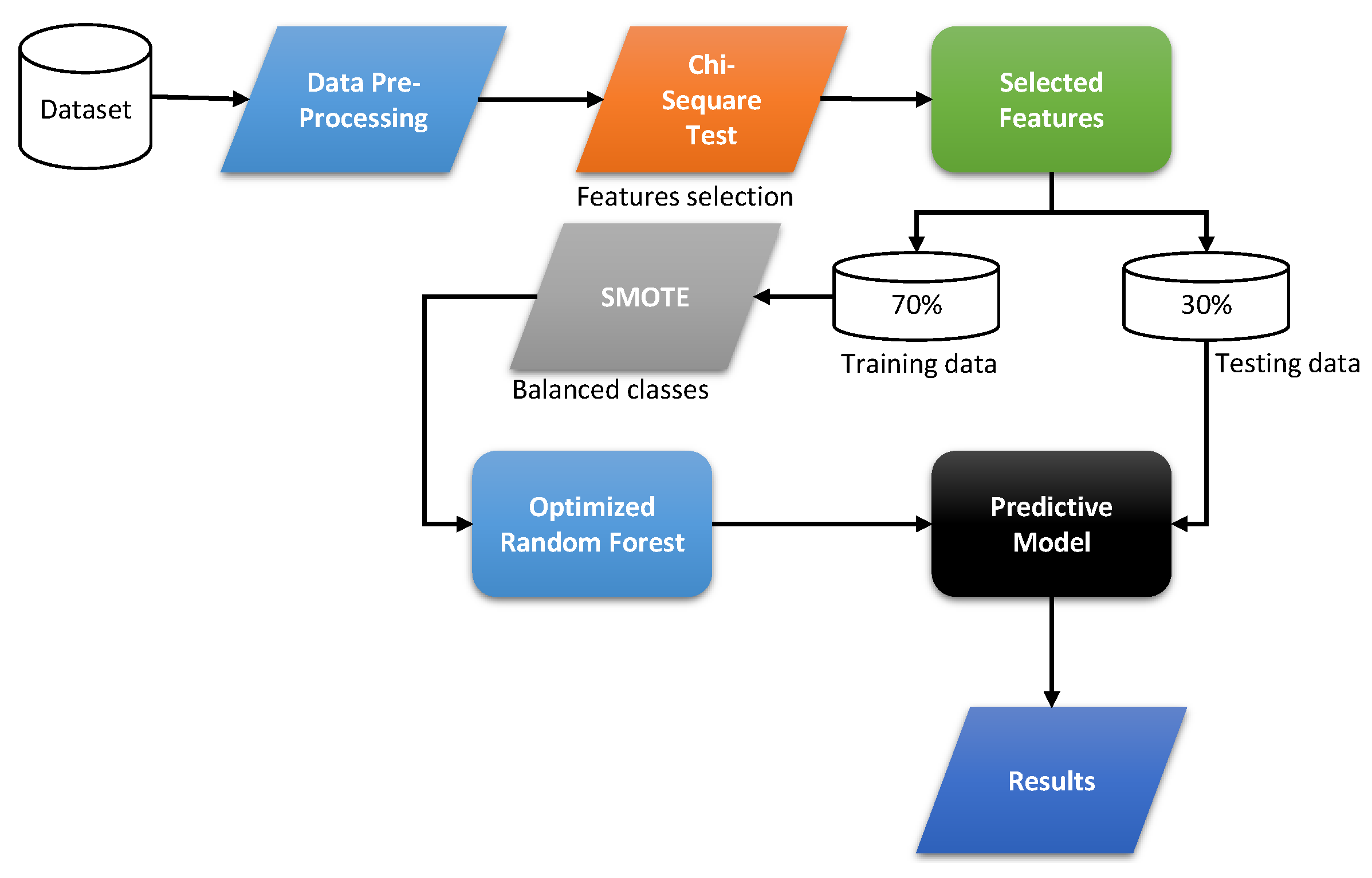
2.2. Proposed Work
In this study, we developed a diagnostic system for the early detection of mortality risk in heart disease. The proposed diagnostic system uses a dataset of 55 features based on daily lifestyle factors, medical history, and biochemical test results. The newly developed system consists of two main components. One of the main components is to select useful features from the dataset that can help predict the cause of cardiac mortality. The second component, on the other hand, is a classifier that predicts cardiac mortality. To select features from the given dataset, we used a static chi-square model (
), whereas, for the classification problem, we used a random forest classifier (RF), fine-tuning the hyperparameters of the RF using the grid search algorithm [
23]. The working of the newly developed system is given in
Figure 2.
For this study, we used the (
) model to rank the features from the dataset to eliminate irrelevant features. In feature selection, (
) computes the statistics between the non-negative feature
and the class. The model performs the (
) test, which analyzes the degree of dependence between the features and the class. As a result, the model can exclude features more likely to be class-independent, as these features could be considered unimportant for classification. The features are sorted in the first phase depending on the (
) test score. Then, we search for ideal features
from the scored features. For information on feature selection and discretization using (
) statistics, the reader is referred to [
24]. Mathematical feature selection based on the (
)-test is described as follows:
From
Table 2, we can calculate the statistical score
for positive and negative classes for the ℧ instances of the binary classification problem of heart mortality. In
Table 2,
represents the number of instances that do not have feature
, ℧−
denotes the number of instances that do not contain feature
,
represents the positive instances, and the number of positive instances can be represented from ℧−
. The main purpose of the
test is to measure the expected count, i.e., C, and the observed count, i.e., B, which are derived from each other. Assuming that
,
,
, and
represent the observed values and
,
,
, and
denote the expected values, then the predicted values based on the null hypothesis of independent events can be calculated as follows:
From Equation (
1),
,
, and
can be computed. For the general formulation of the
score, we have
From solving the equations, we obtain a simple form of Equation (
3)
After feature ranking from Equation (
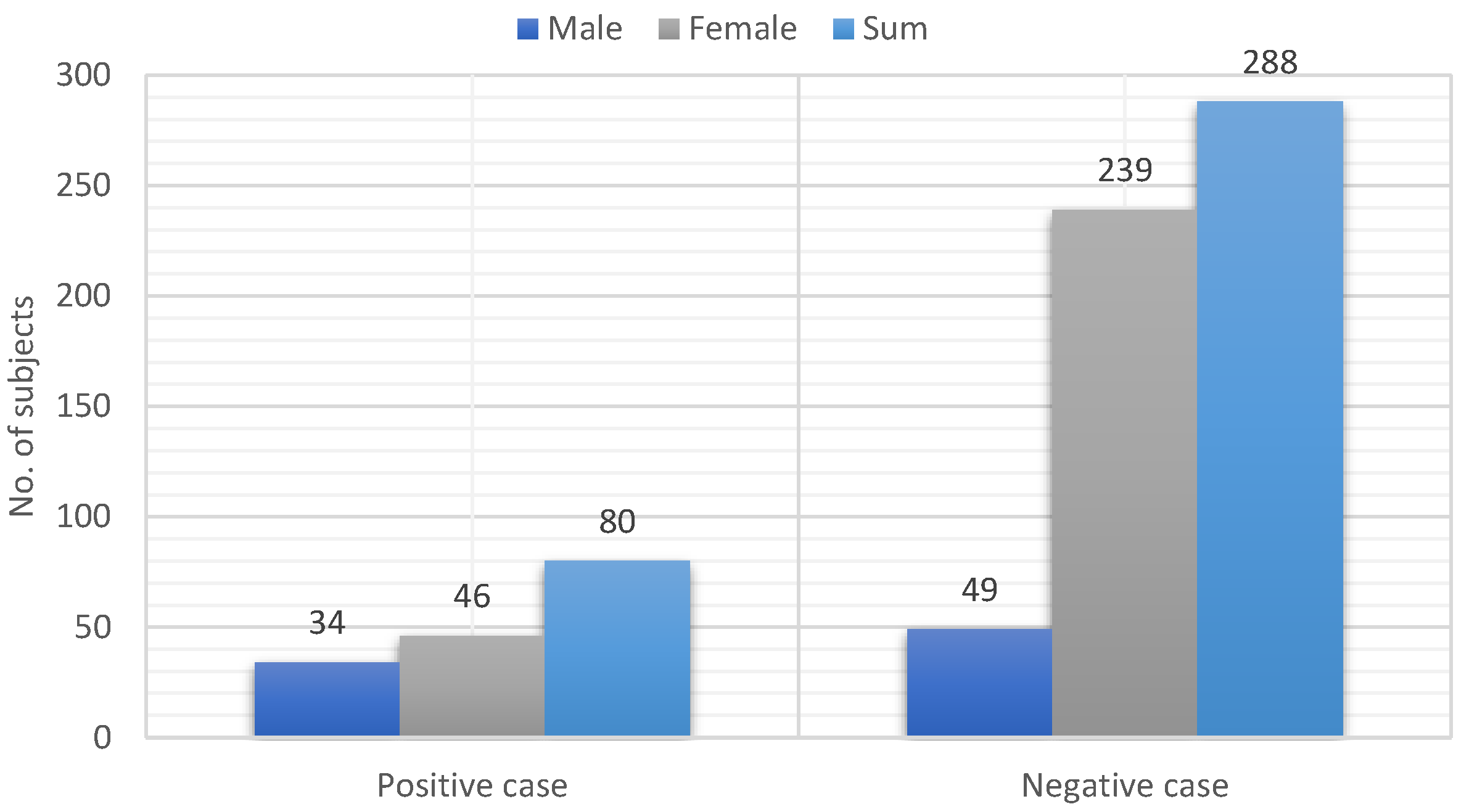
4), the highly ranked (selected) features (or a subset of the features) are input into RF for classification. However, before the classification phase, it was found that the number of class instances in the dataset was highly imbalanced. To overcome the problem of bias in machine learning, we employed the synthetic minority oversampling technique (SMOTE). SMOTE achieves balanced classes in the data by enriching the training data with synthetic minority class samples, resulting in balanced classes and optimized training processes. It is important to note that SMOTE should be applied to the training data after data partitioning and not to the entire dataset before partitioning to avoid superficial performance caused by having copies of samples from the test dataset in the training dataset [
25]. Unlike other oversampling methods, SMOTE works in the feature space rather than the data space [
26] by synthesizing minority class samples by generating new samples along the line connecting any or all of the nearest neighbors of the k minority class. Using a holdout validation technique, we divided the dataset into two halves for training and testing to balance between classes. Seventy percent of the dataset was used for training, and thirty percent for ML model testing. After utilizing SMOTE, we only balanced the classes in the training data, which had 396 samples for each class and 198 samples total (positive and negative). In this study, we used the Python software package and the imbalance-learn package to implement the SMOTE technique [
27].
After balancing the dataset, the RF model was used; here, the formulation of the RF model is reproduced as follows: RF is an ensemble model q(s, t) in which n is a uniformly distributed irregular vector. Each tree contributes to determining the most abundant class at input s. For an input sample of size P, where P represents an instance in the training set, p samples are taken from each instance. After this, F features are used to sample f features. This process is randomly repeated n times, resulting in n training sets, denoted as
,
,
…,
. The decision tree
,
,
…,
is generated from the corresponding training sets. Each tree in the forest, except for shear, is fully mature. Many decision trees have contributed to the development of the random forest classification algorithm. The number of decision trees, E, and the depth of each tree, D, are two important hyperparameters for classification [
28]. These parameters determine the number of trees forming the forest and the maximum depth of each tree, respectively. For the objective of this study, we used a grid search algorithm to determine the values of E and D that maximize the efficiency of the RF model. In addition, a random forest model was created, and a new sample is included. In addition, the decision tree examines the new sample to determine its category. The final classification of a sample can be determined based on the votes cast by all decision trees within the forest. The trees that result from the formation of the random forest are called bootstrap trees. This is because they are created by resampling by reverting to the training data. Bootstrap is a simple and useful solution for model integration using the replacement method [
29]. The training set is used to obtain a set number of samples for bootstrap sampling. The number of samples is returned to the training set after sampling. The extracted samples are assembled into a new batch of bootstrap samples. There is also the possibility that the sampled samples will be resampled after being returned to the training set. For this reason, it is best to test the samples that have already been taken. As an example, consider a random sample of d samples. Using
and
, we can calculate the probability that the sample will or will not be captured each time. If the random sample is run D times, then the probability of the selected sample is given as
and D converges to
∞ and
converges to
. There will be a mirror sample. In addition, the
instance will break into new samples. Out-of-bag instances are data that are missed during extraction. An out-of-bag instance is called an OOB error. This problem can be expressed mathematically as follows:
From Equation (
5),
ı denotes the error value for testing
, where
stands for the number of OOB instances and is acknowledged as a class of each data. From Equation (
5),
ı denotes the error value for testing
, where
stands for the number of OOB instances and is acknowledged as a class of each data.
The Gini index is used to build a decision tree, which is then used to determine the model’s impurity level of the model using the CART method. A lower Gini index value indicates fewer contaminants. The Gini index is lower when there are fewer impurities. For the classification problem, the probability of the
Nth category is
for
N categories, and the mathematical formulation of the Gini index is as follows:
The Gini index is used for the feature selection in the decision tree, and the mathematical formula for this is given in Equation (
7):
The highest Gini index value is selected for the split attribute and the node for the split condition. In the case of overfitting, the decision tree is mirrored. Pre- and post-pruning procedures can reduce the overfitting rate [
30]. Pre-pruning can lead to the premature development of decision trees but post-pruning can produce greater results. In addition, the selection is made without pruning. We apply a subset of features selected from the
statistical model to the RF method for classification. The best RF hyperparameters for this subset of features are found using the grid search approach. Then, another group of features is input to the RF algorithm. The grid search algorithm is used to research the optimized values of the hyperparameters from RF. The best hyperparameters from RF, such as the number of edges (E) and the depth of the tree (D), are searched out. The
method is used for each subset of features created. Finally, the subset of features with the best predictive accuracy of cardiac mortality is selected and published.