Fruiting Characteristics and Molecular-Assisted Identification of Korla Fragrant Pear Bud Mutation Materials
Abstract
1. Introduction
- (1)
- Is there any difference in the fruiting characteristics between Korla fragrant pear bud mutant material and Korla fragrant pears?
- (2)
- Are there any differences in the structural characteristics of sepal (cylindrical) cells between Korla fragrant pear bud mutation materials and Korla fragrant pears?
- (3)
- Does the degree of variation between Korla fragrant pear bud mutation materials and Korla fragrant pears affect population evolution?
2. Materials and Methods
2.1. Experimental Site and Plant Materials
2.2. Investigation of Fruiting Characteristics
2.3. Observation of the Anatomical Structure of the Calyx Tube
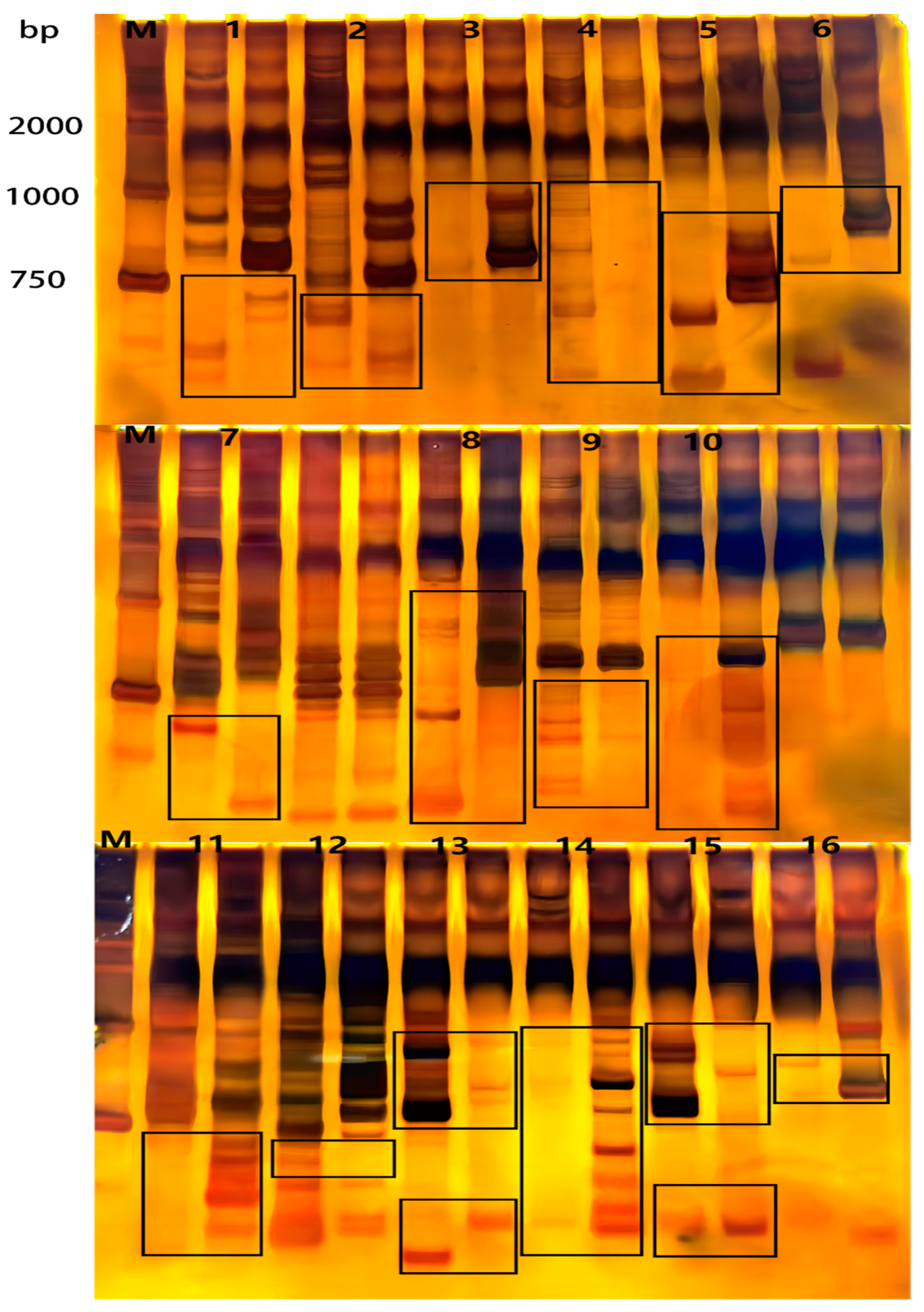
2.4. Genotyping by Sequencing
2.5. SSR Molecular Marker Primer Screening
2.6. Statistical Analysis
3. Results
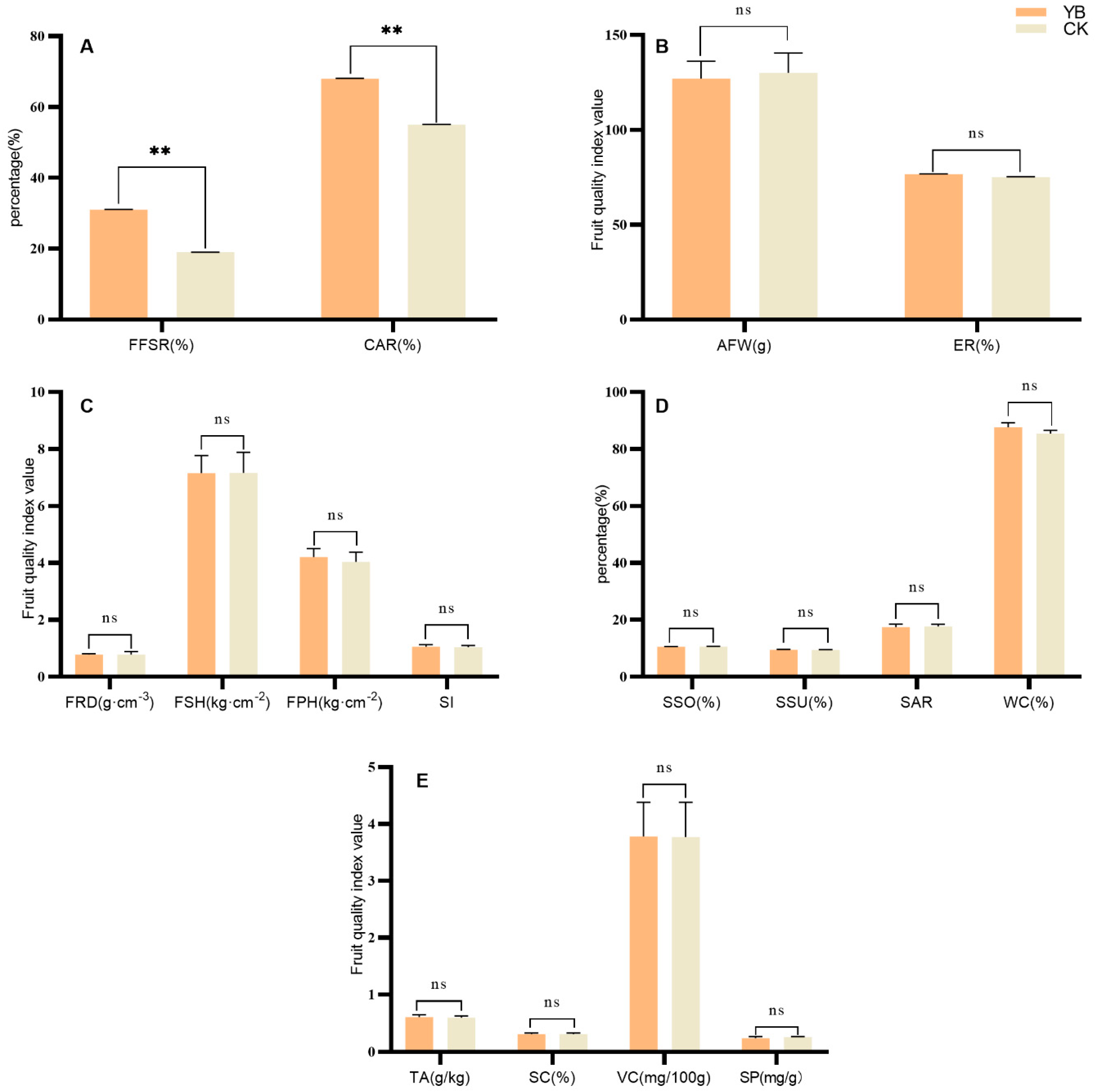
3.1. Comparison of Fruiting Characteristics of Korla Fragrant Pear and Its Bud Mutation Materials
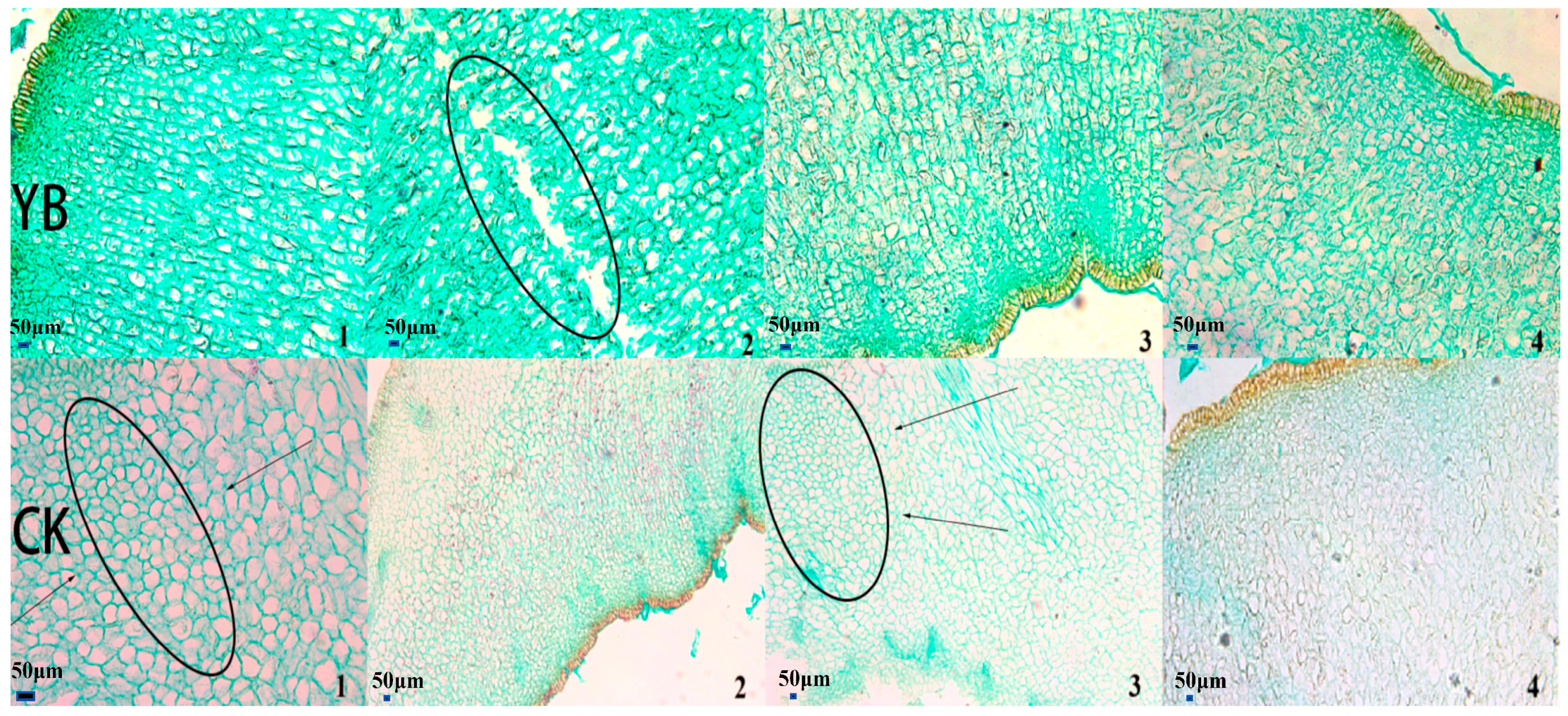
3.2. Korla Fragrant Pear and Its Bud Mutation Material Sepal (Cylinder) Shedding Cell Morphology Structure
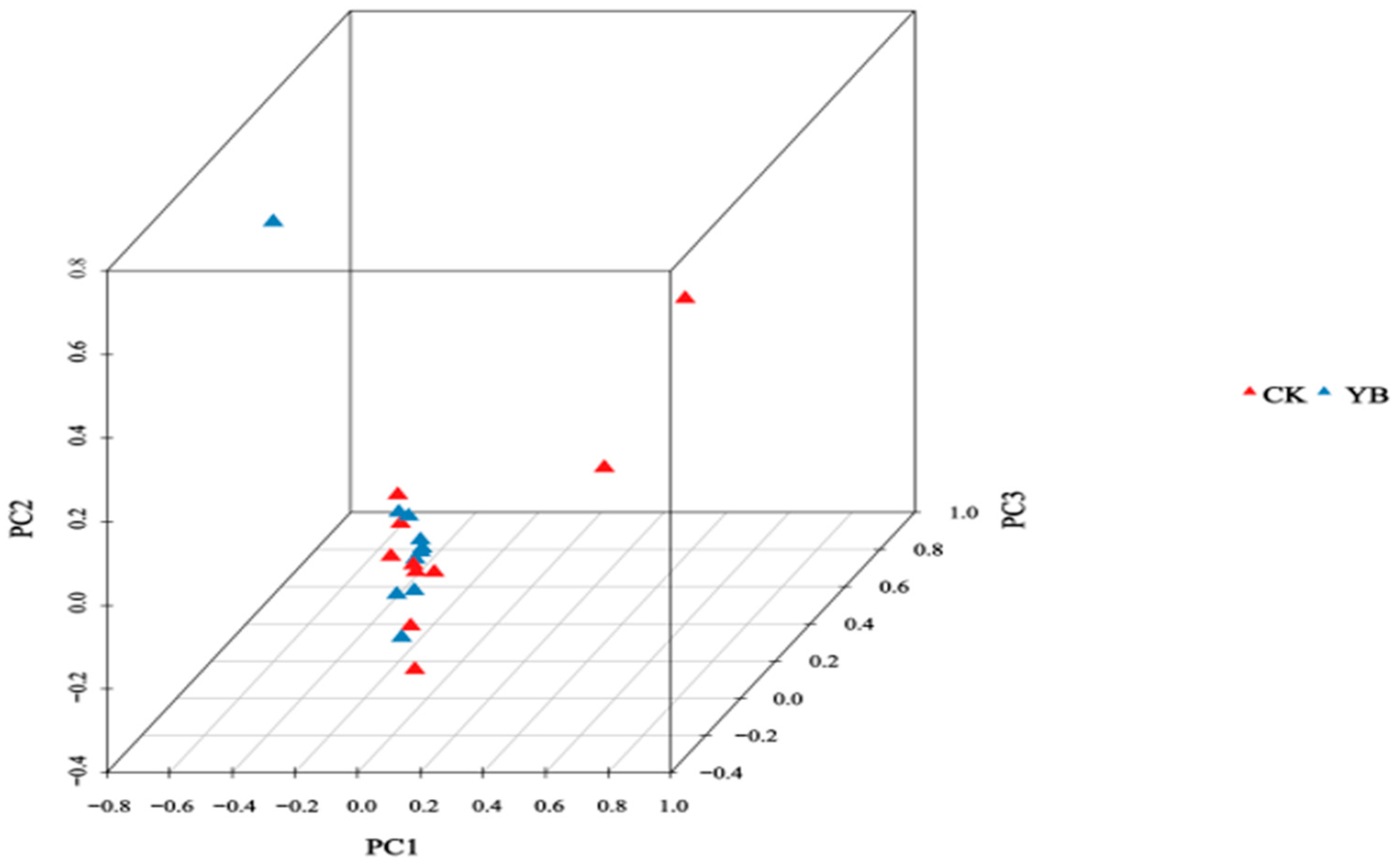
3.3. GBS Detection of Korla Fragrant Pear and Its Bud Mutation Material
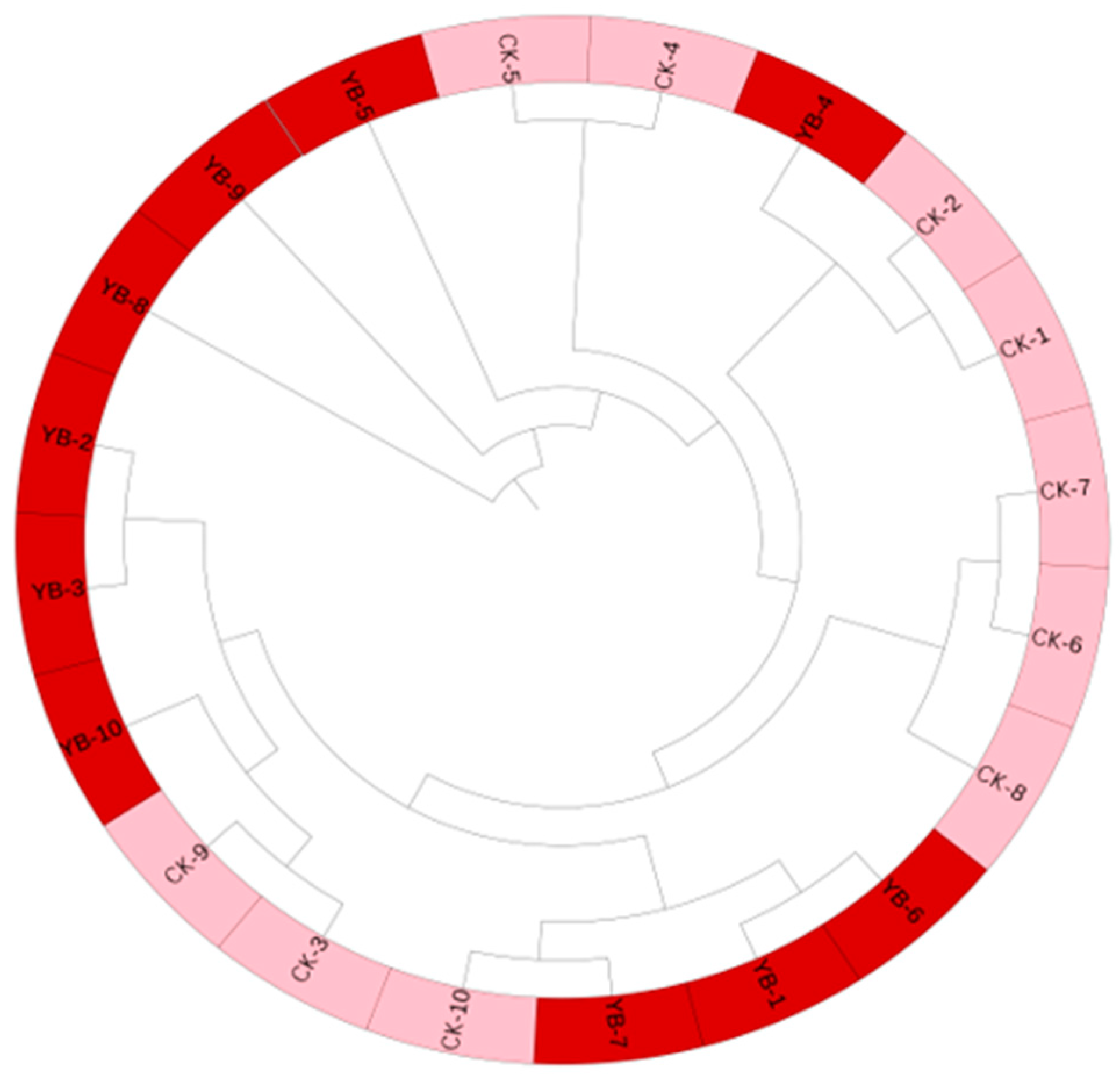
3.4. SSR Molecular Markers of Korla Fragrant Pear and Its Bud Mutation Materials
4. Discussion
4.1. Fruiting Characteristics Analysis of Korla Fragrant Pear Bud Mutation Material
4.2. Anatomical Structure Analysis of Sepals and Calyx Tube of Korla Fragrant Pear Bud Mutation Material
4.3. Genetic Structure Analysis of Korla Fragrant Pear Bud Mutation Materials
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, J.Q.; Lü, J.H.; He, Z.S.; Zhang, F.; Zhang, S.L.; Zhang, H.P. Investigations into the production of volatile compounds in Korla fragrant pears (Pyrus sinkiangensis Yu). Food Chem. 2020, 302, 125337. [Google Scholar] [CrossRef]
- Zhang, X.; Li, B.; Duan, R.; Han, C.; Wang, L.; Yang, J.; Wang, L.; Wang, S.; Su, Y.; Xue, H. Transcriptome Analysis Reveals Roles of Sucrose in Anthocyanin Accumulation in ‘Kuerle Xiangli’ (Pyrus sinkiangensis Yü). Genes 2022, 13, 1064. [Google Scholar] [CrossRef]
- Li, Q.; Sun, M.; Liu, Y.; Liu, B.; Bianchi, F.J.J.A.; van der Werf, W.; Lu, Y. High Pollination Deficit and Strong Dependence on Honeybees in Pollination of Korla Fragrant Pear, Pyrus sinkiangensis. Plants 2022, 11, 1734. [Google Scholar] [CrossRef]
- Li, Y.; Huang, Y.; Long, H.; Yu, Z.; Huang, M.; Liu, X. Biodiversity and Oenological Property Analysis of Non-Saccharomyces Yeasts Isolated from Korla Fragrant Pears (Pyrus sinkiangensis Yu). Fermentation 2022, 8, 388. [Google Scholar] [CrossRef]
- Han, S.J.; Zhao, J.F.; Liu, Y.; Xi, L.Q.; Liao, J.A.; Liu, X.Y.; Su, G.D. Effects of green manure planting mode on the quality of Korla fragrant pears (Pyrus sinkiangensis Yu). Front. Plant Sci. 2022, 13, 1027595. [Google Scholar] [CrossRef]
- Qi, X.X.; Wu, J.; Wang, L.F.; Cao, Y.F.; Tian, L.M.; Dong, X.G.; Zhang, S.L. Identifying the candidate genes involved in the calyx abscission process of ‘Kuerlexiangli’ (Pyrus sinkiangensis Yu) by digital transcript abundance measurements. BMC Genom. 2013, 14, 727. [Google Scholar] [CrossRef]
- Zhou, L.; Li, C.J.; Niu, J.X.; Pei, M.S.; Cao, F.J.; Quan, S.W. Identification of miRNAs involved in calyx persistence in Korla fragrant pear (Pyrus sinkiangensis Yu) by high-throughput sequencing. Sci. Hortic. 2018, 240, 344–353. [Google Scholar] [CrossRef]
- Wang, B.H.; Sun, X.X.; Dong, F.Y.; Zhang, F.; Niu, J.X. Cloning and expression analysis of an MYB gene associated with calyx persistence in Korla fragrant pear. Plant Cell Rep. 2014, 33, 1333–1341. [Google Scholar] [CrossRef]
- Chen, Y.; Jin, M.; Wu, C.Y.; Bao, J.P. Effects of Plant Growth Regulators on the Endogenous Hormone Content of Calyx Development in ‘Korla’ Fragrant Pear. HortScience 2022, 57, 497–503. [Google Scholar] [CrossRef]
- Ma, H.C.; Wen, X.; Qi, M.; Li, J. Effects of different reagent treatments on the microstructure of calyx tube with or without calyx of Korla fragrant pear. J. Fruit. Sci. 2011, 28, 518–520+551. [Google Scholar] [CrossRef]
- Stopar, M.; Tojnko, S. Small fruit appearance on ‘Fuji/M. 9’apples thinned by the most known thinning agents. Gronn Kunnskap 2005, 9, 1–4. [Google Scholar]
- Zhang, S.K.; Abreq, N.Y.Z.; Wang, S.P.; Wang, Y.T.; Sun, S.Z.; Fan, G.Q. Effects of Uniconazole on Branch and Leaf Growth and Fruit Quality of Korla Fragrant Pear. Xinjiang Agric. Sci. 2020, 57, 1674–1680. [Google Scholar]
- McCarthy, B.L. Trends in organic and green food consumption in China: Opportunities and challenges for regional Australian exporters. J. Econ. Soc. Policy 2015, 1, 6–31. [Google Scholar]
- Nuttavuthisit, K.; Thøgersen, J. The Importance of Consumer Trust for the Emergence of a Market for Green Products: The Case of Organic Food. J. Bus. Ethics 2017, 140, 323–337. [Google Scholar] [CrossRef]
- Mohd, S.N.; Majeed, A.; Mohd, S.N. Impact of consumption values on consumers purchase of organic food and green environmental concerns. Soc. Responsib. J. 2022, 18, 1128–1141. [Google Scholar] [CrossRef]
- Datta, S.K. Breeding of ornamentals: Success and technological status. Nucleus 2022, 65, 107–128. [Google Scholar] [CrossRef]
- Huang, J.Q.; Zhang, G.; Li, Y.H.; Lyu, M.J.; Zhang, H.; Zhang, N.; Chen, R. Integrative genomic and transcriptomic analyses of a bud sport mutant ‘Jinzao Wuhe’ with the phenotype of large berries in grapevines. PeerJ 2023, 11, e14617. [Google Scholar] [CrossRef]
- Li, D.L.; Jing, M.M.; Dai, X.H.; Chen, Z.H.; Ma, C.M.; Chen, J.J. Current status of pineapple breeding, industrial development, and genetics in China. Euphytica 2022, 218, 85. [Google Scholar] [CrossRef]
- Chen, X.Y.; Niu, Y.Y.; Ding, X.; Mansur, N.; You, L.Y.; Liao, K. Analysis on the Difference of Fruit Tissue Structure between Korla Fragrant Pear and Its Bud Mutation Sha 01. Xinjiang Agric. Sci. 2019, 56, 1063–1071. [Google Scholar] [CrossRef]
- Niu, Y.Y.; Zhou, W.Q.; Chen, X.Y.; Fan, G.Q.; Zhang, S.K.; Liao, K. Genome size and chromosome ploidy identification in pear germplasm represented by Asian pears—Local pear varieties. Sci. Hortic. 2020, 265, 109202. [Google Scholar] [CrossRef]
- Yali, W.; Mitiku, T. Mutation breeding and its importance in modern plant breeding. J. Plant Sci. 2022, 10, 64–70. [Google Scholar] [CrossRef]
- Reyes, V.P.; Kitony, J.K.; Nishiuchi, S.; Makihara, D.; Doi, K. Utilization of Genotyping-by-Sequencing (GBS) for Rice Pre-Breeding and Improvement: A Review. Life 2022, 12, 1752. [Google Scholar] [CrossRef] [PubMed]
- Nyirahabimana, F.; Shimira, F.; Zahid, G. Recent status of Genotyping by Sequencing (GBS) Technology in cucumber (Cucumis sativus L.): A review. Mol. Biol. Rep. 2022, 49, 5547–5554. [Google Scholar] [CrossRef]
- Peterson, G.W.; Dong, Y.; Horbach, C.; Fu, Y.B. Genotyping-By-Sequencing for Plant Genetic Diversity Analysis: A Lab Guide for SNP Genotyping. Diversity 2014, 6, 665–680. [Google Scholar] [CrossRef]
- Wang, N.; Yuan, Y.; Wang, H. Applications of genotyping-by-sequencing (GBS) in maize genetics and breeding. Sci. Rep. 2020, 10, 16308. [Google Scholar] [CrossRef]
- Kitony, J.K.; Sunohara, H.; Tasaki, M.; Mori, J.-I.; Shimazu, A.; Reyes, V.P.; Yasui, H.; Yamagata, Y.; Yoshimura, A.; Yamasaki, M. Development of an Aus-Derived Nested Association Mapping (Aus-NAM) Population in Rice. Plants 2021, 10, 1255. [Google Scholar] [CrossRef] [PubMed]
- Pereira, D.L.; Vilanova, S.; Fita, A.; Prohens, J.; Rodríguez, B.A. Genetic diversity, population structure, and relationships in a collection of pepper (Capsicum spp.) landraces from the Spanish centre of diversity revealed by genotyping-by-sequencing (GBS). Hortic. Res. 2019, 6, 54. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.N. Genetic diversity and population structure of a Camelina sativa spring panel. Front. Plant Sci. 2019, 10, 184. [Google Scholar] [CrossRef]
- Liang, X.; Gao, M.; Amanullah, S.; Guo, Y.; Liu, X.; Xu, H.; Luan, F. Identification of QTLs linked with watermelon fruit and seed traits using GBS-based high-resolution genetic mapping. Sci. Hortic. 2022, 303, 111237. [Google Scholar] [CrossRef]
- Kim, W.J.; Yang, B.; Kim, D.G.; Kim, S.H.; Lee, Y.J.; Kim, J.; Baek, S.H.; Kang, S.Y.; Ahn, J.W.; Choi, Y.J. Genotyping-by-Sequencing Analysis Reveals Associations between Agronomic and Oil Traits in Gamma Ray-Derived Mutant Rapeseed (Brassica napus L.). Plants 2024, 13, 1576. [Google Scholar] [CrossRef]
- Khadgi, A.; Weber, C.A. Genome-Wide Association Study (GWAS) for Examining the Genomics Controlling Prickle Production in Red Raspberry (Rubus idaeus L.). Agronomy 2021, 11, 27. [Google Scholar] [CrossRef]
- Ramesh, P.; Mallikarjuna, G.; Sameena, S.; Kumar, A.; Gurulakshmi, K.; Reddy, B.V.; Sekhar, A.C. Advancements in molecular marker technologies and their applications in diversity studies. J. Biosci. 2020, 45, 1–15. [Google Scholar] [CrossRef]
- Hong, Y.B. Genetic diversity and distinctness based on morphological and SSR markers in peanut. Agron. J. 2021, 113, 4648–4660. [Google Scholar] [CrossRef]
- Zhang, J. Genetic diversity analysis and variety identification using SSR and SNP markers in melon. BMC Plant Biol. 2023, 23, 39. [Google Scholar] [CrossRef] [PubMed]
- Nickravesh, M.H.; Kourosh, V.; Fatemeh, A.; Erica, A.; Di, P.; Reza, A.; Keith, W.; Mohammad, M.A. Reliable Propagation of Persian Walnut Varieties Using SSR Marker-based True-to-type Validation. HortScience 2023, 58, 64–66. [Google Scholar] [CrossRef]
- Sugiura, A.; Kataoka, I.; Tomana, T. Use of refractometer to determine soluble solids of astringent fruits of Japanese persimmon (Diospyros kaki L.). J. Hortic. Sci. 1983, 58, 241–246. [Google Scholar] [CrossRef]
- Mattila, P.; Kumpulainen, J. Determination of free and total phenolic acids in plant-derived foods by HPLC with diode-array detection. J. Agric. Food Chem. 2002, 50, 3660–3667. [Google Scholar] [CrossRef]
- Ren, J.; Li, J.F.; Zhang, J.; Liu, J.F.; Xu, X.Y.; Jiang, J.B. A method for rapid determination of total sugar content based on 3,5-dinitrosalicylic acid colorimetry. Heilongjiang Sci. 2017, 8, 66–69. [Google Scholar]
- Zhang, M.L.; Guo, M.H.; Chen, D.H. Discussion on the Methods of Moisture Determination in Different Food. Mod. Agric. Sci. Technol. 2009, 21, 291–293. [Google Scholar]
- Elgailani, I.E.H.; Gad Elkareem, M.A.M.; Noh, E.A.A.; Adam, O.E.A.; Alghamdi, A.M.A. Comparison of two methods for the determination of vitamin c (ascorbic acid) in some fruits. Am. J. Chem. 2017, 2, 1–7. [Google Scholar] [CrossRef]
- Qu, C.X.; Shen, S.D.; Wang, X.F.; Cui, Y.H.; Song, W.P. Method research of measuring soluble protein contents of plant rough extraction using coomassie brilliant blue. J. Suzhou Univ. (Nat. Sci. Ed.) 2006, 2, 82–85. [Google Scholar]
- Liu, X.Y.; Li, L.; Zong, M.; Cai, Y.P. Contents and Distributions of Stone Cell and Their Effects on Fruit Quality of Pear. J. Anhui Agric. Univ. 2004, 31, 104–106. [Google Scholar] [CrossRef]
- Feng, Y.; Ning, R.; Wang, Z.; He, Y.; Hu, Y.; Sun, L.; Liu, Z. A Study on the Causes of Apomixis in Malus shizongensis. Horticulturae 2023, 9, 926. [Google Scholar] [CrossRef]
- Wang, H.; Hao, X.; Zhang, W.; Guo, Y.; Zhao, X.; Li, Y.; He, W.; Cai, S.; Song, X. Functional Verification of the Four Splice Variants from Ajania purpurea NST1 in Transgenic Tobacco. Horticulturae 2023, 9, 916. [Google Scholar] [CrossRef]
- Yong, Y.; Yuan, J.; Jin, X.; Huang, Y.; Zhang, Z.; Chen, Y.; Zhang, M. Analysis of Aroma Volatiles from Michelia crassipes Flower and Its Changes in Different Flower Organs during Flowering. Horticulturae 2023, 9, 442. [Google Scholar] [CrossRef]
- Qiu, J.; Gao, C.; Wei, H.; Wang, B.; Hu, Y.; Guo, Z.; Long, L.; Yang, L.; Li, H. Flowering Biology of Rhododendron pulchrum. Horticulturae 2021, 7, 508. [Google Scholar] [CrossRef]
- Torkamaneh, D.; Boyle, B.; St, C.J.; Légaré, G.; Pomerleau, S.; Belzile, F. NanoGBS: A miniaturized procedure for GBS library preparation. Front. Genet. 2020, 11, 67. [Google Scholar] [CrossRef]
- Rachappanavar, V.; Padiyal, A.; Sharma, J.K.; Negi, N. Analytical Pipelines for the GBS Analysis. Genotyping Seq. Crop Improv. 2022, 8, 161–187. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A. The Sequence Alignment/Map format and SAMtool. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Li, R.Q.; Zhu, H.M.; Ruan, J.; Qian, W.B.; Fang, X.D.; Shi, Z.B.; Li, Y.R.; Li, S.T.; Shan, G.; Karsten, K.; et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010, 20, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Kõressaar, T.; Lepamets, M.; Kaplinski, L.; Raime, K.; Andreson, R.; Remm, M. Primer3_masker: Integrating masking of template sequence with primer design software. Bioinformatics 2018, 34, 1937–1938. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Song, Y.; Li, L.T.; Khan, M.A.; Li, X.G.; Korban, S.S.; Wu, J.; Zhang, S.L. Construction of a High-Density Simple Sequence Repeat Consensus Genetic Map for Pear (Pyrus spp.). Plant Mol. Biol. Rep. 2015, 33, 316–325. [Google Scholar] [CrossRef]
- Hu, X.; Li, M.; Zhang, F.P.; Chen, H. Influence of Starvation on the Structure of Gut-Associated Bacterial Communities in the Chinese White Pine Beetle (Dendroctonus armandi). Forests 2016, 7, 126. [Google Scholar] [CrossRef]
- Pastorelli, R.; Agnelli, A.E.; De Meo, I.; Graziani, A.; Paletto, A.; Lagomarsino, A. Analysis of Microbial Diversity and Greenhouse Gas Production of Decaying Pine Logs. Forests 2017, 8, 224. [Google Scholar] [CrossRef]
- Li, C.L.; Xu, P.Y.; Zhou, A.T.; Song, J.L.; Wu, Y.X.; He, T.M. A Preliminary Study of Apomictic Characteristics of ‘Mianli’ (Pyrus sinkiangensis Yü). HortScience 2022, 57, 818–826. [Google Scholar] [CrossRef]
- Claessen, H.; Keulemans, W.; Poel, B.V.D.; Storme, N.D. Finding a Compatible Partner: Self-Incompatibility in European Pear (Pyrus communis); Molecular Control, Genetic Determination, and Impact on Fertilization and Fruit Set. Front. Plant Sci. 2019, 10, 407. [Google Scholar] [CrossRef] [PubMed]
- Losada, J.M.; Herrero, M. Flower strategy and stigma performance in the apple inflorescence. Sci. Hortic. 2013, 150, 283–289. [Google Scholar] [CrossRef]
- Li, X.C.; Tao, H.; Zhang, S.M.; Liu, H.Y. The Impact and Prediction of Climate Change on First-flowering Date of Korla Fragrant Pear. Chin. J. Agrometeorol. 2012, 33, 119–123. [Google Scholar]
- Xu, Y.T.; Zhang, Q.P.; Zhang, X.; Wang, J.; Ayup, M.; Yang, B.; Guo, C.M.; Gong, P.; Dong, W.X. The proteome reveals the involvement of serine/threonine kinase in the recognition of self- incompatibility in almond. J. Proteom. 2022, 256, 104505. [Google Scholar] [CrossRef]
- Matsumoto, D.; Yamane, H.; Abe, K.; Tao, R. Identification of a Skp1-Like Protein Interacting with SFB, the Pollen S Determinant of the Gametophytic Self-Incompatibility in Prunus. Plant Physiol. 2012, 159, 1252–1262. [Google Scholar] [CrossRef]
- Zhao, W.; Baldwin, E.A.; Bai, J.; Plotto, A.; Irey, M. Comparative analysis of the transcriptomes of the calyx abscission zone of sweet orange insights into the huanglongbing-associated fruit abscission. Hortic. Res. 2019, 6, 831–845. [Google Scholar] [CrossRef]
- Tian, J.; Wen, Y.; Zhang, F.; Sai, J.Y.; Zhang, Y.; Li, W.S. Effects of Endogenous Hormones and Sugars on Fruit Size Driven by Cell Division between Korla Fragrant Pear and Its Bud Mutation. HortScience Horts 2021, 56, 881–888. [Google Scholar] [CrossRef]
- Ma, L.; Li, C.J.; Niu, J.X.; Pei, M.S.; Cao, F.J.; Quan, S.W. Cloning and identification of novel miRNAs in the flower organs of Korla fragrant pear at anthesis. J. Hortic. Sci. Biotechnol. 2019, 94, 1–12. [Google Scholar] [CrossRef]
- Pei, M.S.; Niu, J.X.; Li, C.J.; Cao, F.J.; Quan, S.W. Identification and expression analysis of genes related to calyx persistence in Korla fragrant pear. BMC Genom. 2016, 17, 132. [Google Scholar] [CrossRef]
- Gong, X.; Bao, J.P.; Chen, J.; Qi, K.J.; Xie, Z.H.; Rui, W.K.; Hao, G.W.; Katsuhiro, S.; Shahrokh, K.; Zhang, S.L.; et al. Candidate proteins involved in the calyx abscission process of ‘Kuerlexiangli’ (Pyrus sinkiangensis Yu) identified by iTRAQ analysis. Acta Physiol. Plant 2020, 42, 112. [Google Scholar] [CrossRef]
- Liu, S.Z.; Liu, J.; Zhang, Y.; Peng, Y.J. Mass Fraction of Sugar and Acid and Flavor Index of Pear Cultivars(Pyurs bretschneideri) at Different Ripening Stages. Acta Agric. Boreali-Occident. Sin. 2015, 24, 97–102. [Google Scholar]
- Guo, N.; Zhang, Y.J.; Sun, X.; Fan, H.H.; Gao, J.S.; Chao, Y.P.; Liu, A.F.; Yu, X.T.; Cai, Y.P.; Lin, Y. Genome-wide identification of differentially expressed miRNAs induced by ethephon treatment in abscission layer cells of cotton (Gossypium hirsutum). Gene 2018, 676, 263–268. [Google Scholar] [CrossRef]
- Liao, W.B.; Wang, G.; Li, Y.Y.; Wang, B.; Zhang, P.; Peng, M. Reactive oxygen species regulate leaf pulvinus abscission zone cell separation in response to water-deficit stress in cassava. Sci. Rep. 2016, 6, 21542. [Google Scholar] [CrossRef]
- Xiao, X.; Zhu, M.J.; Liu, Y.S.; Zheng, J.R.; Cui, Y.P.; Xiong, C.D.; Liu, J.J.; Chen, J.; Cai, H.W. Phenotypical and gene co-expression network analyses of seed shattering in divergent sorghum (Sorghum spp.). Crop J. 2023, 11, 478–489. [Google Scholar] [CrossRef]
- Mckim, S.M.; Stenvik, G.E.; Butenko, M.A.; Kristiansen, W.; Cho, S.K.; Hepworth, S.R.; Aalen, R.B.; Haughn, G.W. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development 2008, 135, 1537–1546. [Google Scholar] [CrossRef] [PubMed]
- Shimon, M.; Sonia, P.H.; Srivignesh, S.; Vijay Selvaraj, K.S.; Shaul, B.; Ron, O.; Bettina, K.; Michael, S.R.; Cai, Z.J.; Amnon, L. Microarray Analysis of the Abscission-Related Transcriptome in the Tomato Flower Abscission Zone in Response to Auxin Depletion. Plant Physiol. 2010, 154, 1929–1956. [Google Scholar] [CrossRef]
- Hoban, S.; Archer, F.I.; Bertola, L.D.; Bragg, J.G.; Breed, M.F.; Bruford, M.W.; Hunter, M.E. Global genetic diversity status and trends: Towards a suite of Essential Biodiversity Variables (EBVs) for genetic composition. Biol. Rev. 2022, 97, 1511–1538. [Google Scholar] [CrossRef] [PubMed]
- Al Salameen, F.; Habibi, N.; Al Amad, S.; Al Doaij, B. Genetic Diversity of Rhanterium eppaposumOliv. Populations in Kuwait as Revealed by GBS. Plants 2022, 11, 1435. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.S.M.F.; Sanders, D.; Mishra, A.K.; Joshi, V. Genetic Diversity and Population Structure Analysis of the USDA Olive Germplasm Using Genotyping-By-Sequencing (GBS). Genes 2021, 12, 2007. [Google Scholar] [CrossRef] [PubMed]
- Delfini, J.; Moda, C.V.; Dos, S.N.J.; Ruas, P.M.; Sant, A.G.C.; Gepts, P.; Gonçalves, L.S.A. Population structure, genetic diversity and genomic selection signatures among a Brazilian common bean germplasm. Sci. Rep. 2021, 11, 2964. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Hou, L.; Zhang, Z.; Pang, X.; Li, Y. Genetic diversity, population structure, and linkage disequilibrium of a core collection of Ziziphus jujuba assessed with genome-wide SNPs developed by genotyping-by-sequencing and SSR markers. Front. Plant Sci. 2017, 8, 575. [Google Scholar] [CrossRef] [PubMed]
- Hussain, H.; Nisar, M. Assessment of plant genetic variations using molecular markers: A review. J. Appl. Biol. Biotechnol. 2020, 8, 99–109. [Google Scholar] [CrossRef]
- Yin, J.; Zhao, H.; Wu, X.; Ma, Y.; Zhang, J.; Li, Y.; Xu, Z. SSR marker based analysis for identification and of genetic diversity of non-heading Chinese cabbage varieties. Front. Plant Sci. 2023, 14, 1112748. [Google Scholar] [CrossRef]
- Olejnik, A.; Parkitna, K.; Kozak, B.; Florczak, S.; Matkowski, J.; Nowosad, K. Assessment of the Genetic Diversity of Chrysanthemum Cultivars Using SSR Markers. Agronomy 2021, 11, 2318. [Google Scholar] [CrossRef]
- Castellana, S. Genetic characterization and molecular fingerprint of traditional Umbrian tomato (Solanum lycopersicum L.) landraces through SSR markers and application for varietal identification. Genet. Resour. Crop Evol. 2020, 67, 1–14. [Google Scholar] [CrossRef]





| Genome of Reference | Number of Samples | Filter Condition | Total Number of SNP | Total Number of SNP after Filtering |
|---|---|---|---|---|
| Pyrus bretschneideri | 20 | Dp4-miss 0.4-maf 0.01 | 713,605 | 114,911 |
| Population (Variety) | Observed Heterozygosity (Ho) | Expected Heterozygosity (He) | Genetic Diversity Index (pi) |
|---|---|---|---|
| YB | 0.7757 a | 0.4276 a | 0.4545 a |
| CK | 0.7726 a | 0.4285 a | 0.4560 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, X.; Zhang, C.; Sun, H.; Wang, S.; Cui, Y.; Zhao, L. Fruiting Characteristics and Molecular-Assisted Identification of Korla Fragrant Pear Bud Mutation Materials. Appl. Sci. 2024, 14, 6589. https://doi.org/10.3390/app14156589
Yang X, Zhang C, Sun H, Wang S, Cui Y, Zhao L. Fruiting Characteristics and Molecular-Assisted Identification of Korla Fragrant Pear Bud Mutation Materials. Applied Sciences. 2024; 14(15):6589. https://doi.org/10.3390/app14156589
Chicago/Turabian StyleYang, Xian’an, Cuifang Zhang, Haichang Sun, Shiwei Wang, Yutong Cui, and Long Zhao. 2024. "Fruiting Characteristics and Molecular-Assisted Identification of Korla Fragrant Pear Bud Mutation Materials" Applied Sciences 14, no. 15: 6589. https://doi.org/10.3390/app14156589
APA StyleYang, X., Zhang, C., Sun, H., Wang, S., Cui, Y., & Zhao, L. (2024). Fruiting Characteristics and Molecular-Assisted Identification of Korla Fragrant Pear Bud Mutation Materials. Applied Sciences, 14(15), 6589. https://doi.org/10.3390/app14156589





