DeepSarc-US: A Deep Learning Framework for Assessing Sarcopenia Using Ultrasound Images
Abstract
1. Introduction
- Three CNN- and three ViT-based models are proposed to estimate QMT using ultrasound images acquired in a clinical setting.
- A strategy is proposed for optimizing the training of DL models to estimate QMT more accurately, especially when limited data are available.
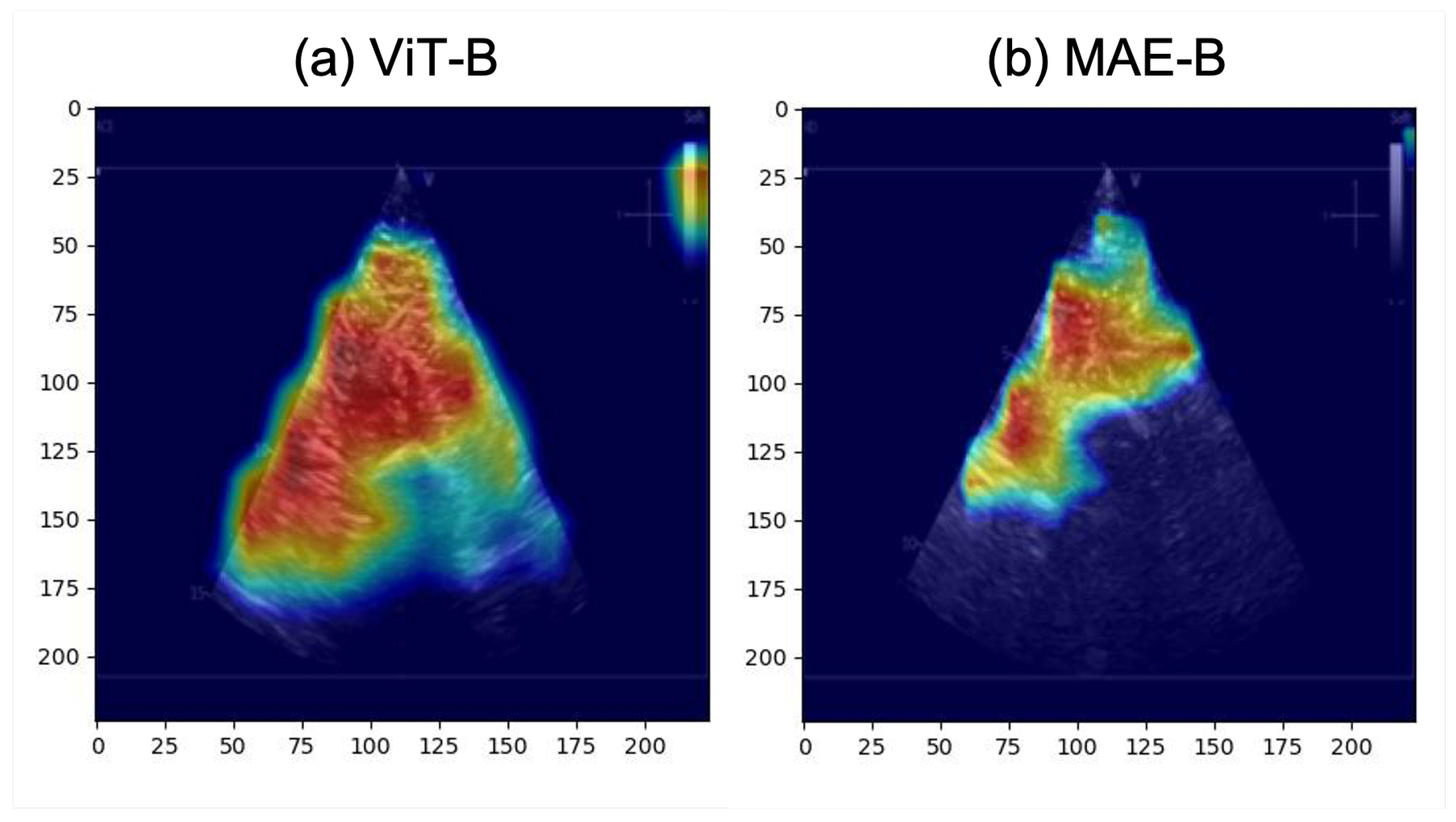
- The activation maps are explored to provide clinicians with real-time feedback. This feedback can potentially be used to help clinicians collect better US images to help DL models estimate QMT more accurately.
- To the best of our knowledge, it is shown for the first time that DL can be used to automatically estimate QMT from US images taken from phased array probes.
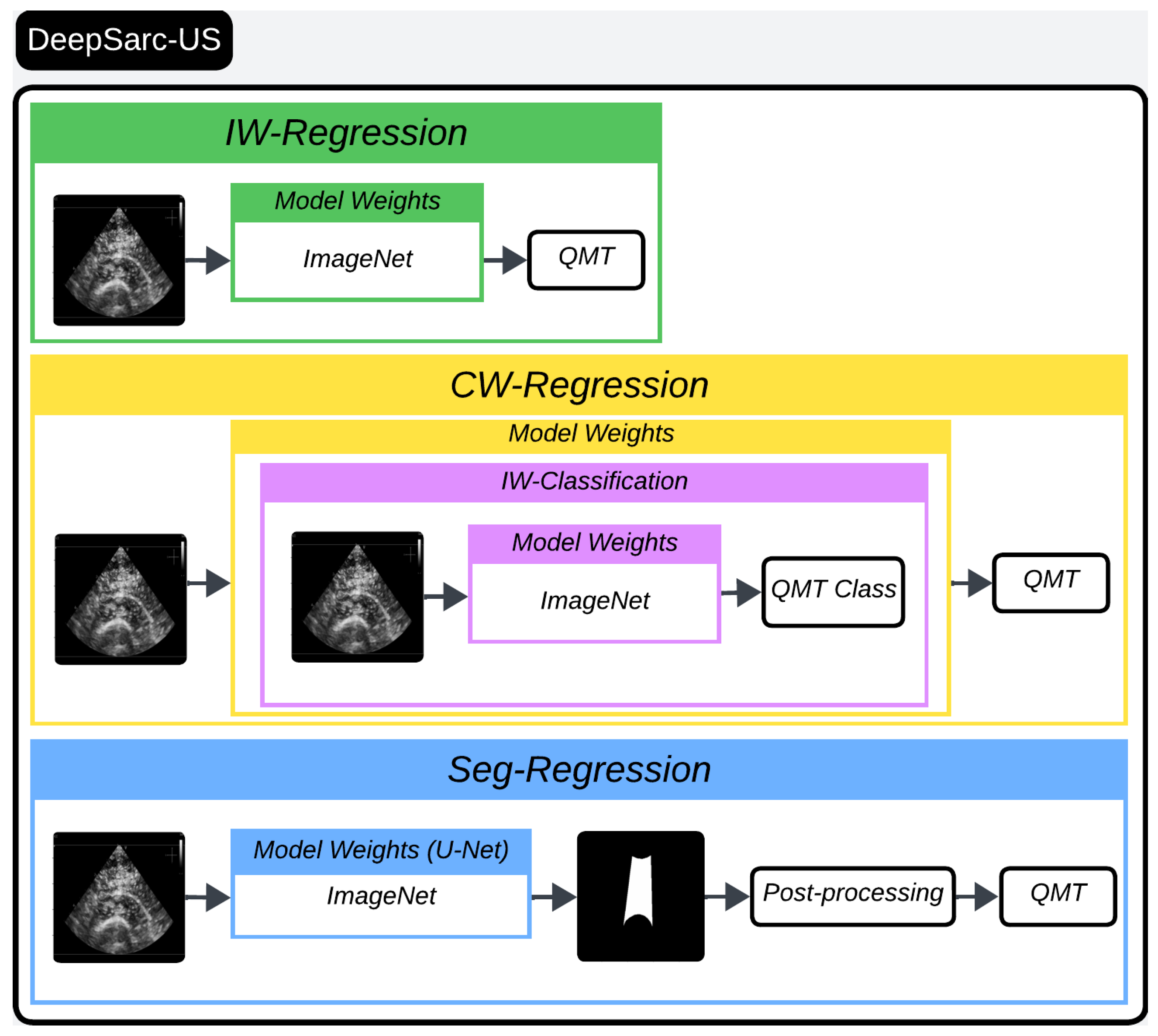
2. Methods
2.1. Dataset
2.2. Experimental Setup
2.2.1. Regression and Classification for QMT Measurements
- Models with transformer and CNN architecture would achieve good results in predicting QMT.
- Regression models with pre-trained weights experimentally derived from classification training runs would outperform the same models with pre-trained weights from ImageNet.
- Classification models with pre-trained weights experimentally derived from regression training runs would outperform the same models with pre-trained weights from ImageNet.
- Activation maps that correctly highlighted the anatomical structures of interest would be more likely to correspond to accurate predictions of QMT.
- Training Regression Models for QMT Estimation:
- Training Classification Models for QMT Estimation and Activation Map Visualization:
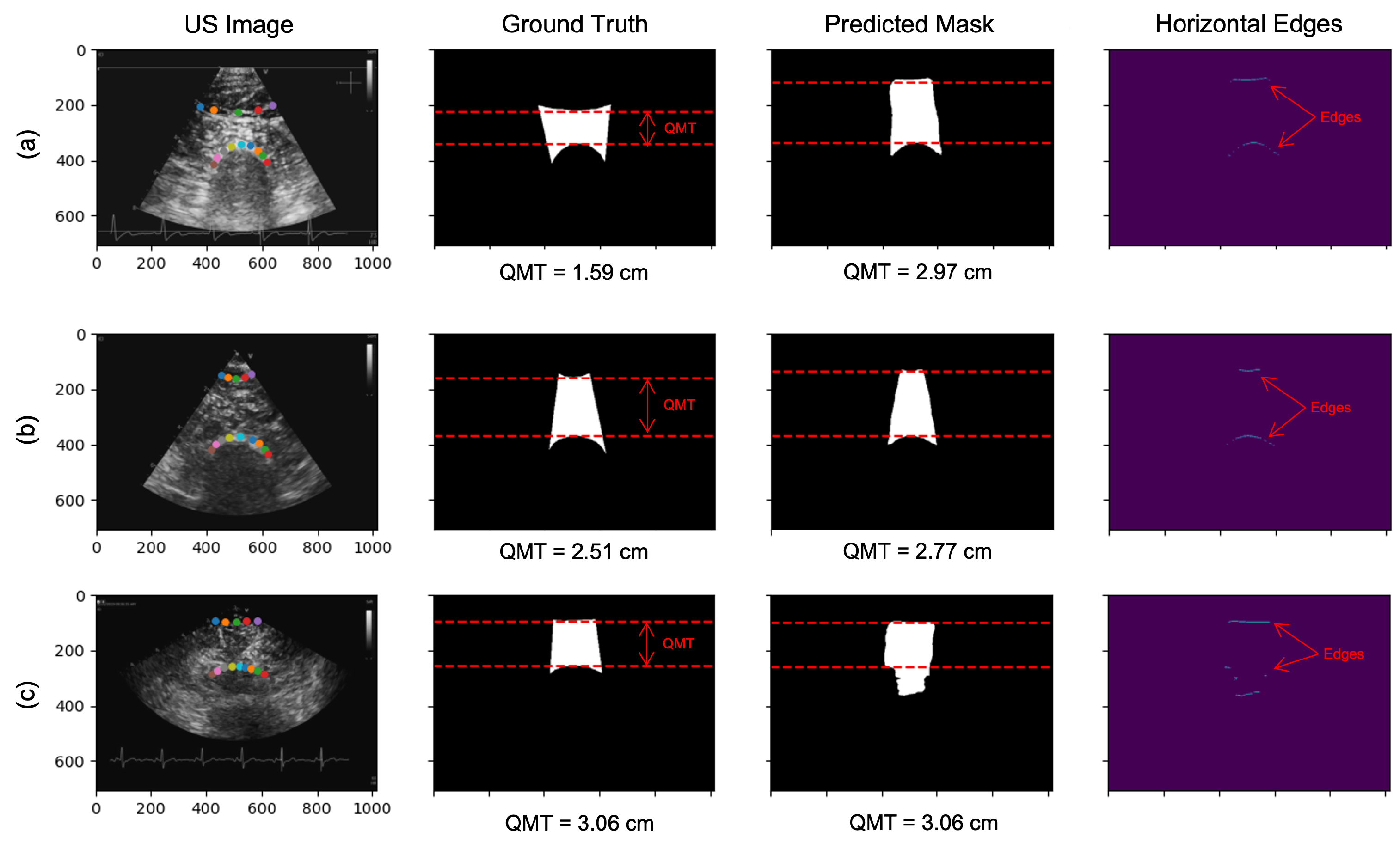
2.2.2. Segmentation for QMT Measurement
3. Results
- IW-Regression: This denotes the regression model utilizing ImageNet pre-trained weights (I referring to ImageNet weights). For instance, IW-Regression ResNet101 signifies the fine-tuned ResNet101 with ImageNet weights specifically tailored for the regression task of QMT.
- IW-Classification: This represents the classification model leveraging ImageNet pre-trained weights. Similarly, IW-Classification ResNet101 signifies the ResNet101 with ImageNet weights fine-tuned for the classification of QMT.
- CW-Regression: This designates the regression model fine-tuned using IW-Classification model weights. For example, CW-Regression ResNet101 is the ResNet101 initially initialized with ImageNet then fine-tuned for the classification of QMT (as denoted by IW-Classification), and subsequently fine-tuned once more for the regression task of QMT.
- RW-Classification: This signifies the classification model fine-tuned using IW-Regression model weights. For example, RW-Classification ResNet101 is the ResNet101 initially initialized with ImageNet and fine-tuned for the regression of QMT (as denoted by IW-Regression) and then further fine-tuned for the classification task of QMT.
- Seg-Regression: This denotes the measurement of QMT through a post-processing step applied to the predicted segmentation masks.
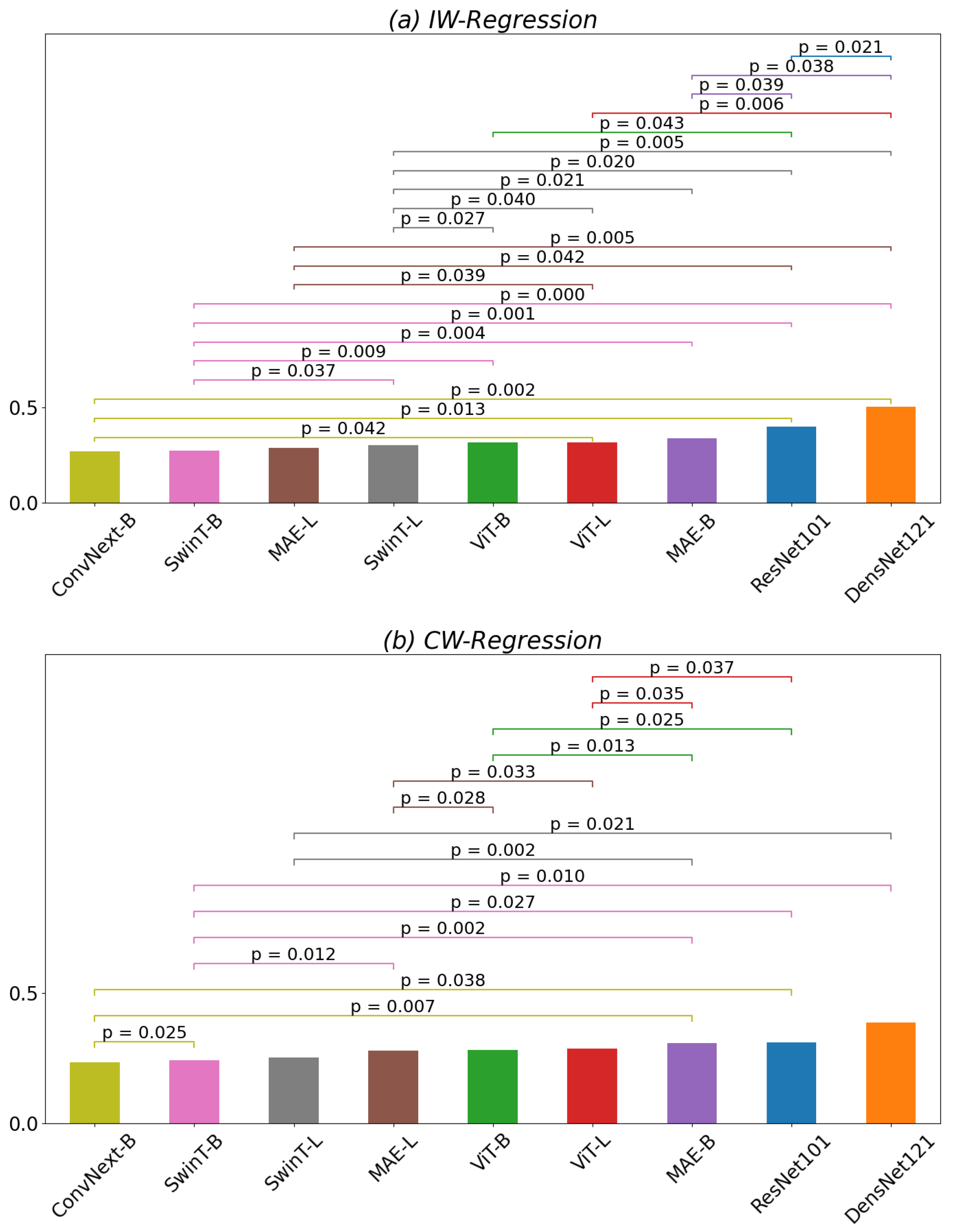
3.1. Regression of QMT
3.2. Classification of QMT
3.3. Segmentation of QMT
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Morley, J.E.; Vellas, B.; Van Kan, G.A.; Anker, S.D.; Bauer, J.M.; Bernabei, R.; Cesari, M.; Chumlea, W.; Doehner, W.; Evans, J.; et al. Frailty consensus: A call to action. J. Am. Med. Dir. Assoc. 2013, 14, 392–397. [Google Scholar] [CrossRef]
- Afilalo, J.; Alexander, K.P.; Mack, M.J.; Maurer, M.S.; Green, P.; Allen, L.A.; Popma, J.J.; Ferrucci, L.; Forman, D.E. Frailty assessment in the cardiovascular care of older adults. J. Am. Coll. Cardiol. 2014, 63, 747–762. [Google Scholar] [CrossRef] [PubMed]
- Hoogendijk, E.O.; Afilalo, J.; Ensrud, K.E.; Kowal, P.; Onder, G.; Fried, L.P. Frailty: Implications for clinical practice and public health. Lancet 2019, 394, 1365–1375. [Google Scholar] [CrossRef] [PubMed]
- Gwyther, H.; Shaw, R.; Dauden, E.A.J.; D’Avanzo, B.; Kurpas, D.; Bujnowska-Fedak, M.; Kujawa, T.; Marcucci, M.; Cano, A.; Holland, C. Understanding frailty: A qualitative study of European healthcare policy-makers’ approaches to frailty screening and management. BMJ Open 2018, 8, e018653. [Google Scholar] [CrossRef]
- Damluji, A.A.; Forman, D.E.; Van Diepen, S.; Alexander, K.P.; Page, R.L.; Hummel, S.L.; Menon, V.; Katz, J.N.; Albert, N.M.; Afilalo, J.; et al. Older adults in the cardiac intensive care unit: Factoring geriatric syndromes in the management, prognosis, and process of care: A scientific statement from the American Heart Association. Circulation 2020, 141, e6–e32. [Google Scholar] [CrossRef] [PubMed]
- Afilalo, J.; Joshi, A.; Mancini, R. If you cannot measure frailty, you cannot improve it. JACC Heart Fail. 2019, 7, 303–305. [Google Scholar] [CrossRef]
- Lee, S.H.; Gong, H.S. Measurement and interpretation of handgrip strength for research on sarcopenia and osteoporosis. J. Bone Metab. 2020, 27, 85. [Google Scholar] [CrossRef]
- Fountotos, R.; Munir, H.; Goldfarb, M.; Lauck, S.; Kim, D.; Perrault, L.; Arora, R.; Moss, E.; Rudski, L.G.; Bendayan, M.; et al. Prognostic value of handgrip strength in older adults undergoing cardiac surgery. Can. J. Cardiol. 2021, 37, 1760–1766. [Google Scholar] [CrossRef]
- Bibas, L.; Saleh, E.; Al-Kharji, S.; Chetrit, J.; Mullie, L.; Cantarovich, M.; Cecere, R.; Giannetti, N.; Afilalo, J. Muscle mass and mortality after cardiac transplantation. Transplantation 2018, 102, 2101–2107. [Google Scholar] [CrossRef]
- Cruz-Jentoft, A.; Michel, J.P. Sarcopenia: A useful paradigm for physical frailty. Eur. Geriatr. Med. 2013, 4, 102–105. [Google Scholar] [CrossRef]
- Zuckerman, J.; Ades, M.; Mullie, L.; Trnkus, A.; Morin, J.F.; Langlois, Y.; Ma, F.; Levental, M.; Morais, J.A.; Afilalo, J. Psoas muscle area and length of stay in older adults undergoing cardiac operations. Ann. Thorac. Surg. 2017, 103, 1498–1504. [Google Scholar] [CrossRef] [PubMed]
- Dodds, R.; Sayer, A.A. Sarcopenia and frailty: New challenges for clinical practice. Clin. Med. 2016, 16, 455. [Google Scholar] [CrossRef] [PubMed]
- Kojima, G.; Iliffe, S.; Walters, K. Frailty index as a predictor of mortality: A systematic review and meta-analysis. Age Ageing 2018, 47, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Church, S.; Rogers, E.; Rockwood, K.; Theou, O. A scoping review of the Clinical Frailty Scale. BMC Geriatr. 2020, 20, 393. [Google Scholar] [CrossRef]
- Blanc-Durand, P.; Schiratti, J.B.; Schutte, K.; Jehanno, P.; Herent, P.; Pigneur, F.; Lucidarme, O.; Benaceur, Y.; Sadate, A.; Luciani, A.; et al. Abdominal musculature segmentation and surface prediction from CT using deep learning for sarcopenia assessment. Diagn. Interv. Imaging 2020, 101, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Joshi, A.; Mancini, R.; Probst, S.; Abikhzer, G.; Langlois, Y.; Morin, J.F.; Rudski, L.G.; Afilalo, J. Sarcopenia in Cardiac Surgery: Dual X-ray Absorptiometry Study from the McGill Frailty Registry. Am. Heart J. 2021, 239, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft, A.J.; Bahat, G.; Bauer, J.; Boirie, Y.; Bruyère, O.; Cederholm, T.; Cooper, C.; Landi, F.; Rolland, Y.; Sayer, A.A.; et al. Sarcopenia: Revised European consensus on definition and diagnosis. Age Ageing 2019, 48, 16–31. [Google Scholar] [CrossRef] [PubMed]
- Wijntjes, J.; van Alfen, N. Muscle ultrasound: Present state and future opportunities. Muscle Nerve 2021, 63, 455–466. [Google Scholar] [CrossRef] [PubMed]
- Stock, M.S.; Thompson, B.J. Echo intensity as an indicator of skeletal muscle quality: Applications, methodology, and future directions. Eur. J. Appl. Physiol. 2021, 121, 369–380. [Google Scholar] [CrossRef]
- Stringer, H.J.; Wilson, D. The role of ultrasound as a diagnostic tool for sarcopenia. J. Frailty Aging 2018, 7, 258–261. [Google Scholar] [CrossRef]
- Bian, P.; Zhang, X.; Liu, R.; Li, H.; Zhang, Q.; Dai, B. Deep-Learning-Based Color Doppler Ultrasound Image Feature in the Diagnosis of Elderly Patients with Chronic Heart Failure Complicated with Sarcopenia. J. Healthc. Eng. 2021, 2021, 2603842. [Google Scholar] [CrossRef] [PubMed]
- Pintelas, E.; Livieris, I.E.; Barotsis, N.; Panayiotakis, G.; Pintelas, P. An Autoencoder Convolutional Neural Network Framework for Sarcopenia Detection Based on Multi-frame Ultrasound Image Slices. In Proceedings of the IFIP International Conference on Artificial Intelligence Applications and Innovations, Crete, Greece, 25–27 June 2021; pp. 209–219. [Google Scholar]
- Sobral, C.; Silva, J.S.; André, A.; Santos, J.B. Sarcopenia Diagnosis: Deep Transfer Learning versus Traditional Machine Learning. 2010. Available online: https://recpad2021.uevora.pt/wp-content/uploads/2020/10/RECPAD_2020_paper_2.pdf (accessed on 23 July 2024).
- Marzola, F.; van Alfen, N.; Doorduin, J.; Meiburger, K.M. Deep learning segmentation of transverse musculoskeletal ultrasound images for neuromuscular disease assessment. Comput. Biol. Med. 2021, 135, 104623. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11976–11986. [Google Scholar]
- Hassanien, M.A.; Singh, V.K.; Puig, D.; Abdel-Nasser, M. Predicting breast tumor malignancy using deep ConvNeXt radiomics and quality-based score pooling in ultrasound sequences. Diagnostics 2022, 12, 1053. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Macruz, F.; Wu, D.; Bridge, C.; McKinney, S.; Al Saud, A.A.; Sharaf, E.; Pely, A.; Danset, P.; Duffy, T.; et al. Point-of-care AI-assisted stepwise ultrasound pneumothorax diagnosis. Phys. Med. Biol. 2023, 68, 205013. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Liu, Y.; Zhao, J.; Wang, R.; Li, C.; Luo, Q.; Shen, C. A novel wavelet-transform-based convolution classification network for cervical lymph node metastasis of papillary thyroid carcinoma in ultrasound images. Comput. Med Imaging Graph. 2023, 109, 102298. [Google Scholar] [CrossRef] [PubMed]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- He, K.; Chen, X.; Xie, S.; Li, Y.; Dollár, P.; Girshick, R. Masked autoencoders are scalable vision learners. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 16000–16009. [Google Scholar]
- Li, J.; Chen, J.; Tang, Y.; Wang, C.; Landman, B.A.; Zhou, S.K. Transforming medical imaging with Transformers? A comparative review of key properties, current progresses, and future perspectives. Med. Image Anal. 2023, 85, 102762. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Hu, H.; Lin, Y.; Yao, Z.; Xie, Z.; Wei, Y.; Ning, J.; Cao, Y.; Zhang, Z.; Dong, L.; et al. Swin transformer v2: Scaling up capacity and resolution. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 12009–12019. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 10012–10022. [Google Scholar]
- Gheflati, B.; Rivaz, H. Vision transformers for classification of breast ultrasound images. In Proceedings of the 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Scotland, UK, 11–15 July 2022; pp. 480–483. [Google Scholar]
- Li, L.; Wu, Z.; Liu, J.; Wang, L.; Jin, Y.; Jiang, P.; Feng, J.; Wu, M. Cross-Attention Based Multi-Scale Feature Fusion Vision Transformer For Breast Ultrasound Image Classification. In Proceedings of the 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Las Vegas, NV, USA, 6–8 December 2022; pp. 1616–1619. [Google Scholar]
- Sun, J.; Wu, B.; Zhao, T.; Gao, L.; Xie, K.; Lin, T.; Sui, J.; Li, X.; Wu, X.; Ni, X. Classification for thyroid nodule using ViT with contrastive learning in ultrasound images. Comput. Biol. Med. 2023, 152, 106444. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, Y.; Liu, P. Automatic Recognition of Standard Liver Sections Based on Vision-Transformer. In Proceedings of the 2022 IEEE 16th International Conference on Anti-Counterfeiting, Security, and Identification (ASID), Xiamen, China, 2–4 December 2022; pp. 1–4. [Google Scholar]
- Liu, X.; Almekkawy, M. Ultrasound Super Resolution using Vision Transformer with Convolution Projection Operation. In Proceedings of the 2022 IEEE International Ultrasonics Symposium (IUS), Venice, Italy, 10–13 October 2022; pp. 1–4. [Google Scholar]
- Muhtaseb, R.; Yaqub, M. EchoCoTr: Estimation of the Left Ventricular Ejection Fraction from Spatiotemporal Echocardiography. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2022: 25th International Conference, Singapore, 18–22 September 2022; Proceedings, Part IV. Springer: Berlin/Heidelberg, Germany, 2022; pp. 370–379. [Google Scholar]
- Xia, M.; Yang, H.; Qu, Y.; Guo, Y.; Zhou, G.; Zhang, F.; Wang, Y. Multilevel structure-preserved GAN for domain adaptation in intravascular ultrasound analysis. Med. Image Anal. 2022, 82, 102614. [Google Scholar] [CrossRef]
- Qu, X.; Lu, H.; Tang, W.; Wang, S.; Zheng, D.; Hou, Y.; Jiang, J. A VGG attention vision transformer network for benign and malignant classification of breast ultrasound images. Med. Phys. 2022, 49, 5787–5798. [Google Scholar] [CrossRef]
- Shen, X.; Wang, L.; Zhao, Y.; Liu, R.; Qian, W.; Ma, H. Dilated transformer: Residual axial attention for breast ultrasound image segmentation. Quant. Imaging Med. Surg. 2022, 12, 4512. [Google Scholar] [CrossRef] [PubMed]
- Liljequist, D.; Elfving, B.; Skavberg Roaldsen, K. Intraclass correlation—A discussion and demonstration of basic features. PLoS ONE 2019, 14, e0219854. [Google Scholar] [CrossRef] [PubMed]
- Koo, T.K.; Li, M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. (IJCV) 2015, 115, 211–252. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Sun, D.; Yang, X.; Liu, M.Y.; Kautz, J. Models matter, so does training: An empirical study of cnns for optical flow estimation. IEEE Trans. Pattern Anal. Mach. Intell. 2019, 42, 1408–1423. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, X.; Su, H.; Zhu, J. A comprehensive survey of continual learning: Theory, method and application. IEEE Trans. Pattern Anal. Mach. Intell. 2024, 46, 5362–5383. [Google Scholar] [CrossRef]
- Ilg, E.; Mayer, N.; Saikia, T.; Keuper, M.; Dosovitskiy, A.; Brox, T. Flownet 2.0: Evolution of optical flow estimation with deep networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 2462–2470. [Google Scholar]
- Kingma, D.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal loss for dense object detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Chefer, H.; Gur, S.; Wolf, L. Transformer interpretability beyond attention visualization. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 19–25 June 2021; pp. 782–791. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Bengio, Y.; Louradour, J.; Collobert, R.; Weston, J. Curriculum learning. In Proceedings of the 26th Annual International Conference on Machine Learning, Montreal, QC, Canada, 14–18 June 2009; pp. 41–48. [Google Scholar]


| QMT Measurement (cm) | ICC (2,1) | 95% CI | ICC (2,3) | 95% CI | |||
|---|---|---|---|---|---|---|---|
| Rater 1 | Rater 2 | Rater 3 | Rater 4 | ||||
| 3.29 ± 1.07 | 2.94 ± 0.86 | 2.99 ± 0.87 | 3.24 ± 1.12 | 0.83 | [0.77–0.87] | 0.95 | [0.93–0.96] |
| Median Absolute Error (cm) | |||
|---|---|---|---|
| Model | IW-Regression | CW-Regression | Delta |
| ResNet101 | 0.40 | 0.30 | |
| DensNet121 | 0.50 | 0.38 | * |
| ConvNext-B | 0.27 | 0.23 | |
| ViT-B | 0.31 | 0.27 | * |
| ViT-L | 0.32 | 0.29 | |
| MAE-B | 0.34 | 0.31 | |
| MAE-L | 0.28 | 0.28 | |
| SwinT-B | 0.27 | 0.24 | * |
| SwinT-L | 0.30 | 0.25 | * |
| All | 0.28 | 0.25 | ** |
| Accuracy (%) | |||
|---|---|---|---|
| Model | IW-Classification | RW-Classification | Delta |
| ResNet101 | 36.99 | 30.14 | * |
| DensNet121 | 39.73 | 38.36 | |
| ConvNext-B | 31.51 | 32.88 | |
| ViT-B | 42.47 | 38.36 | ** |
| ViT-L | 41.10 | 43.84 | |
| MAE-B | 38.36 | 36.99 | |
| MAE-L | 41.10 | 41.10 | |
| SwinT-B | 41.10 | 36.99 | * |
| SwinT-L | 43.84 | 42.47 | |
| All | 43.84 | 41.10 | |
| Median of Absolute Errors (cm) | ||
|---|---|---|
| Seg-Regression | IW-Regression | CW-Regression |
| 0.13 | 0.28 ** | 0.25 ** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Behboodi, B.; Obrand, J.; Afilalo, J.; Rivaz, H. DeepSarc-US: A Deep Learning Framework for Assessing Sarcopenia Using Ultrasound Images. Appl. Sci. 2024, 14, 6726. https://doi.org/10.3390/app14156726
Behboodi B, Obrand J, Afilalo J, Rivaz H. DeepSarc-US: A Deep Learning Framework for Assessing Sarcopenia Using Ultrasound Images. Applied Sciences. 2024; 14(15):6726. https://doi.org/10.3390/app14156726
Chicago/Turabian StyleBehboodi, Bahareh, Jeremy Obrand, Jonathan Afilalo, and Hassan Rivaz. 2024. "DeepSarc-US: A Deep Learning Framework for Assessing Sarcopenia Using Ultrasound Images" Applied Sciences 14, no. 15: 6726. https://doi.org/10.3390/app14156726
APA StyleBehboodi, B., Obrand, J., Afilalo, J., & Rivaz, H. (2024). DeepSarc-US: A Deep Learning Framework for Assessing Sarcopenia Using Ultrasound Images. Applied Sciences, 14(15), 6726. https://doi.org/10.3390/app14156726






