K. Invasive cancerous area detection in Non-Muscle invasive bladder cancer whole slide images. In Proceedings of the 2022 IEEE 14th Image, Video, and Multidimensional Signal Processing Workshop (IVMSP), Nafplio, Greece, 26–29 June 2022; pp. 1–5. [Google Scholar]
- Tosta, T.A.A.; de Faria, P.R.; Neves, L.A.; do Nascimento, M.Z. Computational normalization of H&E-stained histological images: Progress, challenges and future potential. Artif. Intell. Med. 2019, 95, 118–132. [Google Scholar]
- Bandi, P.; Geessink, O.; Manson, Q.; Van Dijk, M.; Balkenhol, M.; Hermsen, M.; Bejnordi, B.E.; Lee, B.; Paeng, K.; Zhong, A.; et al. From detection of individual metastases to classification of lymph node status at the patient level: The camelyon17 challenge. IEEE Trans. Med. Imaging 2018, 38, 550–560. [Google Scholar] [CrossRef] [PubMed]
- Salvi, M.; Acharya, U.R.; Molinari, F.; Meiburger, K.M. The impact of pre-and post-image processing techniques on deep learning frameworks: A comprehensive review for digital pathology image analysis. Comput. Biol. Med. 2021, 128, 104129. [Google Scholar] [CrossRef]
- Vijh, S.; Saraswat, M.; Kumar, S. A new complete color normalization method for H&E stained histopatholgical images. Appl. Intell. 2021, 51, 7735–7748. [Google Scholar]
- Roy, S.; kumar Jain, A.; Lal, S.; Kini, J. A study about color normalization methods for histopathology images. Micron 2018, 114, 42–61. [Google Scholar] [CrossRef]
- Ionescu, B.; Müller, H.; Drăgulinescu, A.M.; Popescu, A.; Idrissi-Yaghir, A.; García Seco de Herrera, A.; Andrei, A.; Stan, A.; Storås, A.M.; Abacha, A.B.; et al. ImageCLEF 2023 Highlight: Multimedia Retrieval in Medical, Social Media and Content Recommendation Applications. In Proceedings of the European Conference on Information Retrieval, Dublin, Ireland, 2–6 April 2023; pp. 557–567. [Google Scholar]
- Zheng, Y.; Jiang, Z.; Zhang, H.; Xie, F.; Ma, Y.; Shi, H.; Zhao, Y. Size-scalable content-based histopathological image retrieval from database that consists of WSIs. IEEE J. Biomed. Health Inform. 2017, 22, 1278–1287. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, W.; Dundar, M.; Badve, S.; Zhang, S. Towards large-scale histopathological image analysis: Hashing-based image retrieval. IEEE Trans. Med. Imaging 2014, 34, 496–506. [Google Scholar] [CrossRef]
- Vanegas, J.A.; Arevalo, J.; González, F.A. Unsupervised feature learning for content-based histopathology image retrieval. In Proceedings of the 2014 12th International Workshop on Content-Based Multimedia Indexing (CBMI), Klagenfurt, Austria, 18–20 June 2014; pp. 1–6. [Google Scholar]
- Sukhia, K.N.; Riaz, M.M.; Ghafoor, A.; Ali, S.S.; Iltaf, N. Content-based histopathological image retrieval using multi-scale and multichannel decoder based LTP. Biomed. Signal Process. Control 2019, 54, 101582. [Google Scholar] [CrossRef]
- Riasatian, A.; Babaie, M.; Maleki, D.; Kalra, S.; Valipour, M.; Hemati, S.; Zaveri, M.; Safarpoor, A.; Shafiei, S.; Afshari, M.; et al. Fine-tuning and training of densenet for histopathology image representation using tcga diagnostic slides. Med. Image Anal. 2021, 70, 102032. [Google Scholar] [CrossRef]
- Silva-Rodríguez, J.; Colomer, A.; Sales, M.A.; Molina, R.; Naranjo, V. Going deeper through the Gleason scoring scale: An automatic end-to-end system for histology prostate grading and cribriform pattern detection. Comput. Methods Programs Biomed. 2020, 195, 105637. [Google Scholar] [CrossRef]
- Shaban, M.T.; Baur, C.; Navab, N.; Albarqouni, S. Staingan: Stain style transfer for digital histological images. In Proceedings of the 2019 Ieee 16th international symposium on biomedical imaging (Isbi 2019), Venice, Italy, 8–11 April 2019; pp. 953–956. [Google Scholar]
- Macenko, M.; Niethammer, M.; Marron, J.S.; Borland, D.; Woosley, J.T.; Guan, X.; Schmitt, C.; Thomas, N.E. A method for normalizing histology slides for quantitative analysis. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 1107–1110. [Google Scholar]
- Vahadane, A.; Peng, T.; Sethi, A.; Albarqouni, S.; Wang, L.; Baust, M.; Steiger, K.; Schlitter, A.M.; Esposito, I.; Navab, N. Structure-preserving color normalization and sparse stain separation for histological images. IEEE Trans. Med. Imaging 2016, 35, 1962–1971. [Google Scholar] [CrossRef]
- BenTaieb, A.; Hamarneh, G. Adversarial stain transfer for histopathology image analysis. IEEE Trans. Med. Imaging 2017, 37, 792–802. [Google Scholar] [CrossRef]
- Zanjani, F.G.; Zinger, S.; Bejnordi, B.E.; van der Laak, J.A.; de With, P.H. Stain normalization of histopathology images using generative adversarial networks. In Proceedings of the 2018 IEEE 15th International symposium on biomedical imaging (ISBI 2018), Washington, DC, USA, 4–7 April 2018; pp. 573–577. [Google Scholar]
- Pérez-Bueno, F.; Serra, J.G.; Vega, M.; Mateos, J.; Molina, R.; Katsaggelos, A.K. Bayesian K-SVD for H and E blind color deconvolution. Applications to stain normalization, data augmentation and cancer classification. Comput. Med. Imaging Graph. 2022, 97, 102048. [Google Scholar] [CrossRef]
- Tellez, D.; Litjens, G.; Bándi, P.; Bulten, W.; Bokhorst, J.M.; Ciompi, F.; Van Der Laak, J. Quantifying the effects of data augmentation and stain color normalization in convolutional neural networks for computational pathology. Med. Image Anal. 2019, 58, 101544. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Shi, X.; Zhang, Y.; Wu, D.; Guizani, M. Deep feature learning for medical image analysis with convolutional autoencoder neural network. IEEE Trans. Big Data 2017, 7, 750–758. [Google Scholar] [CrossRef]
- Ahn, E.; Kumar, A.; Fulham, M.; Feng, D.; Kim, J. Unsupervised domain adaptation to classify medical images using zero-bias convolutional auto-encoders and context-based feature augmentation. IEEE Trans. Med. Imaging 2020, 39, 2385–2394. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Gach, H.M.; Kim, S.; Lee, S.J.; Motai, Y. Autoencoder-inspired convolutional network-based super-resolution method in MRI. IEEE J. Transl. Eng. Health Med. 2021, 9, 1800113. [Google Scholar] [CrossRef] [PubMed]
- Daoud, M.I.; Saleh, A.; Hababeh, I.; Alazrai, R. Content-based Image Retrieval for Breast Ultrasound Images using Convolutional Autoencoders: A Feasibility Study. In Proceedings of the 2019 3rd International Conference on Bio-engineering for Smart Technologies (BioSMART), Paris, France, 24–26 April 2019; pp. 1–4. [Google Scholar] [CrossRef]
- Tabatabaei, Z.; Colomer, A.; Engan, K.; Oliver, J.; Naranjo, V. Self-supervised learning of a tailored Convolutional Auto Encoder for histopathological prostate grading. In Proceedings of the 2023 31st European Signal Processing Conference (EUSIPCO), Helsinki, Finland, 4–8 September 2023; pp. 980–984. [Google Scholar]
- Zheng, Y.; Jiang, Z.; Zhang, H.; Xie, F.; Shi, J.; Xue, C. Adaptive color deconvolution for histological WSI normalization. Comput. Methods Programs Biomed. 2019, 170, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Kalra, S.; Tizhoosh, H.R.; Choi, C.; Shah, S.; Diamandis, P.; Campbell, C.J.; Pantanowitz, L. Yottixel–an image search engine for large archives of histopathology whole slide images. Med. Image Anal. 2020, 65, 101757. [Google Scholar] [CrossRef]
- Cao, Z.; Long, M.; Wang, J.; Yu, P.S. Hashnet: Deep learning to hash by continuation. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 5608–5617. [Google Scholar]
- Tabatabaei, Z.; Colomer, A.; Moll, J.O.; Naranjo, V. Toward More Transparent and Accurate Cancer Diagnosis With an Unsupervised CAE Approach. IEEE Access 2023, 11, 143387–143401. [Google Scholar] [CrossRef]
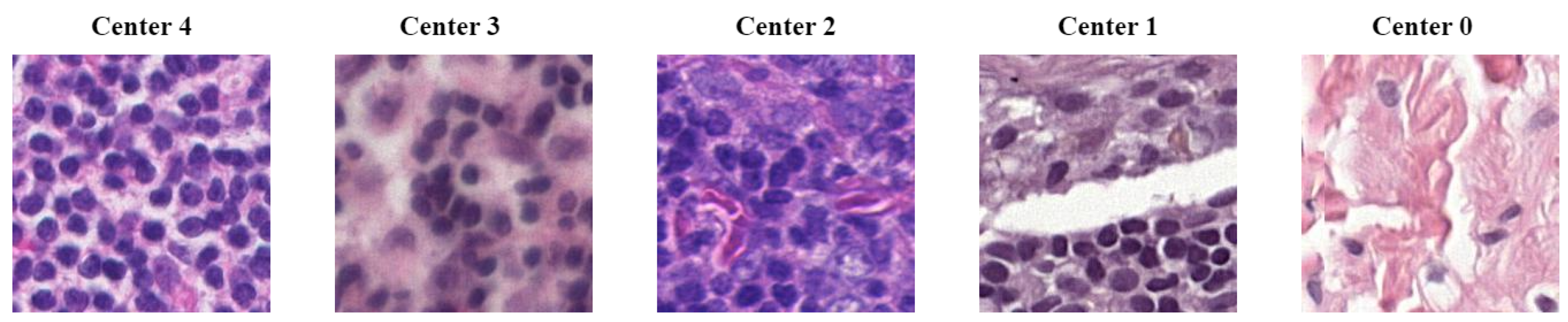

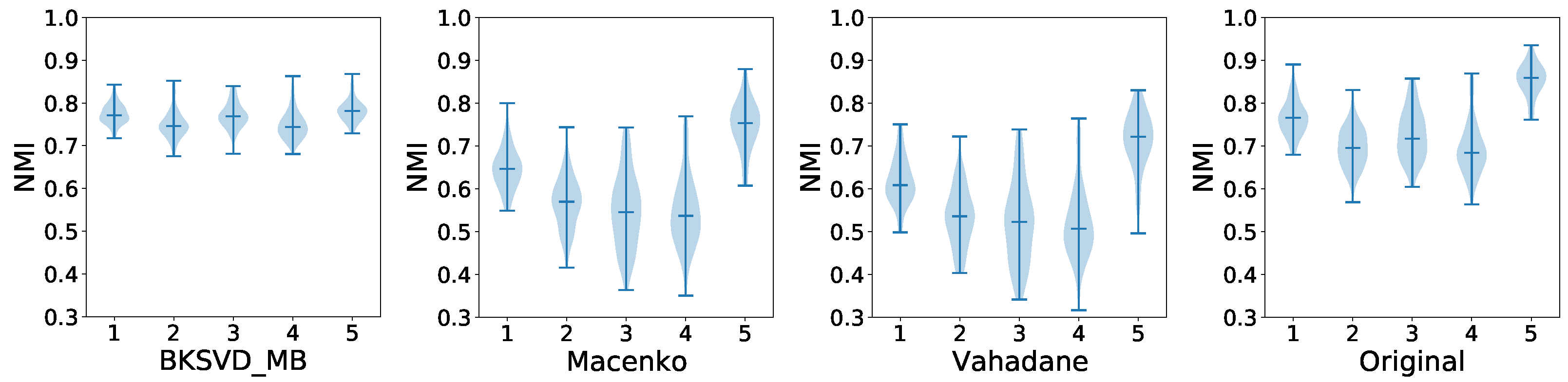
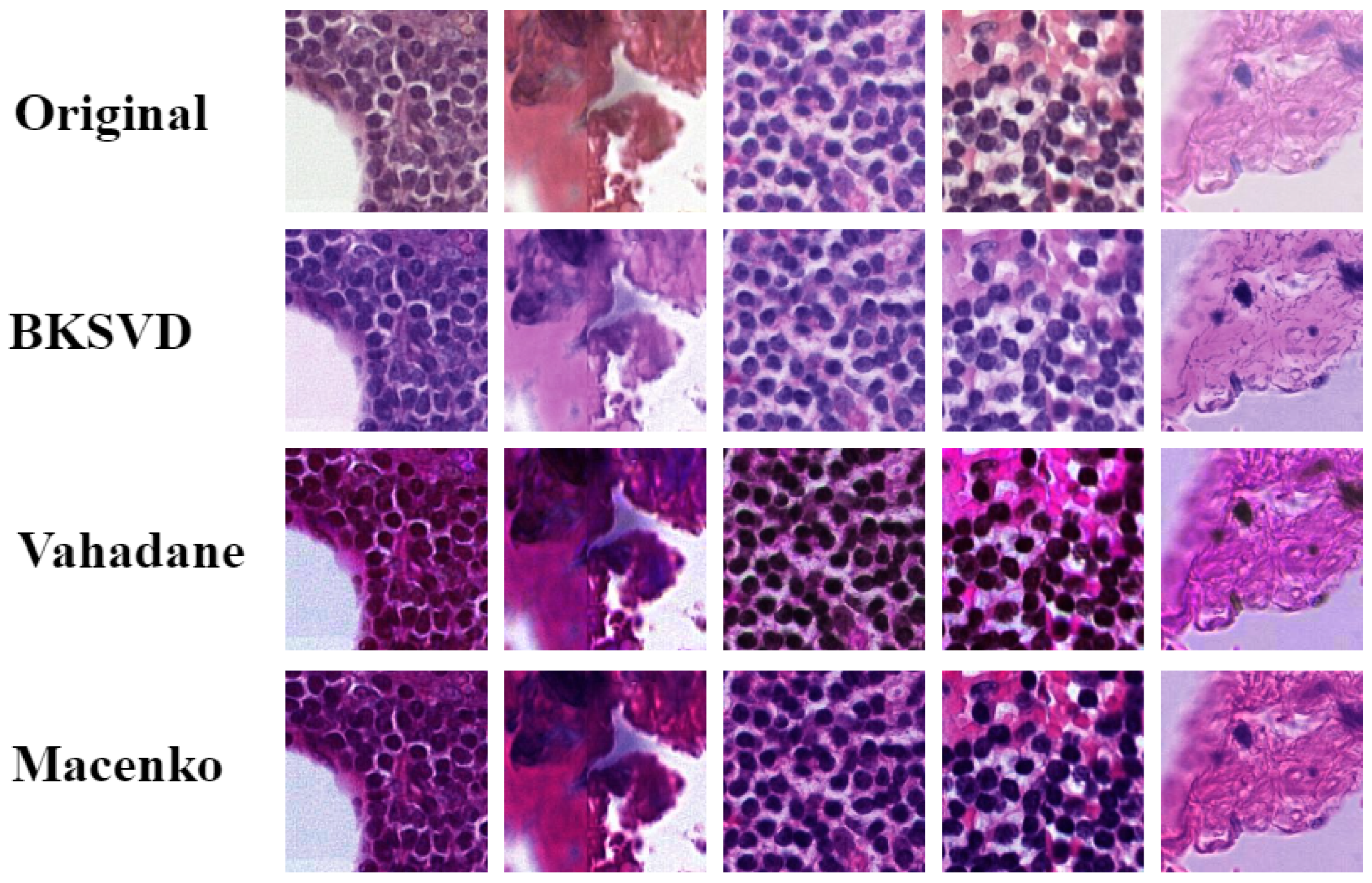

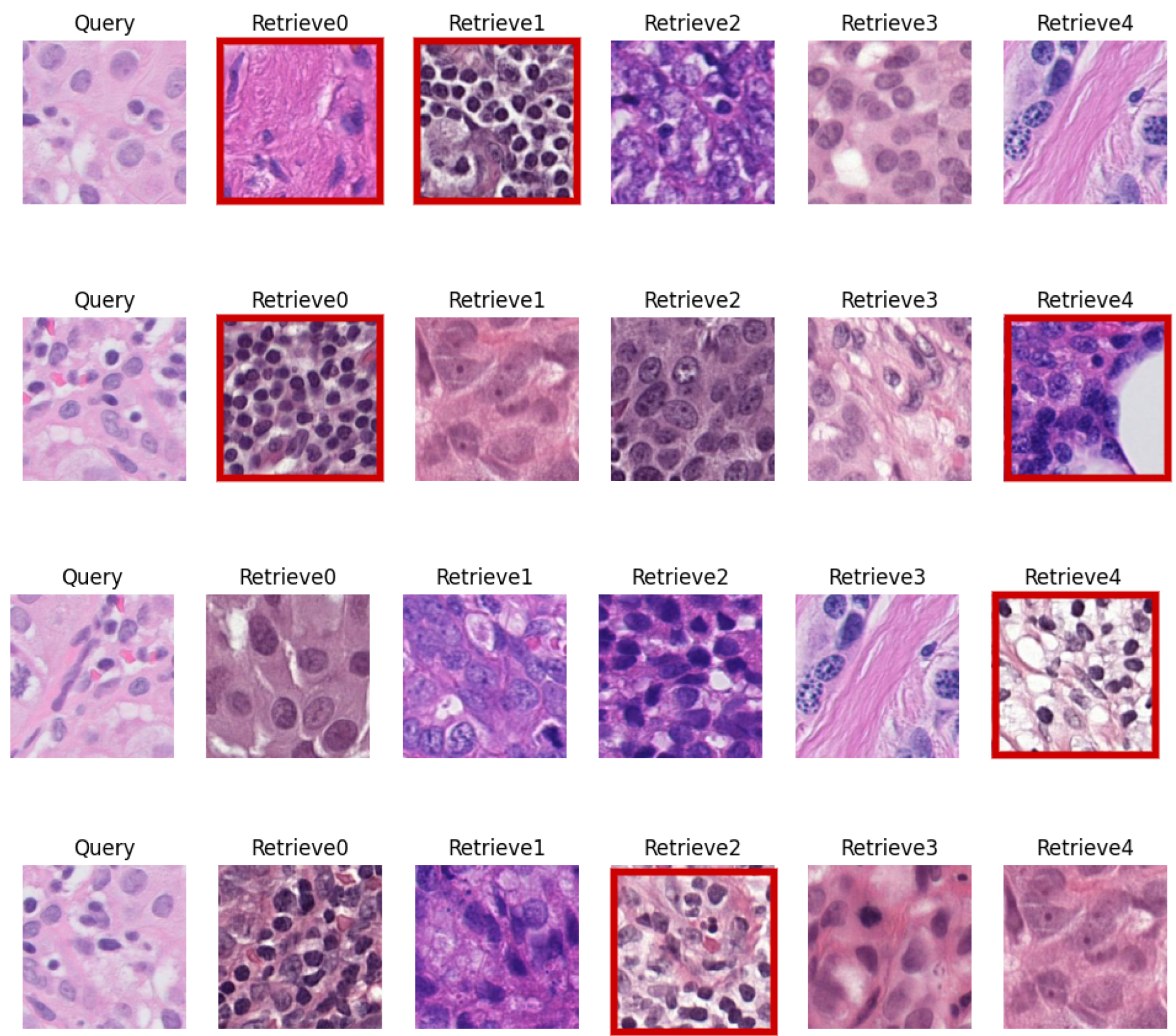
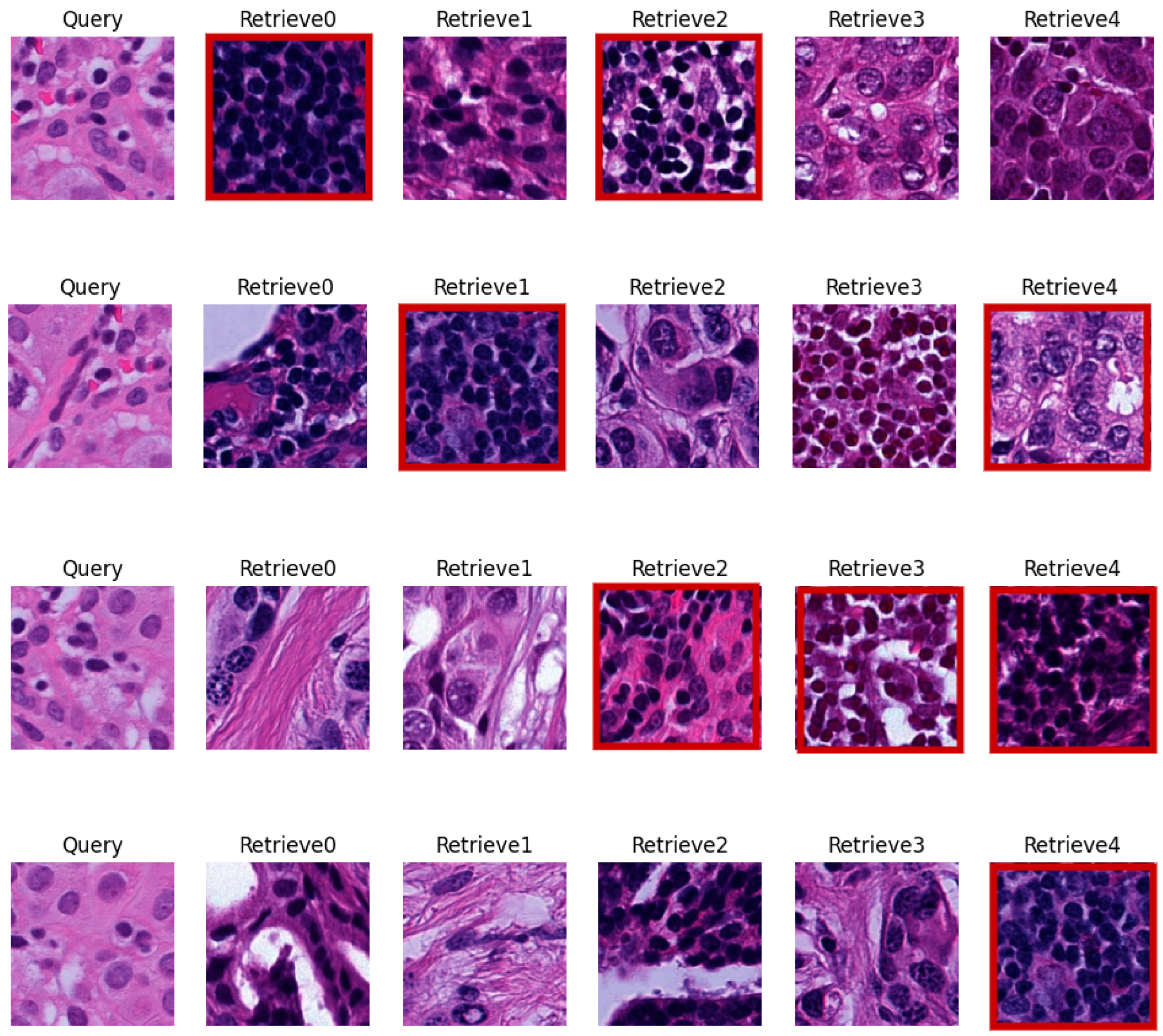
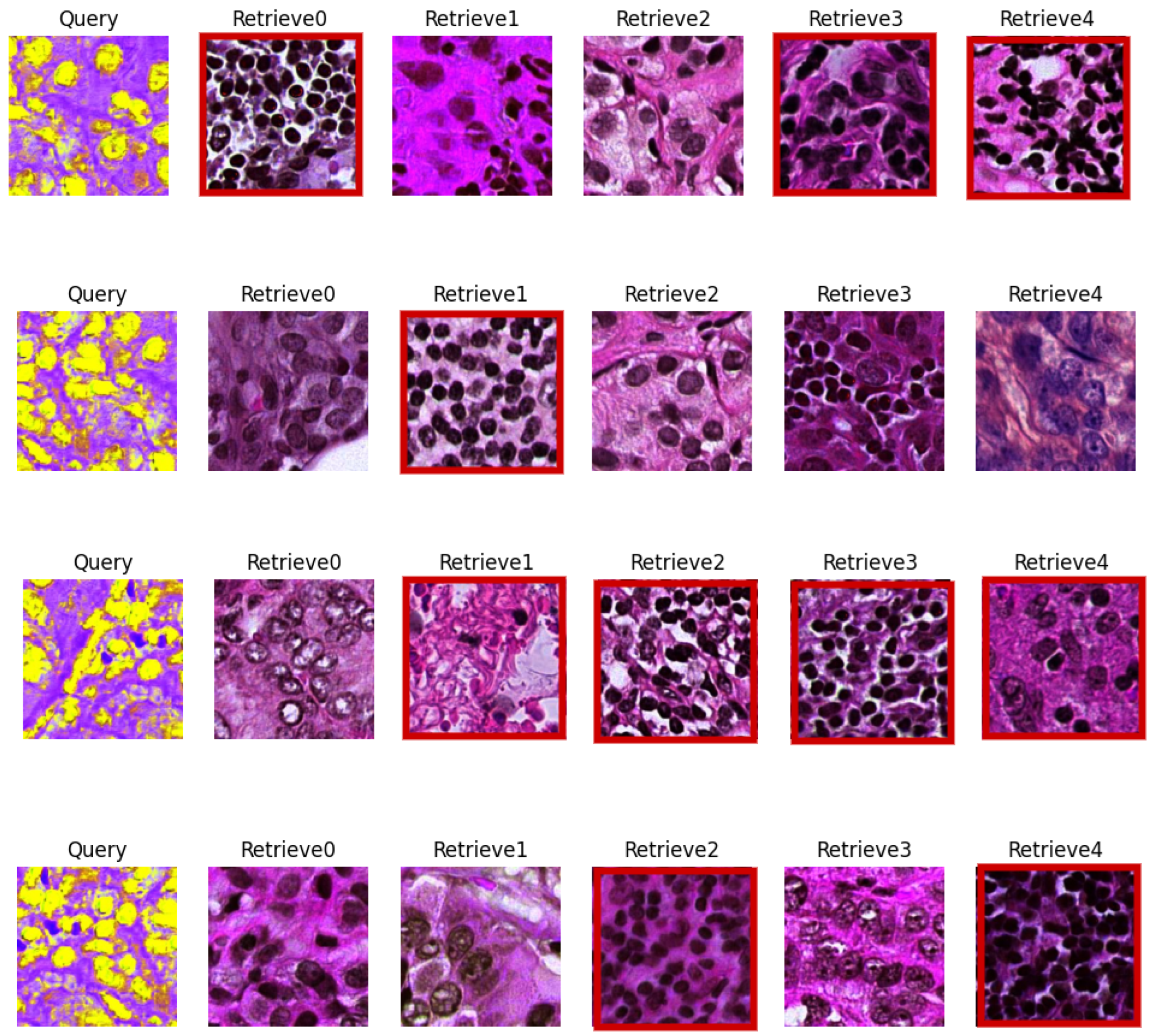

| CN Techniques | BKSVD [36] | Vah [33] | Mac [32] |
|---|---|---|---|
| PSNR | 19.54 | 12.74 | 13.80 |
| K | Original | BKSVD | Vah [33] | Mac [32] |
|---|---|---|---|---|
| 3 | 0.73 | 0.91 | 0.66 | 0.68 |
| 5 | 0.83 | 0.97 | 0.79 | 0.81 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabatabaei, Z.; Pérez Bueno, F.; Colomer, A.; Moll, J.O.; Molina, R.; Naranjo, V. Advancing Content-Based Histopathological Image Retrieval Pre-Processing: A Comparative Analysis of the Effects of Color Normalization Techniques. Appl. Sci. 2024, 14, 2063. https://doi.org/10.3390/app14052063
Tabatabaei Z, Pérez Bueno F, Colomer A, Moll JO, Molina R, Naranjo V. Advancing Content-Based Histopathological Image Retrieval Pre-Processing: A Comparative Analysis of the Effects of Color Normalization Techniques. Applied Sciences. 2024; 14(5):2063. https://doi.org/10.3390/app14052063
Chicago/Turabian StyleTabatabaei, Zahra, Fernando Pérez Bueno, Adrián Colomer, Javier Oliver Moll, Rafael Molina, and Valery Naranjo. 2024. "Advancing Content-Based Histopathological Image Retrieval Pre-Processing: A Comparative Analysis of the Effects of Color Normalization Techniques" Applied Sciences 14, no. 5: 2063. https://doi.org/10.3390/app14052063
APA StyleTabatabaei, Z., Pérez Bueno, F., Colomer, A., Moll, J. O., Molina, R., & Naranjo, V. (2024). Advancing Content-Based Histopathological Image Retrieval Pre-Processing: A Comparative Analysis of the Effects of Color Normalization Techniques. Applied Sciences, 14(5), 2063. https://doi.org/10.3390/app14052063






