Abstract
Breast cancer is a leading cause of cancer-related deaths among women both worldwide and in Poland. Consequently, ensuring equitable access to diagnostic tests for all populations is crucial, alongside the urgent need to develop new, minimally invasive methods for early cancer detection with a particular focus on metastasis. Long noncoding RNAs (lncRNAs) have emerged as critical regulators of cancer metastasis. This review aims to investigate the potential of lncRNAs as novel biomarkers for breast cancer, focusing on their mechanisms, clinical relevance, and therapeutic implications. A comprehensive literature search was conducted using PubMed and Google Scholar databases, targeting publications from 2013 to 2024. Keywords included “lncRNA”, “biomarker”, “breast cancer”, “metastasis”, “prognosis”, and “diagnosis”. A total of 111 articles were selected based on their relevance and quality. Recent studies have identified numerous lncRNAs such as HOTAIR, MALAT1, BANCR, NEAT1, H19, and GAS5 as key regulators of various metastatic processes in breast cancer. They can be both upregulated and downregulated. Clinical studies have shown that abnormal lncRNA expression correlates with poor prognosis, higher metastatic potential, and therapy resistance in breast cancer patients. LncRNAs have significant potential as novel biomarkers for breast cancer metastasis because of their regulatory roles in metastasis-related processes and detectability in body fluids. Further research is essential to validate these findings in larger clinical studies and to develop lncRNA-based diagnostic and therapeutic tools, ultimately improving patient outcomes in breast cancer.
1. Introduction
1.1. Breast Cancer
Breast cancer (BC) is the most common malignancy in women worldwide and the most frequent cause of cancer-related death. The factors such as the type and size of the tumor, histological grade, lymph node metastasis (LNM), estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2/neu) affect the prognosis and response to the treatment of cancer [1]. Regarding the median overall survival (OS) in accordance with molecular subtypes, patients with luminal A tumors achieve the longest survival with 26–40 months compared with the other subtypes like luminal B (19–32 months), HER2-enriched (8–32 months), and triple-negative tumors (10–22 months). The presence of BC metastasis at the time of diagnosis is strongly associated with the patients’ mortality. The hormone HER-2-targeted therapies are significantly improving the survival of patients with metastatic BC. Nevertheless, early detection of BC metastasis is pivotal to improve patients’ outcomes and provide care at the earliest possible stage [2].
1.2. Epidemiology of Breast Cancer in Poland
BC is the most common malignant tumor among women in Poland. In recent years, there has been an increase in the number of cases due to better diagnostics and lifestyle changes. In Poland, BC mainly affects women over 50. Most BC deaths occur in this age group, reflecting global trends. In 2020, Poland reported 17,511 new cases of BC among women, accounting for 24% of all new female cancer cases. The standardized incidence rate was 84.4 per 100,000 women. BC was also the leading cause of cancer-related deaths in women, with 6.956 fatalities, representing 15% of all female cancer deaths. The standardized mortality rate was 32.9 per 100,000 women [3]. The detection of BC in Poland has improved with the introduction of screening programs in 2006. However, participation in these programs remains relatively low (around 40% in 2010), affecting the effectiveness of early-stage cancer detection. Consequently, advanced-stage cancers are still frequently diagnosed.
1.3. Urban–Rural Differences
Studies show significant disparities in BC mortality and detection between urban and rural areas in Poland. Women in rural areas have less access to diagnostics and treatment, resulting in poorer health outcomes. In 2016, the difference in years of life lost due to BC between urban and rural women was 21%, highlighting the disadvantage faced by rural women. This gap is further exacerbated by socioeconomic factors and lower participation in screening programs. Improved healthcare infrastructure and targeted screening programs in rural areas are essential to reduce these disparities and mortality rates [4,5].
1.4. New Potential Biomarkers
Given the rising incidence of BC both globally and in Poland, it is crucial to address disparities in diagnostic access and more importantly, to identify new biomarkers for early detection, particularly for early metastasis. One promising avenue is the exploration of molecules such as long noncoding RNAs (lncRNAs). Many research studies have revealed that lncRNAs play a pivotal role in the early prediction of BC progression. Each of the processes of the metastasis cascade is regulated by changes in gene expression at epigenetic, transcriptional, and post-transcriptional levels. LncRNA has the ability to regulate multiple signaling pathways associated with BC progression, making it a promising biomarker for early BC metastasis [6]. The objective of this review was to investigate the potential of long noncoding RNAs (lncRNAs) as novel biomarkers for BC, with a focus on their mechanisms, clinical relevance, and therapeutic implications.
2. Review Methods
To conduct a comprehensive review on the role of lncRNAs as biomarkers for BC metastasis, we searched the literature using PubMed and Google Scholar databases. The search included articles published between January 2013 and March 2024. We utilized specific keywords such as “lncRNA”, “biomarker”, “breast cancer”, “metastasis”, “prognosis”, and “diagnosis” to identify relevant studies. Filters were applied to only include articles published in English within the specified date range. We focused mostly on original research articles that discussed lncRNAs as biomarkers for BC metastasis, provided mechanistic insights, clinical correlations, or potential therapeutic implications. Articles not related to BC or metastasis, non-peer-reviewed articles, and editorials, as well as commentaries studies, were excluded. Each selected article was meticulously assessed based on its abstract, which served as the primary criterion for exclusion during the initial screening. Full-text articles were then reviewed to confirm the relevance and quality of the study, with particular attention given to their clinical implications. A total of 111 articles were ultimately selected for inclusion in the review.
3. Molecular and Functional Insights into Breast Cancer Metastasis
3.1. Mutational Background of Breast Cancer
BC is characterized by a heterogeneous and complex mutational landscape that significantly influences its development, progression, and clinical behavior. This landscape encompasses a wide range of genetic alterations, including point mutations, insertions, deletions, copy number variations, and chromosomal rearrangements. These mutations can occur in oncogenes, tumor suppressor genes, and genes involved in DNA repair, contributing to the diversity of BC subtypes and their responses to therapy. One of the most well-studied aspects of BC genetics is the presence of mutations in the BRCA1 and BRCA2 genes, which are crucial for DNA repair through homologous recombination. Germline mutations in these high-penetrance genes significantly increase the risk of developing BC and are associated with hereditary breast and ovarian cancer syndromes. BRCA1/2 mutations often lead to defects in DNA repair mechanisms, resulting in genomic instability and a higher likelihood of accumulating additional mutations that drive tumorigenesis [7]. Mutations in other high-penetrance genes such as TP53, PTEN, PALB2, and CHEK2 also contribute to hereditary breast cancer syndromes, each implicating distinct molecular pathways in tumorigenesis [8]. For example, TP53 mutations, which are common in TNBC, lead to the loss of its function in regulating cell cycle arrest, apoptosis, and genomic stability, thereby promoting cancer cell survival and proliferation [9]. In addition to germline mutations, somatic mutations acquired during the patient’s lifetime further complicate the genomic architecture of BC. These mutations frequently involve key oncogenes and tumor suppressor genes, including PIK3CA, TP53, and HER2, driving oncogenic signaling pathways, genomic instability, and tumor heterogeneity. PIK3CA mutations are prevalent in hormone receptor-positive BC and predict responsiveness to PI3K inhibitors, while HER2 amplifications inform the use of HER2-targeted therapies [10]. The mutational landscape also includes alterations in genes such as GATA3, MAP3K1, CDH1, and PTEN, each contributing to various oncogenic processes. For instance, mutations in GATA3, a transcription factor involved in luminal cell differentiation, are prevalent in the luminal A subtype of BC and are associated with better prognosis. Conversely, PTEN mutations, leading to the activation of the PI3K/AKT pathway, are often found in more aggressive forms of BC and are linked to unfavorable outcomes [11].
LncRNAs play a pivotal role in modulating the effects of genetic mutations and contribute to the complexity of the mutational landscape in BC. These lncRNAs can interact with mutated genes and pathways, influencing their expression and activity. As discussed in subsequent sections of this review, lncRNAs can impact the behavior of mutated oncogenes and tumor suppressor genes, thereby altering the progression and characteristics of BC. This intricate interplay between lncRNAs and genetic mutations underscores the importance of understanding lncRNA functions in the context of BC pathogenesis [12].
3.2. A Brief Biogenesis of lncRNAs
LncRNA is a type of noncoding RNA defined as a molecule longer than 200 nucleotides that does not encode proteins. The length of lncRNA is comparable to that of mRNA, although it contains fewer exons. The biogenesis of lncRNA occurs primarily in the cell nucleus, where it is transcribed by RNA polymerase II (Pol II) from genomic DNA. Once transcribed, lncRNAs undergo various post-transcriptional processing steps, including 5′ capping, splicing, and polyadenylation, similar to protein-coding mRNAs. Some lncRNAs exist in the cytoplasm, but they are not translated [13]. A classification of lncRNAs is shown in Figure 1.
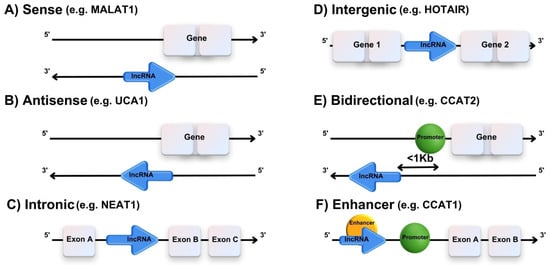
Figure 1.
Classification of lncRNAs based on genomic location. (A) Transcribed from the same DNA strand as neighboring protein-coding genes. (B) Transcribed from the opposite strand, affecting mRNA stability, splicing, or translation. (C) Located within protein-coding gene introns, regulating host or nearby genes through splicing and serving as precursors for small RNAs. (D) Found between genes, acting as epigenetic regulators by modifying chromatin states and interacting with chromatin complexes. (E) Transcribed near a gene’s promoter in the opposite direction, regulating nearby genes by affecting transcription initiation or chromatin structure. (F) Originating from enhancer regions, facilitating enhancer-promoter interactions, transcription, and transcription factor positioning. Image created with Canva Pro, https://www.canva.com/pro/ (accessed on 25 July 2024).
LncRNAs interact with DNA, RNA, or proteins, impacting the structure and function of these molecules. Within the nucleus, they guide chromatin-modifying complexes or transcription factors, while in the cytosol, they regulate mRNA stability or compete for protein expression machinery access. Numerous studies have demonstrated that lncRNA plays a role mainly in the formation, invasion, progression, and metastases of BC, particularly in triple-negative breast cancer (TNBC). Only a small number of lncRNAs are responsible for inhibiting these processes. Furthermore, an increasing number of studies are investigating the relationship between lncRNAs and drug resistance, revealing that lncRNAs play a significant role in carcinogenesis [14].
3.3. Role of lncRNAs Involved in Breast Cancer Metastasis Processes
In the complex process of BC metastasis, the pivotal roles of lncRNAs are highlighted, especially in the dissemination of cancer cells to regional lymph nodes (LNs), which significantly influences disease progression and patient outcomes. At the transcriptional level, lncRNAs serve as decoys, guides, scaffold molecules, and enhancers to modulate gene expression. For instance, Hox Transcript Antisense Intergenic (HOTAIR) acts as a scaffold for Polycomb Repressive Complex 2 (PRC2), facilitating the epigenetic silencing of metastasis-suppressor genes such as E-cadherin. Additionally, guiding lncRNAs like KCNQ1OT1 interact with transcriptional co-regulators to modulate gene transcription. Moreover, scaffolding lncRNA Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) and enhancing lncRNALnc-SLC4A1-140 participate in forming ribonucleoprotein complexes and stabilizing enhancer–promoter interactions, respectively, further influencing gene expression profiles associated with metastasis [15]. At the post-transcriptional level, lncRNAs regulate mRNA stability, mRNA splicing, protein translation, and protein stability. For example, MALAT1 modulates alternative splicing patterns by interacting with splicing factors, while growth arrest-specific 5 (GAS5) stabilizes mRNA molecules by forming RNA duplexes. Furthermore, lncRNA Regulator of Reprogramming (lncRNA-ROR) and lncRNA Interacting partner (lincIN) regulate protein translation, while anti-differentiation noncoding RNA (ANCR) and lncRNA Low Expression in Tumor (lncRNA-LET) contribute to protein stability by facilitating ubiquitination and degradation. Epigenetically, lncRNAs such as HOTAIR and linc-ROR regulate histone modifications and chromatin remodeling, while others like H19 and XIST control DNA methylation, thereby shaping the epigenetic landscape conducive to LNM [13]. The Epithelial–Mesenchymal Transition (EMT) is a pivotal process in BC metastasis, where epithelial cells acquire traits typical of mesenchymal cells, enhancing their ability to migrate and invade surrounding tissues. LncRNAs play a crucial role in regulating genes critical for the EMT by interacting with protein complexes, chromatin-modifying enzymes, and transcription factors. Moreover, they modulate signaling pathways such as Wnt, TGF-β, or NF-κB, which are pivotal for the EMT, thus impacting the metastatic spread of BC cells. Consequently, lncRNAs emerge as key regulators in driving or suppressing the EMT process, influencing the metastatic potential of BC [16].
4. The Role of lncRNAs beyond Metastasis
While this review focuses on the role of lncRNAs in breast cancer metastasis, it is crucial to acknowledge their broader functions that extend beyond metastatic processes. LncRNAs are involved in various cellular activities, including gene expression regulation, epigenetic modifications, and interactions with other noncoding RNAs and proteins, impacting cancer initiation, progression, and treatment response.
4.1. Tumor Environment
LncRNAs profoundly influence the tumor microenvironment (TME) by modulating interactions between cancer cells and surrounding stromal and immune cells. UCA1 activates the Wnt/β-catenin signaling pathway, crucial for creating a supportive milieu for cancer cells [17]. Linc-ROR regulates hypoxia-inducible factors (HIFs), aiding cancer cells in adapting to low-oxygen conditions within the TME, thereby promoting survival and metastasis [18]. HOTAIR enhances the activation of cancer-associated fibroblasts (CAFs), which secrete cytokines and growth factors that drive tumor progression [19]. MALAT1 and BANCR influences the extracellular matrix (ECM) by regulating matrix metalloproteinases (MMPs), enzymes that degrade ECM components, facilitating tumor invasion and metastasis establishment [2].
4.2. Cancer Stem Cell Maintenance
Cancer stem cells (CSCs) are a subpopulation of cancer cells characterized by their ability to self-renew, differentiate, and drive tumorigenesis and metastasis. LncRNAs are crucial in maintaining CSCs through various mechanisms. The aberrant expression of X-inactive specific transcript (XIST) is identified as a key regulator of BC CSCs, significantly impacting their spheroid/colony-forming capacity, tumor growth, and tumor-initiating potential. XIST promotes CSC self-renewal via STAT3 activation and the expression of key CSC factors by functioning as a nuclear sponge for miRNA let-7a-2-3p, thereby enhancing IL-6 production in a paracrine manner [20]. HOTAIR is essential for BC CSC self-renewal and tumor propagation by recruiting the PRC2 protein complex to inhibit IκBα expression, thereby activating NF-κB signaling and promoting c-Myc and Cyclin D1 expression [21]. Another example is the novel long noncoding RNA lnc030, which is crucial for maintaining BC CSC stemness and promoting tumorigenesis. This is achieved by stabilizing squalene epoxidase (SQLE) mRNA through interaction with poly(rC)-binding protein 2 (PCBP2). It enhances cholesterol synthesis, which in turn activates the PI3K/Akt signaling pathway, governing BC CSC properties [22].
4.3. Immune Modulation
LncRNAs were found to modulate the immune response in BC. Shen et al. identified 11 lncRNAs, including LINC01010, LINC00668, and LINC02418, that were associated with immune cell infiltration within the tumor microenvironment. These lncRNAs influenced immune cell behavior, cytokine production, and antigen presentation, thereby affecting tumor progression and patient prognosis [23]. A low expression of another lncRNA Cell leukemia/lymphoma 6 (TCL6) was found to be associated with worse prognosis in PR-negative BC patients and served as an independent factor for poorer survival, particularly in luminal B BC, by regulating immune-related pathways and correlating with various immune infiltrating cells or immune checkpoint molecules [24]. Wang et al. studied the relation between TINCR and the upregulation of PD-L1. The research revealed that TINCR upregulated USP20 expression through the dual mechanism. In the cytoplasm, TINCR functions, through the ceRNA regulatory mechanism, as a sponge for miR-199a-5p, increasing the stability of USP20 mRNA and thereby promoting the expression of PD-L1 by blocking its ubiquitination. In the nucleus, TINCR may attract DNMT1 in order to promote methylation and, as a result, suppress miR-199a-5p transcription. Decreased miR-199a-5p level weakens its inhibition of USP20 mRNA stability and consequently may lead to the upregulation of PD-L1 expression. Researchers emphasize the importance of a potential combination of TINCR knockdown and PD-L1 inhibitor as a target for BC immunotherapy [25].
4.4. Drug Resistance
LncRNAs have emerged as critical players in the development of drug resistance in BC, significantly impacting the efficacy of chemotherapy, hormone therapy, and targeted therapies. For instance, elevated levels of HOTAIR increase ER protein levels and chromatin occupancy, thus potentiating ER downstream gene regulation and promoting tamoxifen-resistant cell growth [26]. In addition, silencing HOTAIR in doxorubicin-resistant BC cells reduces cell proliferation and increases apoptosis by inhibiting the PI3K/AKT/mTOR pathway, suggesting that targeting HOTAIR could be an effective strategy to overcome drug resistance in BC [27]. Furthermore, one of the key mechanisms of chemoresistance associated with MALAT1 is doxorubicin resistance, which mediates by targeting miR-570-3p [28]. The high expression of lncRNA H19 is linked to paclitaxel resistance in ERα-positive BC cells by inhibiting pro-apoptotic genes BIK and NOXA, suggesting that the ERα–H19–BIK signaling axis is crucial in promoting chemoresistance [29]. Another study focused on identifying biomarkers to predict the response to neoadjuvant chemotherapy in patients with locally advanced BC within luminal B-like phenotype. GATA3-AS1 was the only lncRNA that was overexpressed in all nonresponder patients. This study revealed GATA3-AS1 as the first potential molecular biomarker with a use in clinical practice in the prediction of neoadjuvant chemotherapy treatment response in BC [30].
5. Potential lncRNA-Based Biomarkers
Numerous studies have confirmed the pivotal role of lncRNAs in BC metastasis, highlighting their complex mechanisms of action and impact on various signaling pathways. They can be either upregulated or downregulated, as shown in Figure 2, and serve as promising markers for metastasis. Below, we present the most significant ones.
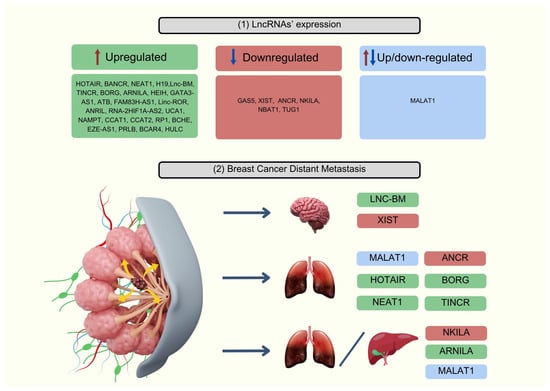
Figure 2.
(1) Expression of potential lncRNA-based biomarkers for breast cancer and (2) their association with distant metastasis. Image created with Canva Pro, https://www.canva.com/pro/ (accessed on 25 July 2024).
5.1. HOTAIR
HOTAIR is a 2.158-nucleotide-long lncRNA expressed from the HOXC locus on chromosome 12q13.13. The role of HOTAIR in normal development is multifaceted, encompassing its involvement in gene regulation, scaffold formation, and cell cycle progression. Numerous studies have confirmed a strong association between HOTAIR expression and metastasis. They revealed an overexpression of HOTAIR within BC cells and tumor tissues, closely linked to both cell proliferation and the dissemination of cancer cells. HOTAIR emerged as a pioneering lncRNA identified to facilitate tumor progression and linked with unfavorable outcomes in BC cases [31].
Sørensen et al. demonstrated that HOTAIR expression showed a bimodal distribution in primary BC, with metastatic BC patients displaying higher levels of HOTAIR. Additionally, individuals with elevated HOTAIR level had worse survival outcomes compared to those with low expression, particularly among ER-positive patients. These findings suggested that HOTAIR expression might serve as an independent biomarker for predicting metastasis risk in ER-positive BC patients but not in ER-negative BC [6]. On the contrary, a study by Gökmen-Polar et al. demonstrated that elevated HOTAIR expression correlated with metastatic progression, particularly in ER-negative BC. However, its prognostic relevance appeared restricted to ER-negative, LN-positive tumors. The observed differences might stem from variations in cohort sizes, the ratio of ER-positive to ER-negative patients, and the endpoints selected for both analyses [32]. Collina et al. assessed the in situ expression of HOTAIR in numerous case series of patients with TNBC by employing ISH techniques with an RNA probe. It revealed a strong correlation between elevated HOTAIR levels and the metastasis of tumor cells to the LNs. Furthermore, they identified a direct association between HOTAIR expression and androgen receptor (AR) expression, suggesting a potential role of HOTAIR in regulating the AR pathway in TNBC [14]. The potential of HOTAIR as an indicator of BC metastasis was also confirmed by the study conducted by Tao et al., wherein its expression was significantly elevated both in peripheral blood mononuclear and cancerous tissue samples from patients with BC migration to the LNs. The results also suggested that G protein-Coupled Estrogen Receptor (GPER) regulates E2-induced HOTAIR levels in BC cells, and E2/GPER enhances these levels through miR-148a. Hence, in ER-negative BC, E2-ER increases HOTAIR by binding to ERE in its promoter, while in TNBC, E2-GPER boosts HOTAIR by inhibiting miR-148a, which recognizes the sequence within HOTAIR [33]. Zheng et.al found that HOTAIR expression in plasma significantly correlated with ER level and LNM. Following surgery, patients exhibited a marked decrease in HOTAIR levels, which moderately correlated with its expression in tumor tissues. Notably, plasma HOTAIR demonstrated superior diagnostic sensitivity and specificity compared to Carcinoembryonic Antigen (CEA) and Carbohydrate Antigen 153 (CA153); meanwhile, combining these three markers allowed for increased diagnostic power [34]. The integration of various studies highlights HOTAIR’s significant role in promoting BC progression through multiple pathways, particularly EMT regulation, critical for metastasis to LNs and other organs. Elevated levels of HOTAIR triggered by TGF-β1 stimulate the EMT, whereas its suppression through siRNA prevents this transition and reduces the colony-forming ability of colon and BC cells [9]. The activity of various miRNAs, particularly in BC, significantly influences the modulation of the EMT process and subsequent metastasis induction in BC. HOTAIR serves as a mediator of hypoxia-induced metastasis, promoting the formation of vasculogenic mimicry and enhancing TNBC cell migration. Hypoxia increases HOTAIR expression, but its siRNA-mediated silencing prevents the development of hypoxia-induced vasculogenic mimicry and increases miR-204 levels, impacting cell migration. Additionally, the knockdown of HOTAIR alters cytoskeletal organization by reducing Focal Adhesion Kinase (FAK) expression. In BC stem cells derived from MCF-7 and MB-231 cell lines, HOTAIR indirectly suppresses miR-7, resulting in increased expression of SETDB1, STAT3, c-Myc, twist, and miR-9. This cascade of events leads to the downregulation of E-cadherin, promoting the EMT [35].
5.2. MALAT1
Following HOTAIR, one of the most extensively studied lncRNAs in BC is MALAT1. It is located on chromosome 11q13.1 and was originally identified as a transcript associated with lung cancer metastasis but was later found to be overexpressed in other types of cancers as well, including BC. MALAT-1 regulates gene expression by interacting with gene ends and participating in active transcriptional elongation. It also influences pre-mRNA alternative splicing through the control of serine/arginine splicing factor phosphorylation and acts as a post-transcriptional regulator through a competing endogenous RNA (ceRNA) mechanism. By using miRNA-responsive elements, MALAT-1 communicates with mRNAs and pseudogenes, leading to changes in cell behavior like invasion and metastasis. Another mechanism involves BC exosomes promoting cell proliferation, with MALAT1 inducing cell proliferation through exosomes [36]. The crucial role of MALAT1 expression in the EMT process has been confirmed in several studies. Wang et al. observed a reciprocal repression between MALAT1 and miR-204, with Zinc Finger E-Box Binding Homeobox 2 (ZEB2) identified as a downstream target of miR-204, mediating MALAT1’s function through the miR-204/ZEB2 axis. ZEB2 can trigger the EMT by directly suppressing E-cadherin. Moreover, higher MALAT1 expression correlated with poorer overall survival, especially in patients with advanced tumors [16].
Huang et al. showed that reducing MALAT1 expression in MCF-7 cells significantly inhibited proliferation, migration, and tube formation. They also found a negative correlation between miR-145 and MALAT1 expression in BC tissues. Furthermore, the knockdown of MALAT1 significantly decreased the expression of Vascular Endothelial Growth Factor (VEGF) [37]. Despite the consistent elevation of MALAT1 levels in the luminal subtype and relatively lower levels in TNBC subtypes, Jadaliha et al. demonstrated that MALAT1 promoted proliferation, progression, and metastasis in TNBC as well. Elevated MALAT1 expression was associated with decreased survival in ER-negative, LN-negative patients of the HER2 and TNBC subtypes, with MALAT1 showing independent prognostic significance in TNBC LN-negative patients. These findings suggest the potential of MALAT1 as a predictive marker for metastatic disease onset in LN-negative patients considered low risk for metastasis [38]. Analyzing samples from 240 patients who were histopathologically diagnosed with TNBC, Ou et al. discovered that BTB and CNC Homology 1 (BACH1) and MALAT1 were simultaneously upregulated in cancerous tissues. Individuals with elevated expression levels of MALAT1 and BACH1 displayed characteristics such as larger tumor size, LNM, and advanced TNM. These findings suggest that MALAT1 and BACH1 may play critical roles and serve as potential markers in the carcinogenesis and progression of TNBC [39]. The expression of MALAT1 in triple-negative and HER-2-positive BC positively correlated with the number of metastatic LNs. Furthermore, MALAT1 emerged as an independent prognostic factor for these metastases. It promoted proliferation and invasion via the X-box Binding Protein 1(XBP1)-hypoxia-inducible factor-1α (HIF) pathway in MDA-MB-231 and the HER-2 pathway in MDA-MD-435 [40]. In a study of Miao et al., MALAT1 expression was significantly upregulated in 85.9% of BC tissues collected from 78 patients compared to normal tissues, and its elevation was associated with LNM and adverse 5-year disease-free survival (DFS). Furthermore, serum MALAT1 levels were significantly higher in BC patients compared to those with benign breast disease and showed satisfactory diagnostic efficacy with an AUC of 0.833 [41].
Contrary to the above findings, there are also studies examining the role of MALAT1 not as a promoter but as a suppressor of metastasis in BC. Kim et al. discovered that MALAT1 overexpression inhibited BC metastasis, while MALAT1 deficiency induced them, reversible by reintroducing MALAT1. MALAT1 inhibited the TEA domain family’s transcriptional activity by sequestering the transcription factor. Their rigorous study addressed previous discrepancies by using precise methods, such as a transcriptional terminator insertion to inactivate MALAT1 without affecting neighboring genes [42]. In another study, the researchers investigated the impact of Phosphatase and Tensin Homolog (PTEN) dysregulation on the noncoding transcriptome, identifying MALAT1 as a downstream target of PTEN across both human and mouse models. They characterized a PTEN–miRNA–MALAT1 axis that regulates tumorigenesis, demonstrating that MALAT1 possesses novel tumor-suppressive properties in colon and BC. PTEN-mediated regulation of MALAT1 relies on miRNAs, particularly miR-17 and 20a, which influence the expression of pro-migratory genes like Epithelial Cell Adhesion Molecule (EpCAM) and Integrin Subunit Beta 4 (ITGB4) [43]. Xu et al. revealed that MALAT was downregulated in both tumorous tissue samples and BC cell lines (BT549, MCF7, MDA-MB-453, and MDA-MB-231). They observed that MALAT1 knockdown induced the EMT in BC cells via the PI3K–AKT pathway, leading to enhanced migration and invasion. Furthermore, a lower expression of MALAT1 was strongly associated with axillary LNM and shorter metastatic relapse-free survival [44]. The discrepancies in reports regarding MALAT1 in BC may stem from variations in experimental conditions, sample sizes, patient populations, and the complex expression pattern of various transcript variants of MALAT1. There is a need for large, multicenter studies to validate its role comprehensively. Nonetheless, MALAT1 remains a promising biomarker for BC progression [45].
5.3. BANCR
An important role in metastasis has also been demonstrated for the Abnormally Expressed BRAF-activated Noncoding RNA (BANCR), which spans 693 base pairs and is situated on chromosome 9. It was initially found in melanoma cells in 2012. Subsequently, its pro-oncogenic role was further elucidated in lung cancer, hepatocellular carcinoma, and colorectal cancer. In a study of Lou et al., the expression of BANCR was found to be significantly elevated in BC tissues from 65 patients compared to normal tissues. Furthermore, its higher levels showed significant correlations with TNM staging, size, and LNM. DFS and overall survival were longer in a group of patients with lower BANCR’s level. The knockdown of BANCR led to the inhibition of proliferation in vitro, promoted apoptosis, and suppressed the migration and invasion capacities of MCF-7 cells. Moreover, reducing BANCR expression hindered the EMT process and the expression of matrix metalloproteinase-2 (MMP-2) and matrix metalloproteinase-9 (MMP-9) [2]. Similar results were obtained in the study by Jiang et al., which suggests that BANCR might serve as a promising prognostic marker of BC due to its contribution in clinical progression, proliferation, and metastasis [46].
5.4. NEAT1
Another lncRNA contributing to the intricate landscape of BC is Nuclear-Enriched Abundant Transcript 1 (NEAT1), which plays a role in the initiation and advancement of various cancer types. Research conducted by Jiang et al. showed a notable increase in NEAT1 expression in human BC cells, including MCF-7, MDA-MB-453, MDA-MB-231, and SKBR3 cells, when compared to normal mammary epithelial cells such as MCF-10A. However, the isoforms NEAT1_1 and NEAT1_2 displayed differing expression patterns across various intrinsic subtypes of BC. While NEAT1_2 demonstrated the highest expression levels in HER2-positive and luminal B cancers, NEAT1_1 expression peaked in ER-positive luminal A and luminal B cancers [47].
Shin et al. observed an elevated expression of NEAT1 in blood samples from BC patients, with the highest levels detected in the TNBC subgroup [48]. A strong correlation was demonstrated between NEAT1 overexpression in BC tissues and LNM, TNM staging, and tumor size. Elevated NEAT1 expression was more commonly detected in M1 and III–IV tumors compared to M0 and I–II tumors [49]. A study by Li et al. elucidated the role of Forkhead Box N3 (FOXN3) as a transcriptional repressor associated with the Switch-Insensitive 3A (SIN3A) complex in ER-positive BC cells. NEAT1 emerged as a crucial mediator of FOXN3–SIN3A interactions, contributing to the repression of genes involved in the EMT and promoting BC metastasis. The researchers proved that the overexpression of NEAT1 was strongly correlated with LN positivity among ER+ BC patients [50]. An alternative mechanism through which NEAT1 promotes the EMT involves targeting the miR-211/High-Mobility Group AT-Hook 2 (HMGA2) axis. Given the pivotal role of HMGA2 in the EMT, it is hypothesized that NEAT1 contributes to cellular metastasis by modulating the miR-211/HMGA2 axis [49]. The study of Lo et al. reveals NEAT1’s oncogenic role in BRCA1-related breast cancer, involving its regulation of miR-129-5p and WNT4. NEAT1 overexpression due to BRCA1 deficiency promotes breast tumorigenicity by epigenetically silencing miR-129-5p, leading to increased WNT4 expression and activation of oncogenic signaling [51]. Choudhry et al. highlighted the crucial role of NEAT1 in BC progression under hypoxic conditions, mediated by hypoxia-inducible factor 2 (HIF-2) pathways. NEAT1 induction in hypoxia promotes paraspeckle formation which stores RNA in the nucleus, accelerates tumorigenesis, and correlates with worse survival in BC patients [52].
5.5. H19
The involvement of lncRNA H19 in BC cancer metastasis further underscores the diverse roles of lncRNAs in tumor progression. As a transcript of the H19 gene, located close to the telomeric region of chromosome 11p15.5, H19 undergoes reciprocal imprinting, meaning its expression is regulated in coordination with its neighboring gene of Insulin-like Growth Factor 2 (IGF2). H19 mostly plays a dual role: it acts as a storage site for miR-675, which suppresses specific targets, and it also binds to miRNAs or proteins. However, its role varies across different types of cancers [53].
Lin et al. compared two Single-Nucleotide Polymorphisms (SNPs) in the lncRNA H19, rs217727, and rs2839698 and found that elevated levels of the former were associated with an increased risk of BC, particularly in patients with ER+, HER2-, and ER+/HER2-subtypes [54]. H19 promotes the G1-S cell cycle transition in BC cells by binding to the E2F transcription factor 1 (E2F1). H19 binds to and inhibits S-adenosylhomocysteine hydrolase (SAHH), impacting broad epigenetic alterations and suggesting its involvement in gene methylation dynamics in development and diseases, including BC. Moreover, the overexpression of miR-675-5p in MDA-MB-231 cells downregulates ubiquitin ligase E3 family proteins (c-Cbl and Cbl-b), activating epidermal growth factor receptor (EGFR) and c-Met pathways, thereby enhancing proliferation and metastasis abilities in BC [55]. In a study by Si et al., H19 expression in BC tissue samples was notably higher in cases with LNM compared to those without, and it was also elevated in grades III–IV compared to grades I–II [56]. Zhang et al. observed elevated expression levels of H19 in both BC tissues and plasma samples compared to healthy controls. Plasma H19 levels correlated significantly with ER, PR, HER-2, and LNM. The AUC was estimated to be 0.810 (sensitivity 56.7% and specificity, 86.7%), which exceeded the values obtained for CEA and CA153. Furthermore, postoperative plasma H19 levels were significantly reduced compared to preoperative levels [57]. In another study, similar results showed a plasma H19 AUC of 0.870 (with a sensitivity of 87.0% and specificity of 70.6%). Exosomal H19 expression levels correlated with LNM, distant metastasis, TNM stages, ER, PR, and HER-2 and were diminished post-operation. These findings establish H19 as a promising biomarker for BC metastasis [58].
5.6. GAS5
Further contributing to the diverse roles of lncRNAs in BC and another potential biomarker is GAS5, a member of the 5’-terminal oligopyrimidine (5’-TOP) gene family, located at 1q25.1. It consists of 12 exons and contains a brief open reading frame devoid of protein-coding capacity. It undergoes alternative splicing, resulting in two mature forms of lncRNA: GAS5a and GAS5. The downregulation of its expression was found in various types of neoplasms such as prostatic, lung, gastric cancer, and gliomas [59]. Significantly lower levels of GAS5 were also detected in BC.
In a study by Li et al., GAS5 showed reduced expression in trastuzumab-resistant SKBR-3/Tr cells and BC tissues from patients treated with trastuzumab, leading to increased cell proliferation and counteracting the inhibitory effects of lapatinib on proliferation. GAS5 was found to hinder the growth and invasion of BC by trapping oncogenic miR-21 and triggering the activation of its targets, including PTEN and Programmed Cell Death Protein 4 (PDCD4) [60]. GAS5 also interacts with miR-196a-5p to suppress TNBC progression, while miR-196a-5p targets Forkhead Box Protein O1 (FOXO1), a pivotal regulator of tumor growth and invasion. Notably, FOXO1 also enhances GAS5 transcription, forming a positive feedback loop [61]. Furthermore, decreased levels of lncRNA-GAS5 facilitate BC metastasis to the lungs through the induction of the EMT process. The expression of GAS5 has a significant, yet not fully understood, impact on the processes of autophagy and apoptosis, which are crucial in the metastasis and treatment resistance of cancer cells. GAS5 has the potential to trigger apoptosis in TNBC cells by modulating the miR-378a-5p/Suppressor of Fused (SUFU) signaling pathway [62]. It was found to positively correlate with unc-51-like autophagy activating kinase (ULK) 1/2, key regulators of autophagy. Overexpressing GAS5 inhibited cell proliferation, invasion, and tumor formation in BC cells, partially through autophagy induction [63]. Based on tissue samples obtained from 103 TNBC patients, Li et al. observed a significant decrease in GAS5 levels, which correlated with an aggressive tumor phenotype, including an advanced clinical stage, LNM, and poorer overall survival [61]. In research conducted by Han et al., plasma RNA was extracted from preoperative (n = 90) and postoperative (n = 39) BC patients, along with healthy controls (n = 76). While no significant differences were found between BC patients and controls in GAS5, it correlated with the Ki67 proliferation index in preoperative patients. Postoperatively, GAS5 and H19 levels significantly decreased in 71.8% cases, with lower GAS5 levels associated with high Ki67 proliferation index pre-surgery and positive LNM post-surgery [64].
On the contrary, Toraih et al. found that GAS5 expression was reduced in the serum of BC patients compared to both healthy individuals and non-cancer patients with a predisposition to BC. Moreover, decreased GAS5 levels were linked to unfavorable prognostic indicators such as metastasis and recurrence. The study findings revealed that GAS5 demonstrated a sensitivity of 72.5% and specificity of 78.5%, effectively distinguishing between normal and at-risk individuals [65]. Based on the discussions, GAS5 emerges as a potential marker for BC metastasis due to its downregulation in TNBC tissues and its association with aggressive tumor phenotypes [66]. Further research is needed to elucidate its mechanisms of action and validate its efficacy across diverse patient cohorts [12].
5.7. HEIH
Adding to the understanding of lncRNA functions in BC is High Expression in Hepatocellular Carcinoma (HEIH), which was found to be upregulated in BC, especially in the TNBC subtype. Nafea et al. identified HEIH as a promoter of TNBC progression by inhibiting miR-939-5p, thereby increasing Inducible Nitric Oxide Synthase (NOS2) production. Silencing HEIH enhances the tumor’s immunogenic profile by inducing MHC class I polypeptide-related sequence A/B (MICA/B) and suppressing PD-L1, IL-10, and TNF-α. Additionally, higher levels of HEIH were observed in tumor samples with a higher expression of Ki-67 (>14%), compared to those with low Ki-67 (<14%) [67]. Furthermore, TNBC cells’ proliferation and inhibition of apoptosis were also determined through the miR-4458/SOCS1 axis, with higher HEIH levels associated with larger tumor size and advanced clinical stage [68]. HEIH promotes BC development by regulating miR-200b and activating the Wnt/β-catenin pathway, with its suppression reducing cell viability, migration, and invasion while promoting apoptosis in BC cells [69]. High expression levels of HEIH were strongly correlated with the TNM stage, LNM, and served as an independent indicator of 3-year OS among 160 patients in a study by Chen et al. [70]. Those insights indicate that HEIH is a key factor promoting BC development and metastasis, suggesting the potential use of HEIH both as a biomarker and therapeutic target in BC management.
5.8. TINCR
The terminal differentiation-induced noncoding RNA (TINCR) is a novel epithelial lncRNA associated with tumor proliferation and metastasis [71]. TINCR is highly expressed during and required for epidermal differentiation [72]. Wang et al. examined the role of TINCR in BC. Their study showed that TINCR was correlated with poor prognosis and promoted tumor growth both in vitro and in vivo. An investigation of 120 BC samples revealed that the EGFR level was higher in tissues with high TINCR expression than in low-TINCR-expression tissues, with elevated levels in advanced LNM cases. Since the way lncRNA regulates EGFR expression remained unclear, further studies were performed. The suppression of TINCR caused a decrease in p-JAK2, STAT3, and p-STAT3 relative to their levels in control cells. TINCR is considered to be present in both the nucleus and the cytoplasm of BC cells. The cytoplasm TINCR behaves like ceRNAs and upregulates the EGFR expression by acting like a molecular sponge for miR-503-5p, whereas the nuclear TINCR can regulate the gene expression by functioning as the epigenetic modulators. This investigation also revealed that the regulation of the methylation of miR-503-5p loci and its transcription can be performed by TINCR-recruiting DNA (cytosine-5)-methyltransferase 1 (DNMT1) [73].
5.9. GATA3-AS1
The involvement of GATA binding protein 3 antisense RNA 1 (GATA3-AS1) in BC highlights its potential as a regulatory lncRNA. GATA3-AS1 is a divergent lncRNA gene neighboring GATA3, a regulator of TH2 lineage commitment that enables TH2 cells to transcribe genes encoding cytokines IL-4, IL-5, and IL-13 [74]. GATA3 seems to suppress the breast tumor progression [75]. Zhang et al. demonstrated that GATA3-AS1 was significantly overexpressed in TNBC tissues. Elevated levels of GATA3-AS1 were associated with a larger tumor size, LNM, and advanced TNM stage. The research showed that this lncRNA could inhibit PD-L1 ubiquitination by upregulating COPS5 and thus stabilize the PD-L1 protein. Knowing the upstream molecular mechanism of PD-L1 in TNBC and exploiting the fact that both PD-L1 and GATA3-AS1 are overexpressed in this BC type, this combination remains not only a novel biomarker but also a possible strategy for the immunotherapy of TNBC [76].
5.10. ATB
Another promising lncRNA-based biomarker for BC is Activator of TGF-β (ATB). It is upregulated in BC, particularly in the luminal B, Her-2, and TNBC subtypes. It facilitates the EMT through miR-141-3p suppression or by enhancing the miR-200c/Twist1 pathway [77,78]. Zhayu et al. proved that the levels of ATB and protein kinase Nek9 in serum were significantly associated with the progression of BC, especially the LNM and TNM stages [79]. Moreover, the study by El-Ashmawy et al. demonstrates that serum lncRNA-ATB serves as a superior non-invasive biomarker for the early diagnosis of BC compared to the conventional marker CA15-3 (AUC of 0.844 and 0.738, respectively), with better performance in distinguishing early-stage disease. Additionally, another lncRNA, FAM83H-AS1, correlates with advanced tumor characteristics, including the TNM stage and LNM [80]. A similar study revealed that elevated lncATB levels in BC patients correlated with increased rates of distant metastasis, greater LN involvement, and elevated Ki67 levels, indicating a more aggressive cancer behavior. Additionally, patients with higher lncATB expression experienced worse clinical outcomes, including reduced DFS and OS [78]. These findings highlight lncATB’s potential as a marker for assessing disease severity and predicting patient prognosis in BC.
5.11. Other Potential lncRNA-Based Biomarkers Involved in the Metastatic Progression of Breast Cancer
In addition to the aforementioned lncRNAs, several other lncRNAs have been identified as potential biomarkers that may play significant roles in the metastatic progression of BC. These emerging lncRNAs add further complexity to our understanding of BC metastasis and highlight the diverse mechanisms through which lncRNAs can influence tumor behavior.
One such lncRNA is XIST, which plays a crucial role in the development of brain metastasis in BC by affecting both tumor cells and the tumor microenvironment. XIST downregulation enhances brain metastatic growth and immune suppression, while fludarabine has been identified as a potential therapeutic agent that specifically targets XIST low tumor cells and effectively blocks brain metastasis [81]. Another lncRNA, BM, promotes brain metastases in BC by enhancing the JAK2 kinase activity and STAT3 signaling. This pathway facilitates vascular co-option and macrophage recruitment, which further supports metastasis [82]. ANCR regulates EZH2 stability by enhancing its interaction with CDK1, leading to EZH2 phosphorylation, ubiquitination, and degradation. ANCR’s reduction promotes BC progression and metastasis, while its overexpression suppresses tumor formation and lung metastasis [83]. Moreover, BMP/OP-Responsive Gene (BORG) drives aggressive behaviors and metastasis in BC by enhancing breast cancer stem cell (BCSC) traits through interaction with the E3 SUMO ligase TRIM28. A high BORG expression correlates with increased metastatic potential and poor outcomes, while inhibiting BORG reduces metastasis and impacts BCSC phenotypes [84,85].
It was found that nuclear factor-κB interacting lncRNA (NKILA) expression was significantly reduced in invasive and metastatic breast cancer compared to benign and ductal carcinoma in situ tissues. Lower NKILA levels are associated with advanced disease stages, higher histopathological grades, and poorer patient survival [86]. NKILA’s role in modulating the TGF-β-induced EMT and its inverse relationship with miR-103 and miR-107 suggest that NKILA could serve as a valuable independent prognostic marker for BC progression [87].
In a study by Yang et al., Androgen receptor negatively induced lncRNA (ARNILA) was associated with dismal PFS in TNBC. ARNILA promotes the EMT, invasion, and metastasis to lung and liver by acting as a ceRNA for miR-204, thereby upregulating Sox4 and contributing to TNBC progression [88].
Another study revealed that linc-ROR was upregulated in BC and drove the EMT process, enhancing cancer cell migration, invasion, and lung metastasis in vivo. It functioned as a ceRNA for miR-205, stabilizing EMT-related genes like ZEB2 [89]. Furthermore, a significant difference was detected between the levels of linc-ROR expression and BC LNM [90].
The research by Liu et al. identified that the plasma levels of lncRNAs Antisense noncoding RNA in the INK4 locus (ANRIL), hypoxia-inducible factor 1 alpha antisense (RNA-2HIF1A-AS2), and urothelial carcinoma-associated 1 (UCA1) were significantly higher in patients with TNBC compared to those with non-TNBC and healthy controls. These lncRNAs showed strong diagnostic capabilities for TNBC, with AUC values of 0.785, 0.739, and 0.817, respectively. Elevated levels of these lncRNAs were also linked to increased LNM. The combined TNBCSigLnc-3 score, incorporating ANRIL, HIF1A-AS2, and UCA1, demonstrated the highest diagnostic accuracy with an AUC of 0.934, underscoring their potential as biomarkers for TNBC diagnosis and progression monitoring [91]. Similarly, another study revealed that UCA 1 and AC026904.1 acted synergistically to amplify TGF-β-induced activation of SNAI2, thereby promoting metastatic progression through the EMT process [92]. Moreover, the upregulation of Nicotinamide phosphoribosyltransferase (NAMPT-AS) in TNBC correlates with poor prognosis, larger tumor size, LN involvement, metastasis, and advanced stage. NAMPT-AS promotes tumor progression by epigenetically regulating NAMPT and affecting the mTOR pathway [93].
The involvement of Colon Cancer-Associated Transcript 1 (CCAT1) and CCAT2 in the oncogenesis of TNBC has also been documented. CCAT1 promotes progression by downregulating miR-218 and upregulating Zinc Finger Protein X (ZFX) and serves as an independent predictor of decreased OS and PFS, while CCAT2 upregulates OCT4-PG1 and activates the Notch2 signaling pathway through inhibition of miR-205 [94,95,96]. RP1-5O65, a novel lncRNA, is highly expressed in BC and is associated with poor prognosis, promoting tumor proliferation and metastasis by repressing p27kip1 protein expression and maintaining the EMT and stemness states in cancer cells both in vivo and in vitro [97]. Based on the analysis of 56 BC specimens, another lncRNA, BCHE, was found to have a strong correlation with LNM [98].
Expression of Neuroblastoma-associated transcript 1 (NBAT1), which is downregulated in BC, correlates with LNM and unfavorable patient prognosis. Mechanistically, NBAT1 interacts with the PRC2 complex member EZH2 to modulate gene expression, specifically affecting the WNT signaling pathway inhibitor DKK1 [99]. Another lncRNA downregulated in BC tissues is Taurine-upregulated gene 1 (TUG1), which is downregulated in BC tissues, with its reduced levels strongly correlating LNM with the presence of the tumor suppressor p53 [100].
Ezrin Antisense RNA 1 (EZR-AS1) is upregulated in BC and acts as an oncogene by activating the Wnt/β-catenin signaling pathway, which is involved in regulating cell proliferation and survival [101]. In addition, Progression-Associated lncRNA in Breast Cancer (PRLB) remains a biomarker of metastatic range and shorter survival rate [102].
It is worth mentioning that the knockdown of Breast Cancer Anti-Estrogen Resistance 4 (BCAR4) resulted in a reduction in LNM in the lungs of mice models. BCAR4 enhances BC metastasis by interacting with SNIP1 (Smad Nuclear Interacting Protein 1) and PNUTS (Phosphatase Nuclear Targeting Subunit) in response to CCL21 (Chemokine C-C Motif Ligand 21). This interaction activates the noncanonical Hedgehog/GLI2 signaling pathway, promoting cell migration [103].
Last, but not least, the lncRNA Highly Upregulated in Liver Cancer (HULC) is found to be overexpressed in BC, correlating with increased malignancy and metastasis. HULC promotes tumor progression by directly binding to miR-6754-5p, which regulates the expression of LYPD1 [104]. Zhou et al. proved that HULC promoted metastasis both in vivo and in vitro. Furthermore, HULC acted as a crucial regulator of the IGF1R signaling pathway in BC by directly binding to IGF1R’s intragenic regulatory elements, enhancing its expression and activation of the PI3K/AKT pathway [105].
To provide a comprehensive overview of the key findings related to lncRNAs as potential biomarkers for BC metastasis, Table 1 summarizes the most relevant studies in this area.

Table 1.
Summary of the Most Relevant Studies on lncRNA-Based Biomarkers in Breast Cancer.
6. Clinical Utility
Clinical utility of lncRNAs in BC spans diagnostics, prognostics, and therapeutic interventions, offering new avenues for personalized medicine and improved patient outcomes.
6.1. Diagnostic Biomarkers
The diagnostic capability of lncRNAs is one of the most promising aspects of their clinical utility. Numerous studies have demonstrated their strong association with LNM, distant organ metastasis, tumor size, and cancer stage [90,93,95,97,98,99,100,101,102,104]. LncRNAs can be detected in various biological fluids, primarily blood, making them accessible through minimally invasive procedures. These biomarkers are particularly valuable for screening high-risk populations and monitoring disease recurrence in BC survivors. Research has shown that lncRNAs such as HOTAIR, MALAT1, and H19 exhibit promising diagnostic efficiency, often surpassing standard markers like CEA and CA153. Unlike protein markers, lncRNAs often exhibit tissue-specific expression patterns, which can lead to more accurate diagnoses. Techniques such as quantitative qRT-PCR and next-generation sequencing (NGS) have enabled the sensitive and specific detection of lncRNAs, making them viable candidates for routine clinical testing [80,91].
6.2. Prognostic Indicators
LncRNAs also hold significant promise as prognostic indicators, helping to predict disease outcomes and guide treatment decisions. Prognostic lncRNAs can also help stratify patients based on risk, allowing for more tailored treatment approaches. Patients with high-risk lncRNA profiles may benefit from more aggressive treatments and closer monitoring, while those with low-risk profiles might avoid overtreatment. This stratification enhances the personalization of BC management, improving outcomes and reducing unnecessary side effects [32,45,46,80,95].
6.3. Therapeutic Targets
The therapeutic potential of lncRNAs lies in their ability to regulate critical pathways involved in cancer development and progression. Targeting oncogenic lncRNAs or restoring the function of tumor suppressor lncRNAs can disrupt cancer cell survival and proliferation. For instance, silencing lncRNAs using ASOs or siRNAs has been shown to inhibit tumor growth and metastasis in preclinical models [106]. Moreover, lncRNAs such as MALAT1 and GAS5 can be targeted to overcome drug resistance. MALAT1 has been implicated in the development of resistance to chemotherapeutic agents, and its inhibition could restore sensitivity to these drugs [107]. Conversely, restoring the expression of tumor suppressor lncRNAs like GAS5 could enhance the efficacy of existing treatments by promoting apoptosis and inhibiting cell proliferation [108]. LncRNA-based therapies are still in the early stages of development, but the potential for clinical application is substantial. The specificity of lncRNA-targeted therapies could minimize off-target effects and reduce toxicity compared to traditional chemotherapy. Furthermore, advances in delivery systems, such as nanoparticles and viral vectors, are enhancing the stability and efficiency of lncRNA therapeutics [109].
7. Conclusions
The role of lncRNAs in BC metastasis is a rapidly evolving field that has identified numerous lncRNAs with significant implications for cancer biology. While much progress has been made, several limitations and challenges must be addressed to fully harness the potential of these molecules as biomarkers and therapeutic targets, particularly in aggressive subtypes such as TNBC.
7.1. Limitations and Challenges
One of the primary challenges in lncRNA research is the incomplete understanding of their diverse functional mechanisms. Although numerous lncRNAs, such as HOTAIR, MALAT1, and NEAT1, have been linked to BC metastasis, the detailed molecular pathways through which they exert their effects remain only partially elucidated. This knowledge gap hinders the development of targeted therapies and precise diagnostic tools. For example, while HOTAIR’s role in promoting tumor progression through chromatin remodeling and gene silencing is well documented, the full spectrum of its interactions and regulatory mechanisms needs further exploration [110].
Another considerable hurdle is the variability in lncRNA expression across different patient populations and BC subtypes. This heterogeneity complicates the identification of universal lncRNA biomarkers. Variations in genetic background, environmental influences, and tumor microenvironment factors can lead to inconsistent lncRNA expression profiles. Furthermore, discrepancies in experimental methodologies, such as RNA extraction and analysis techniques contribute to the variability in research findings, making standardization and reproducibility a concern [111].
The stability and detectability of lncRNAs in biological samples also present difficulties. While some lncRNAs are stable and detectable in blood, others degrade rapidly or exist at low concentrations, complicating reliable detection and quantification [34,41,58,80]. This issue is particularly pertinent for developing non-invasive diagnostic tests, which are crucial for early detection and monitoring of metastasis.
Moreover, the diverse functions of lncRNAs adds another layer of complexity. Some lncRNAs, such as H19 and GAS5, can play varying roles in different cellular contexts, sometimes exhibiting opposing effects depending on the disease state and molecular subtype [58,64]. This duality complicates the interpretation of their functions and potential as therapeutic targets. For instance, in TNBC, the absence of hormone receptors and HER2 amplification necessitates a unique approach to leveraging lncRNA profiles for therapeutic and diagnostic purposes [107].
7.2. Future Directions and Application into Clinical Practice
Integration of lncRNAs into clinical practice necessitates robust validation studies and the development of standardized protocols. Large-scale, multicenter clinical trials are essential to confirm the diagnostic, prognostic, and therapeutic efficacy of lncRNAs. Regulatory approvals and the establishment of clinical guidelines will be crucial for the widespread adoption of lncRNA-based applications. Interdisciplinary collaboration among researchers, clinicians, and biotechnologists will drive the translation of lncRNA research into practical clinical tools. Developing diagnostic panels incorporating lncRNAs appears to be a promising approach for enhancing the sensitivity and specificity of BC diagnostics. However, it is sensible to use these panels as part of a broader diagnostic toolkit rather than as standalone tools for monitoring. This combined approach offers the greatest potential for improving diagnostic accuracy, as it leverages the unique expression patterns and stability of lncRNAs while complementing other established diagnostic methods. Personalized medicine, guided by lncRNA profiles, promises improved BC patient outcomes through more accurate diagnoses, tailored treatments, and effective monitoring strategies.
In summary, early detection of BC metastasis is pivotal to improving patients’ outcomes and providing care at the earliest possible stage. LncRNAs have been recognized as vital regulators of multiple signaling pathways involved in processes of invasion and metastasis. Several lncRNAs appear to be upregulated or downregulated during the cancer progression. The availability of serum detection of lncRNAs’ level is a promising minimally invasive method for assessing metastatic risk. However, further studies with the clinical applications of lncRNA should be performed to indicate lncRNA signatures as promising BC biomarkers for the detection of early metastasis.
Author Contributions
Conceptualization: I.H.-M., K.D. and M.K.; methodology: K.D. and M.K.; investigation: K.D. and M.K.; data curation: K.D. and M.K.; writing—original draft preparation: K.D., M.K., T.M.-M. and M.P.-W.; writing—review and editing: M.P.-W., I.H.-M. and T.M.-M.; visualization: K.D. and M.K.; supervision: T.M.-M. and I.H.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Al-thoubaity, F.K. Molecular classification of breast cancer: A retrospective cohort study. Ann. Med. Surg. 2019, 49, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Lou, K.-X.; Li, Z.-H.; Wang, P.; Liu, Z.; Chen, Y.; Wang, X.-L.; Cui, H.-X. Long non-coding RNA BANCR indicates poor prognosis for breast cancer and promotes cell proliferation and invasion. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 1358–1365. [Google Scholar] [CrossRef] [PubMed]
- Wojciechowska, U.; Barańska, K.; Miklewska, M.; Didkowska, J.A. Cancer incidence and mortality in Poland in 2020. Nowotw. J. Oncol. 2023, 73, 129–145. [Google Scholar] [CrossRef]
- Burzyńska, M.; Maniecka-Bryła, I.; Pikala, M. Trends of mortality due to breast cancer in Poland, 2000–2016. BMC Public Health 2020, 20, 120. [Google Scholar] [CrossRef] [PubMed]
- Paszko, A.; Krzyżak, M.J.; Charkiewicz, A.E.; Ziembicka, D.; Żendzian-Piotrowska, M.; Szpak, A.S.; Florek-Łuszczki, M.; Maślach, D. Inequalities in breast cancer incidence and stage distribution between urban and rural female population in Świętokrzyskie Province, Poland. Ann. Agric. Environ. Med. 2019, 26, 159–164. [Google Scholar] [CrossRef]
- Sørensen, K.P.; Thomassen, M.; Tan, Q.; Bak, M.; Cold, S.; Burton, M.; Larsen, M.J.; Kruse, T.A. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res. Treat. 2013, 142, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhang, G.; Li, X.; Ren, C.; Wang, Y.; Li, K.; Mok, H.; Cao, L.; Wen, L.; Jia, M.; et al. Comparison of BRCA versus non-BRCA germline mutations and associated somatic mutation profiles in patients with unselected breast cancer. Aging 2020, 12, 3140–3155. [Google Scholar] [CrossRef] [PubMed]
- Nik-Zainal, S.; Davies, H.; Staaf, J.; Ramakrishna, M.; Glodzik, D.; Zou, X.; Martincorena, I.; Alexandrov, L.B.; Martin, S.; Wedge, D.C.; et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016, 534, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Mitri, Z.I.; Abuhadra, N.; Goodyear, S.M.; Hobbs, E.A.; Kaempf, A.; Thompson, A.M.; Moulder, S.L. Impact of TP53 mutations in Triple Negative Breast Cancer. NPJ Precis. Oncol. 2022, 6, 64. [Google Scholar] [CrossRef]
- Loibl, S.; Gianni, L. HER2-positive breast cancer. Lancet 2017, 389, 2415–2429. [Google Scholar] [CrossRef]
- Staaf, J.; Glodzik, D.; Bosch, A.; Vallon-Christersson, J.; Reuterswärd, C.; Häkkinen, J.; Degasperi, A.; Amarante, T.D.; Saal, L.H.; Hegardt, C.; et al. Whole-genome sequencing of triple-negative breast cancers in a population-based clinical study. Nat. Med. 2019, 25, 1526–1533. [Google Scholar] [CrossRef]
- Arshi, A.; Sharifi, F.S.; Khorramian Ghahfarokhi, M.; Faghih, Z.; Doosti, A.; Ostovari, S.; Mahmoudi Maymand, E.; Ghahramani Seno, M.M. Expression Analysis of MALAT1, GAS5, SRA, and NEAT1 lncRNAs in Breast Cancer Tissues from Young Women and Women over 45 Years of Age. Mol. Ther. Nucleic Acids 2018, 12, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Assaraf, Y.G.; Gacche, R.N. Long non-coding RNA mediated drug resistance in breast cancer. Drug Resist. Updat. 2022, 63, 100851. [Google Scholar] [CrossRef] [PubMed]
- Collina, F.; Aquino, G.; Brogna, M.; Cipolletta, S.; Buonfanti, G.; De Laurentiis, M.; Di Bonito, M.; Cantile, M.; Botti, G. LncRNA HOTAIR up-regulation is strongly related with lymph nodes metastasis and LAR subtype of Triple Negative Breast Cancer. J. Cancer 2019, 10, 2018–2024. [Google Scholar] [CrossRef] [PubMed]
- Pádua Alves, C.; Fonseca, A.S.; Muys, B.R.; Barros e Lima Bueno, R.; Bürger, M.C.; Souza, J.E.; Valente, V.; Zago, M.A.; Silva, W.A., Jr. Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells 2013, 31, 2827–2832. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, Y.; Yang, Z.; Chen, B.; Huang, W.; Liu, Y.; Zhang, Y. MiR-204/ZEB2 axis functions as key mediator for MALAT1-induced epithelial-mesenchymal transition in breast cancer. Tumour Biol. 2017, 39, 1010428317690998. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Wu, C.-H.; Hu, H.-Z. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 2819–2824. [Google Scholar] [PubMed]
- Takahashi, K.; Yan, I.K.; Haga, H.; Patel, T. Modulation of hypoxia-signaling pathways by extracellular linc-RoR. J. Cell Sci. 2014, 127 Pt 7, 1585–1594. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Jia, H.-H.; Xu, Y.-Q.; Zhou, X.; Zhao, X.-H.; Wang, Y.-F.; Song, X.; Zhu, Z.-Y.; Sun, T.; Dou, Y.; et al. Paracrine and epigenetic control of CAF-induced metastasis: The role of HOTAIR stimulated by TGF-ß1 secretion. Mol. Cancer 2018, 17, 5. [Google Scholar] [CrossRef]
- Ma, Y.; Zhu, Y.; Shang, L.; Qiu, Y.; Shen, N.; Wang, J.; Adam, T.; Wei, W.; Song, Q.; Li, J.; et al. LncRNA XIST regulates breast cancer stem cells by activating proinflammatory IL-6/STAT3 signaling. Oncogene 2023, 42, 1419–1437. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Li, P.; Wang, J.; Shu, Y.; Zhong, X.; Gao, Z.; Yang, J.; Jiang, Y.; Zhou, X.; et al. Long noncoding RNA HOTAIR regulates the stemness of breast cancer cells via activation of the NF-κB signaling pathway. J. Biol. Chem. 2022, 298, 102630. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Hou, Y.; Liu, S.; Zhu, P.; Wan, X.; Zhao, M.; Peng, M.; Zeng, H.; Li, Q.; Jin, T.; et al. A Novel Long Non-Coding RNA lnc030 Maintains Breast Cancer Stem Cell Stemness by Stabilizing SQLE mRNA and Increasing Cholesterol Synthesis. Adv. Sci. 2021, 8, 2002232. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Peng, X.; Shen, C. Identification and validation of immune-related lncRNA prognostic signature for breast cancer. Genomics 2020, 112, 2640–2646. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Z.; Chen, M.; Chen, H.; Zhong, Q.; Liang, L.; Li, B. lncRNA TCL6 correlates with immune cell infiltration and indicates worse survival in breast cancer. Breast Cancer 2020, 27, 573–585. [Google Scholar] [CrossRef]
- Wang, Q.; Li, G.; Ma, X.; Liu, L.; Liu, J.; Yin, Y.; Li, H.; Chen, Y.; Zhang, X.; Zhang, L.; et al. LncRNA TINCR impairs the efficacy of immunotherapy against breast cancer by recruiting DNMT1 and downregulating MiR-199a-5p via the STAT1-TINCR-USP20-PD-L1 axis. Cell Death Dis. 2023, 14, 76. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Yang, Y.A.; Zhang, A.; Fong, K.-W.; Kim, J.; Song, B.; Li, S.; Zhao, J.C.; Yu, J. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene 2016, 35, 2746–2755. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Qian, J.; Li, J.; Zhu, C. Knockdown of lncRNA-HOTAIR downregulates the drug-resistance of breast cancer cells to doxorubicin via the PI3K/AKT/mTOR signaling pathway. Exp. Ther. Med. 2019, 18, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Yue, X.; Wu, W.-Y.; Dong, M.; Guo, M. LncRNA MALAT1 promotes breast cancer progression and doxorubicin resistance via regulating miR-570-3p. Biomed. J. 2021, 44 (Suppl. S2), S296–S304. [Google Scholar] [CrossRef] [PubMed]
- Si, X.; Zang, R.; Zhang, E.; Liu, Y.; Shi, X.; Zhang, E.; Shao, L.; Li, A.; Yang, N.; Han, X.; et al. LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget 2016, 7, 81452–81462. [Google Scholar] [CrossRef]
- Contreras-Espinosa, L.; Alcaraz, N.; De La Rosa-Velázquez, I.A.; Díaz-Chávez, J.; Cabrera-Galeana, P.; Rebollar-Vega, R.; Reynoso-Noverón, N.; Maldonado-Martínez, H.A.; González-Barrios, R.; Montiel-Manríquez, R.; et al. Transcriptome Analysis Identifies GATA3-AS1 as a Long Noncoding RNA Associated with Resistance to Neoadjuvant Chemotherapy in Locally Advanced Breast Cancer Patients. J. Mol. Diagn. 2021, 23, 1306–1323. [Google Scholar] [CrossRef]
- Arshi, A.; Raeisi, F.; Mahmoudi, E.; Mohajerani, F.; Kabiri, H.; Fazel, R.; Zabihian-Langeroudi, M.; Jusic, A. A Comparative Study of HOTAIR Expression in Breast Cancer Patient Tissues and Cell Lines. Cell J. 2020, 22, 178–184. [Google Scholar] [CrossRef]
- Gökmen-Polar, Y.; Vladislav, I.T.; Neelamraju, Y.; Janga, S.C.; Badve, S. Prognostic impact of HOTAIR expression is restricted to ER-negative breast cancers. Sci. Rep. 2015, 5, 8765. [Google Scholar] [CrossRef] [PubMed]
- Tao, S.; He, H.; Chen, Q. Estradiol induces HOTAIR levels via GPER-mediated miR-148a inhibition in breast cancer. J. Transl. Med. 2015, 13, 131. [Google Scholar] [CrossRef]
- Zhang, L.; Song, X.; Wang, X.; Xie, Y.; Wang, Z.; Xu, Y.; You, X.; Liang, Z.; Cao, H. Circulating DNA of HOTAIR in serum is a novel biomarker for breast cancer. Breast Cancer Res. Treat. 2015, 152, 199–208. [Google Scholar] [CrossRef]
- Lozano-Romero, A.; Astudillo-de la Vega, H.; Terrones-Gurrola, M.C.D.R.; Marchat, L.A.; Hernández-Sotelo, D.; Salinas-Vera, Y.M.; Ramos-Payan, R.; Silva-Cázares, M.B.; Nuñez-Olvera, S.I.; Hernández-de la Cruz, O.N.; et al. HOX Transcript Antisense RNA HOTAIR Abrogates Vasculogenic Mimicry by Targeting the AngiomiR-204/FAK Axis in Triple Negative Breast Cancer Cells. Noncoding RNA 2020, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhou, H.; Lu, K.; Lu, Y.; Wang, Y.; Feng, T. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. Onco Targets Ther. 2018, 11, 291–299. [Google Scholar] [CrossRef]
- Huang, X.; Xia, Y.; He, G.; Zheng, L.; Cai, Y.; Yin, Y.; Wu, Q. MALAT1 promotes angiogenesis of breast cancer. Oncol. Rep. 2018, 40, 2683–2689. [Google Scholar] [CrossRef] [PubMed]
- Jadaliha, M.; Zong, X.; Malakar, P.; Ray, T.; Singh, D.K.; Freier, S.M.; Jensen, T.; Prasanth, S.G.; Karni, R.; Ray, P.S.; et al. Functional and prognostic significance of long non-coding RNA MALAT1 as a metastasis driver in ER negative lymph node negative breast cancer. Oncotarget 2016, 7, 40418–40436. [Google Scholar] [CrossRef]
- Ou, X.; Gao, G.; Bazhabayi, M.; Zhang, K.; Liu, F.; Xiao, X. MALAT1 and BACH1 are prognostic biomarkers for triple-negative breast cancer. J. Cancer Res. Ther. 2019, 15, 1597–1602. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, B.; Yang, S.; Yu, F.; Yang, H.; Cheng, Q.; Tang, B. Roles of MALAT1 in development and migration of triple negative and Her-2 positive breast cancer. Oncotarget 2017, 9, 2255–2267. [Google Scholar] [CrossRef]
- Miao, Y.; Fan, R.; Chen, L.; Qian, H. Clinical Significance of Long Non-coding RNA MALAT1 Expression in Tissue and Serum of Breast Cancer. Ann. Clin. Lab. Sci. 2016, 46, 418–424. [Google Scholar]
- Kim, J.; Piao, H.-L.; Kim, B.-J.; Yao, F.; Han, Z.; Wang, Y.; Xiao, Z.; Siverly, A.N.; Lawhon, S.E.; Ton, B.N.; et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018, 50, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Kwok, Z.H.; Roche, V.; Chew, X.H.; Fadieieva, A.; Tay, Y. A non-canonical tumor suppressive role for the long non-coding RNA MALAT1 in colon and breast cancers. Int. J. Cancer 2018, 143, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Sui, S.; Zhang, J.; Bai, N.; Shi, Q.; Zhang, G.; Gao, S.; You, Z.; Zhan, C.; Liu, F.; et al. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 4881–4891. [Google Scholar] [PubMed]
- Meseure, D.; Vacher, S.; Lallemand, F.; Alsibai, K.D.; Hatem, R.; Chemlali, W.; Nicolas, A.; De Koning, L.; Pasmant, E.; Callens, C.; et al. Prognostic value of a newly identified MALAT1 alternatively spliced transcript in breast cancer. Br. J. Cancer 2016, 114, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Shi, S.-H.; Li, X.-J.; Sun, L.; Ge, Q.-D.; Li, C.; Zhang, W. Long non-coding RNA BRAF-regulated lncRNA 1 promotes lymph node invasion, metastasis and proliferation, and predicts poor prognosis in breast cancer. Oncol. Lett. 2018, 15, 9543–9552. [Google Scholar] [CrossRef] [PubMed]
- Knutsen, E.; Lellahi, S.M.; Aure, M.R.; Nord, S.; Fismen, S.; Larsen, K.B.; Gabriel, M.T.; Hedberg, A.; Bjørklund, S.S.; Geisler, J.; et al. The expression of the long NEAT1_2 isoform is associated with human epidermal growth factor receptor 2-positive breast cancers. Sci. Rep. 2020, 10, 1277. [Google Scholar] [CrossRef] [PubMed]
- Shin, V.Y.; Chen, J.; Cheuk, I.W.Y.; Siu, M.T.; Ho, C.W.; Wang, X.; Jin, H.; Kwong, A. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019, 10, 270. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, S.; Li, Z.; Long, X.; Guo, Z.; Zhang, G.; Zu, J.; Chen, Y.; Wen, L. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int. J. Biol. Macromol. 2017, 105 Pt 1, 346–353. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Z.; Liu, X.; Cheng, X.; Zhang, Y.; Han, X.; Zhang, Y.; Liu, S.; Yang, J.; Xu, B.; et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J. Clin. Investig. 2017, 127, 3421–3440. [Google Scholar] [CrossRef]
- Lo, P.-K.; Zhang, Y.; Wolfson, B.; Gernapudi, R.; Yao, Y.; Duru, N.; Zhou, Q. Dysregulation of the BRCA1/long non-coding RNA NEAT1 signaling axis contributes to breast tumorigenesis. Oncotarget 2016, 7, 65067–65089. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, H.; Albukhari, A.; Morotti, M.; Haider, S.; Moralli, D.; Smythies, J.; Schödel, J.; Green, C.M.; Camps, C.; Buffa, F.; et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 2015, 34, 4482–4490. [Google Scholar] [CrossRef] [PubMed]
- Peperstraete, E.; Lecerf, C.; Collette, J.; Vennin, C.; Raby, L.; Völkel, P.; Angrand, P.-O.; Winter, M.; Bertucci, F.; Finetti, P.; et al. Enhancement of Breast Cancer Cell Aggressiveness by lncRNA H19 and its Mir-675 Derivative: Insight into Shared and Different Actions. Cancers 2020, 12, 1730. [Google Scholar] [CrossRef]
- Lin, Y.; Fu, F.; Chen, Y.; Qiu, W.; Lin, S.; Yang, P.; Huang, M.; Wang, C. Genetic variants in long noncoding RNA H19 contribute to the risk of breast cancer in a southeast China Han population. Onco Targets Ther. 2017, 10, 4369–4378. [Google Scholar] [CrossRef] [PubMed]
- Vennin, C.; Spruyt, N.; Dahmani, F.; Julien, S.; Bertucci, F.; Finetti, P.; Chassat, T.; Bourette, R.P.; Le Bourhis, X.; Adriaenssens, E. H19 non coding RNA-derived miR-675 enhances tumorigenesis and metastasis of breast cancer cells by downregulating c-Cbl and Cbl-b. Oncotarget 2015, 6, 29209–29223. [Google Scholar] [CrossRef] [PubMed]
- Si, H.; Chen, P.; Li, H.; Wang, X. Long non-coding RNA H19 regulates cell growth and metastasis via miR-138 in breast cancer. Am. J. Transl. Res. 2019, 11, 3213–3225. [Google Scholar]
- Zhang, K.; Luo, Z.; Zhang, Y.; Zhang, L.; Wu, L.; Liu, L.; Yang, J.; Song, X.; Liu, J. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark. 2016, 17, 187–194. [Google Scholar] [CrossRef]
- Zhong, G.; Wang, K.; Li, J.; Xiao, S.; Wei, W.; Liu, J. Determination of Serum Exosomal H19 as a Noninvasive Biomarker for Breast Cancer Diagnosis. Onco Targets Ther. 2020, 13, 2563–2571. [Google Scholar] [CrossRef]
- Dong, L.; Li, G.; Li, Y.; Zhu, Z. Upregulation of Long Noncoding RNA GAS5 Inhibits Lung Cancer Cell Proliferation and Metastasis via miR-205/PTEN Axis. Med. Sci. Monit. 2019, 25, 2311–2319. [Google Scholar] [CrossRef]
- Li, W.; Zhai, L.; Wang, H.; Liu, C.; Zhang, J.; Chen, W.; Wei, Q. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget 2016, 7, 27778–27786. [Google Scholar] [CrossRef]
- Li, S.; Zhou, J.; Wang, Z.; Wang, P.; Gao, X.; Wang, Y. Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p. Biomed. Pharmacother. 2018, 104, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.; Li, M.; Miao, K.; Xu, H. lncRNA GAS5-promoted apoptosis in triple-negative breast cancer by targeting miR-378a-5p/SUFU signaling. J. Cell Biochem. 2020, 121, 2225–2235. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Qian, L.; Tang, X.; Chen, Y.; Zhao, Z.; Zhang, C. Long non-coding RNA growth arrest-specific 5 (GAS5) acts as a tumor suppressor by promoting autophagy in breast cancer. Mol. Med. Rep. 2020, 22, 2460–2468. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Ma, P.; Liu, S.-M.; Zhou, X. Circulating long noncoding RNA GAS5 as a potential biomarker in breast cancer for assessing the surgical effects. Tumour Biol. 2016, 37, 6847–6854. [Google Scholar] [CrossRef] [PubMed]
- Toraih, E.A.; El-Wazir, A.; Ageeli, E.A.; Hussein, M.H.; Eltoukhy, M.M.; Killackey, M.T.; Kandil, E.; Fawzy, M.S. Unleash multifunctional role of long noncoding RNAs biomarker panel in breast cancer: A predictor classification model. Epigenomics 2020, 12, 1215–1237. [Google Scholar] [CrossRef] [PubMed]
- Kanabe, B.O.; Ozaslan, M.; Aziz, S.A.; Al-Attar, M.S.; Kılıç, İ.H.; Khailany, R.A. Expression patterns of LncRNA-GAS5 and its target APOBEC3C gene through miR-103 in breast cancer patients. Cell. Mol. Biol. 2021, 67, 5–10. [Google Scholar] [CrossRef] [PubMed]
- Nafea, H.; Youness, R.A.; Abou-Aisha, K.; Gad, M.Z. LncRNA HEIH/miR-939-5p interplay modulates triple-negative breast cancer progression through NOS2-induced nitric oxide production. J. Cell. Physiol. 2021, 236, 5362–5372. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhou, B.; Lv, Y.; Qian, Q. LncRNA HEIH regulates cell proliferation and apoptosis through miR-4458/SOCS1 axis in triple-negative breast cancer. Hum. Cell 2019, 32, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Meng, R.; Yao, Q.; Wang, H.; Niu, J.; Cui, Y.U.; Chen, S.; Bai, Y. Long non-coding RNA HEIH promotes breast cancer development via negative modulation of microRNA-200b. Die Pharm.-Int. J. Pharm. Sci. 2019, 74, 471–476. [Google Scholar] [CrossRef]
- Chen, C.; Gu, C.; Ren, Q.; Ding, F.; Pan, Q.; Niu, Y.; Ma, D.; Wu, L. lncRNA HEIH, an indicator of high malignancy and poor prognosis, functions as an oncogene in breast cancer. Mol. Med. Rep. 2020, 22, 2869–2877. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Azman, A.A.; Siok-Fong, C.; Rajab, N.F.; Md Zin, R.R.; Ahmad Daud, N.N.; Mohamad Hanif, E.A. The potential roles of lncRNA TINCR in triple negative breast cancer. Mol. Biol. Rep. 2023, 50, 7909–7917. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, J.; You, Z.; Yin, Y.; Liu, L.; Kang, Y.; Li, S.; Ning, S.; Li, H.; Gong, Y.; et al. LncRNA TINCR favors tumorigenesis via STAT3–TINCR–EGFR-feedback loop by recruiting DNMT1 and acting as a competing endogenous RNA in human breast cancer. Cell Death Dis. 2021, 12, 83. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, H.R.; Shaginurova, G.; Kim, L.C.; Chapman, N.; Spurlock, C.F.; Aune, T.M. Divergent lncRNA GATA3-AS1 Regulates GATA3 Transcription in T-Helper 2 Cells. Front. Immunol. 2018, 9, 2512. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Huang, W.; Yang, Y.; Qiu, R.; Zeng, Y.; Hou, Y.; Sun, G.; Shi, H.; Leng, S.; Feng, D.; et al. GATA3 recruits UTX for gene transcriptional activation to suppress metastasis of breast cancer. Cell Death Dis. 2019, 10, 832. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wang, N.; Song, P.; Fu, Y.; Ren, Y.; Li, Z.; Wang, J. LncRNA GATA3-AS1 facilitates tumour progression and immune escape in triple-negative breast cancer through destabilization of GATA3 but stabilization of PD-L1. Cell Prolif. 2020, 53, e12855. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, J.; Jia, S.; Wang, Y.; Kang, Y.; Zhang, W. Down-regulation of lncRNA-ATB inhibits epithelial-mesenchymal transition of breast cancer cells by increasing miR-141-3p expression. Biochem. Cell Biol. 2019, 97, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Li, R.-H.; Chen, M.; Liu, J.; Shao, C.-C.; Guo, C.-P.; Wei, X.-L.; Li, Y.-C.; Huang, W.-H.; Zhang, G.-J. Long noncoding RNA ATB promotes the epithelial-mesenchymal transition by upregulating the miR-200c/Twist1 axe and predicts poor prognosis in breast cancer. Cell Death Dis. 2018, 9, 1171. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Xiong, H.; Zhao, Y.; Dong, Z.; Liu, H.; Shi, F. Study on the Expression of lncRNA ATB and Nek9 in Breast Cancer Patients Based on Q-PCR Technology and Its Relationship with the Disease. Contrast Media Mol. Imaging 2022, 2022, 2634080. [Google Scholar] [CrossRef]
- El-Ashmawy, N.E.; Hussien, F.Z.; El-Feky, O.A.; Hamouda, S.M.; Al-Ashmawy, G.M. Serum LncRNA-ATB and FAM83H-AS1 as diagnostic/prognostic non-invasive biomarkers for breast cancer. Life Sci. 2020, 259, 118193. [Google Scholar] [CrossRef]
- Xing, F.; Liu, Y.; Wu, S.-Y.; Wu, K.; Sharma, S.; Mo, Y.-Y.; Feng, J.; Sanders, S.; Jin, G.; Singh, R.; et al. Loss of XIST in Breast Cancer Activates MSN-c-Met and Reprograms Microglia via Exosomal miRNA to Promote Brain Metastasis. Cancer Res. 2018, 78, 4316–4330. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Liang, K.; Hu, Q.; Li, P.; Song, J.; Yang, Y.; Yao, J.; Mangala, L.S.; Li, C.; Yang, W.; et al. JAK2-binding long noncoding RNA promotes breast cancer brain metastasis. J. Clin. Investig. 2017, 127, 4498–4515. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hou, P.; Fan, D.; Dong, M.; Ma, M.; Li, H.; Yao, R.; Li, Y.; Wang, G.; Geng, P.; et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2017, 24, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Gooding, A.J.; Zhang, B.; Jahanbani, F.K.; Gilmore, H.L.; Chang, J.C.; Valadkhan, S.; Schiemann, W.P. The lncRNA BORG Drives Breast Cancer Metastasis and Disease Recurrence. Sci. Rep. 2017, 7, 12698. [Google Scholar] [CrossRef] [PubMed]
- Parker, K.A.; Gooding, A.J.; Valadkhan, S.; Schiemann, W.P. lncRNA BORG:TRIM28 Complexes Drive Metastatic Progression by Inducing α6 Integrin/CD49f Expression in Breast Cancer Stem Cells. Mol. Cancer Res. MCR 2021, 19, 2068–2080. [Google Scholar] [CrossRef]
- Liu, B.; Sun, L.; Liu, Q.; Gong, C.; Yao, Y.; Lv, X.; Lin, L.; Yao, H.; Su, F.; Li, D.; et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015, 27, 370–381. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Chen, F.; Cui, X.; Yang, L.; Chen, J.; Zhao, J.; Huang, D.; Liu, J.; Yang, L.; Zeng, J.; et al. LncRNA NKILA suppresses TGF-β-induced epithelial-mesenchymal transition by blocking NF-κB signaling in breast cancer. Int. J. Cancer 2018, 143, 2213–2224. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Shen, Y.; Zhang, W.; Jin, J.; Huang, D.; Fang, H.; Ji, W.; Shi, Y.; Tang, L.; Chen, W.; et al. An androgen receptor negatively induced long non-coding RNA ARNILA binding to miR-204 promotes the invasion and metastasis of triple-negative breast cancer. Cell Death Differ. 2018, 25, 2209–2220. [Google Scholar] [CrossRef] [PubMed]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef]
- Chen, Y.-M.; Liu, Y.; Wei, H.-Y.; Lv, K.-Z.; Fu, P. Linc-ROR induces epithelial-mesenchymal transition and contributes to drug resistance and invasion of breast cancer cells. Tumour Biol. 2016, 37, 10861–10870. [Google Scholar] [CrossRef]
- Liu, M.; Xing, L.-Q.; Liu, Y.-J. A three-long noncoding RNA signature as a diagnostic biomarker for differentiating between triple-negative and non-triple-negative breast cancers. Medicine 2017, 96, e6222. [Google Scholar] [CrossRef] [PubMed]
- Li, G.-Y.; Wang, W.; Sun, J.-Y.; Xin, B.; Zhang, X.; Wang, T.; Zhang, Q.-F.; Yao, L.-B.; Han, H.; Fan, D.-M.; et al. Long non-coding RNAs AC026904.1 and UCA1: A “one-two punch” for TGF-β-induced SNAI2 activation and epithelial-mesenchymal transition in breast cancer. Theranostics 2018, 8, 2846–2861. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, N.; Liu, Y.; Su, P.; Liang, Y.; Li, Y.; Wang, X.; Chen, T.; Song, X.; Sang, Y.; et al. Epigenetic Regulation of NAMPT by NAMPT-AS Drives Metastatic Progression in Triple-Negative Breast Cancer. Cancer Res. 2019, 79, 3347–3359. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Li, X.; Fan, Q.; Liu, G.; Yin, J. CCAT1 promotes triple-negative breast cancer progression by suppressing miR-218/ZFX signaling. Aging 2019, 11, 4858–4875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-F.; Liu, T.; Li, Y.; Li, S. Overexpression of long non-coding RNA CCAT1 is a novel biomarker of poor prognosis in patients with breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 9440–9445. [Google Scholar] [PubMed]
- Xu, Z.; Liu, C.; Zhao, Q.; Lü, J.; Ding, X.; Luo, A.; He, J.; Wang, G.; Li, Y.; Cai, Z.; et al. Long non-coding RNA CCAT2 promotes oncogenesis in triple-negative breast cancer by regulating stemness of cancer cells. Pharmacol. Res. 2020, 152, 104628. [Google Scholar] [CrossRef]
- Jia, X.; Shi, L.; Wang, X.; Luo, L.; Ling, L.; Yin, J.; Song, Y.; Zhang, Z.; Qiu, N.; Liu, H.; et al. KLF5 regulated lncRNA RP1 promotes the growth and metastasis of breast cancer via repressing p27kip1 translation. Cell Death Dis. 2019, 10, 373. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-X.; Wei, L.; Zhang, Y.-J.; Hayano, T.; Piñeiro Pereda, M.D.P.; Nakaoka, H.; Li, Q.; Barragán Mallofret, I.; Lu, Y.-Z.; Tamagnone, L.; et al. Long non-coding RNA p10247, high expressed in breast cancer (lncRNA-BCHE), is correlated with metastasis. Clin. Exp. Metastasis 2018, 35, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.; Chu, J.; Wu, Y.; Sun, L.; Lv, X.; Zhu, Y.; Li, J.; Guo, Q.; Gong, C.; Liu, B.; et al. NBAT1 suppresses breast cancer metastasis by regulating DKK1 via PRC2. Oncotarget 2015, 6, 32410–32425. [Google Scholar] [CrossRef]
- Fan, S.; Yang, Z.; Ke, Z.; Huang, K.; Liu, N.; Fang, X.; Wang, K. Downregulation of the long non-coding RNA TUG1 is associated with cell proliferation, migration, and invasion in breast cancer. Biomed. Pharmacother. 2017, 95, 1636–1643. [Google Scholar] [CrossRef]
- Bai, Y.; Zhou, X.; Huang, L.; Wan, Y.; Li, X.; Wang, Y. Long noncoding RNA EZR-AS1 promotes tumor growth and metastasis by modulating Wnt/β-catenin pathway in breast cancer. Exp. Ther. Med. 2018, 16, 2235–2242. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Song, X.; Li, Y.; Sang, Y.; Zhang, N.; Zhang, H.; Liu, Y.; Duan, Y.; Chen, B.; Guo, R.; et al. A novel long non-coding RNA-PRLB acts as a tumor promoter through regulating miR-4766-5p/SIRT1 axis in breast cancer. Cell Death Dis. 2018, 9, 563. [Google Scholar] [CrossRef] [PubMed]
- Xing, Z.; Lin, A.; Li, C.; Liang, K.; Wang, S.; Liu, Y.; Park, P.K.; Qin, L.; Wei, Y.; Hawke, D.H.; et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 2014, 159, 1110–1125. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Zhong, C.; Fu, M.; Li, L.; Wang, F.; Lv, P.; Zhu, M.; Xiong, Y.; Mi, H.; Gu, Y. Long Non-Coding RNA HULC Promotes the Development of Breast Cancer Through Regulating LYPD1 Expression by Sponging miR-6754-5p. Onco Targets Ther. 2019, 12, 10671–10679. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, H.; Sun, T.; Wen, X.; Niu, C.; Li, M.; Li, W.; Hoffman, A.R.; Hu, J.-F.; Cui, J. HULC targets the IGF1R-PI3K-AKT axis in trans to promote breast cancer metastasis and cisplatin resistance. Cancer Lett. 2022, 548, 215861. [Google Scholar] [CrossRef] [PubMed]
- Gong, N.; Teng, X.; Li, J.; Liang, X.-J. Antisense Oligonucleotide-Conjugated Nanostructure-Targeting lncRNA MALAT1 Inhibits Cancer Metastasis. ACS Appl. Mater. Interfaces 2019, 11, 37–42. [Google Scholar] [CrossRef]
- Shaath, H.; Vishnubalaji, R.; Elango, R.; Khattak, S.; Alajez, N.M. Single-cell long noncoding RNA (lncRNA) transcriptome implicates MALAT1 in triple-negative breast cancer (TNBC) resistance to neoadjuvant chemotherapy. Cell Death Discov. 2021, 7, 23. [Google Scholar] [CrossRef]
- Pickard, M.R.; Williams, G.T. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: Implications for chemotherapy. Breast Cancer Res. Treat. 2014, 145, 359–370. [Google Scholar] [CrossRef]
- Vaidya, A.M.; Sun, Z.; Ayat, N.; Schilb, A.; Liu, X.; Jiang, H.; Sun, D.; Scheidt, J.; Qian, V.; He, S.; et al. Systemic Delivery of Tumor-Targeting siRNA Nanoparticles against an Oncogenic LncRNA Facilitates Effective Triple-Negative Breast Cancer Therapy. Bioconjug. Chem. 2019, 30, 907–919. [Google Scholar] [CrossRef]
- Milevskiy, M.J.G.; Al-Ejeh, F.; Saunus, J.M.; Northwood, K.S.; Bailey, P.J.; Betts, J.A.; McCart Reed, A.E.; Nephew, K.P.; Stone, A.; Gee, J.M.W.; et al. Long-range regulators of the lncRNA HOTAIR enhance its prognostic potential in breast cancer. Hum. Mol. Genet. 2016, 25, 3269–3283. [Google Scholar] [CrossRef]
- Zhao, Z.; Guo, Y.; Liu, Y.; Sun, L.; Chen, B.; Wang, C.; Chen, T.; Wang, Y.; Li, Y.; Dong, Q.; et al. Individualized lncRNA differential expression profile reveals heterogeneity of breast cancer. Oncogene 2021, 40, 4604–4614. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).