Integrating PubMed Label Hierarchy Knowledge into a Complex Hierarchical Deep Neural Network †
Abstract
1. Introduction
2. Related Works
3. Methodology
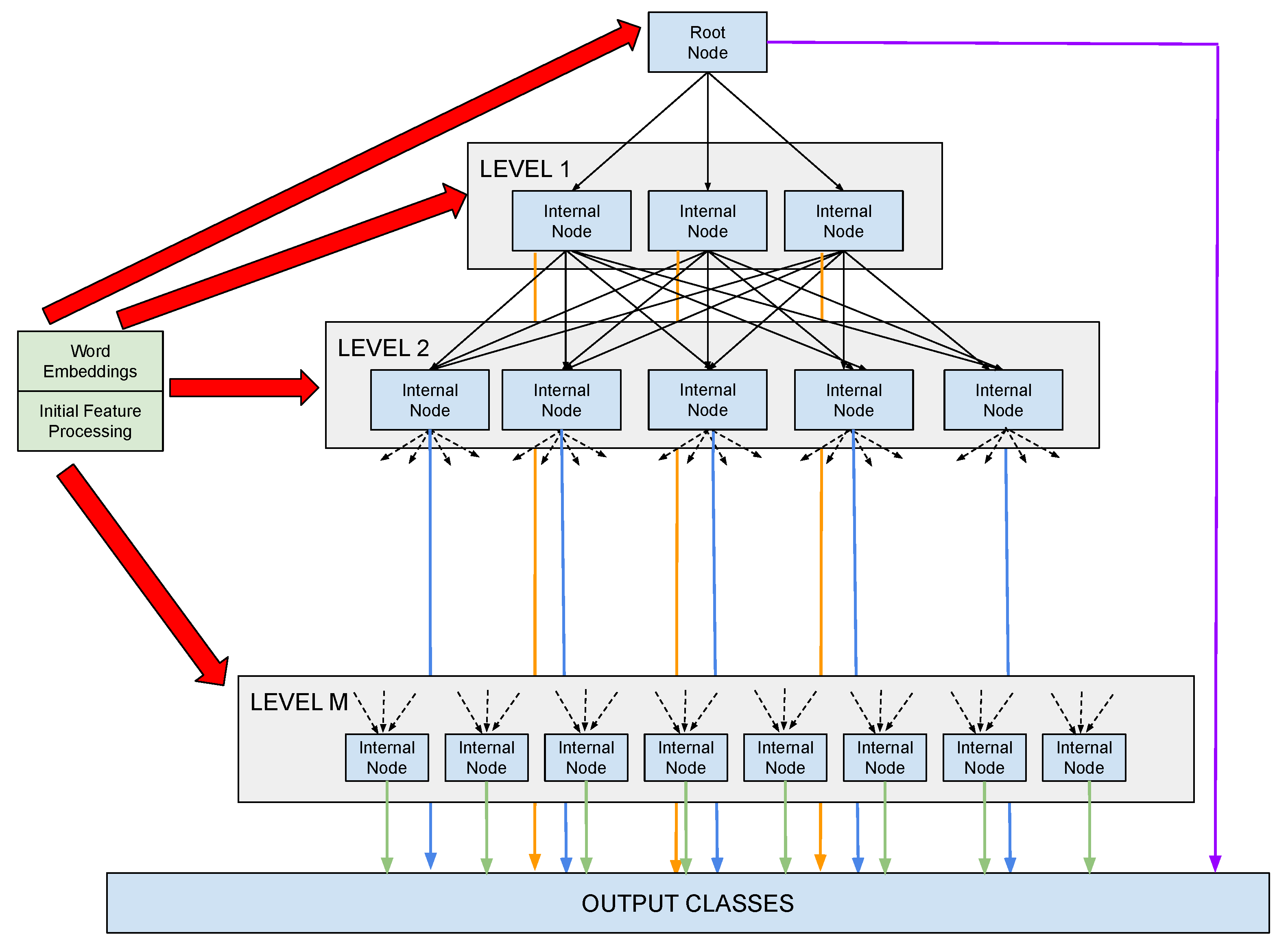
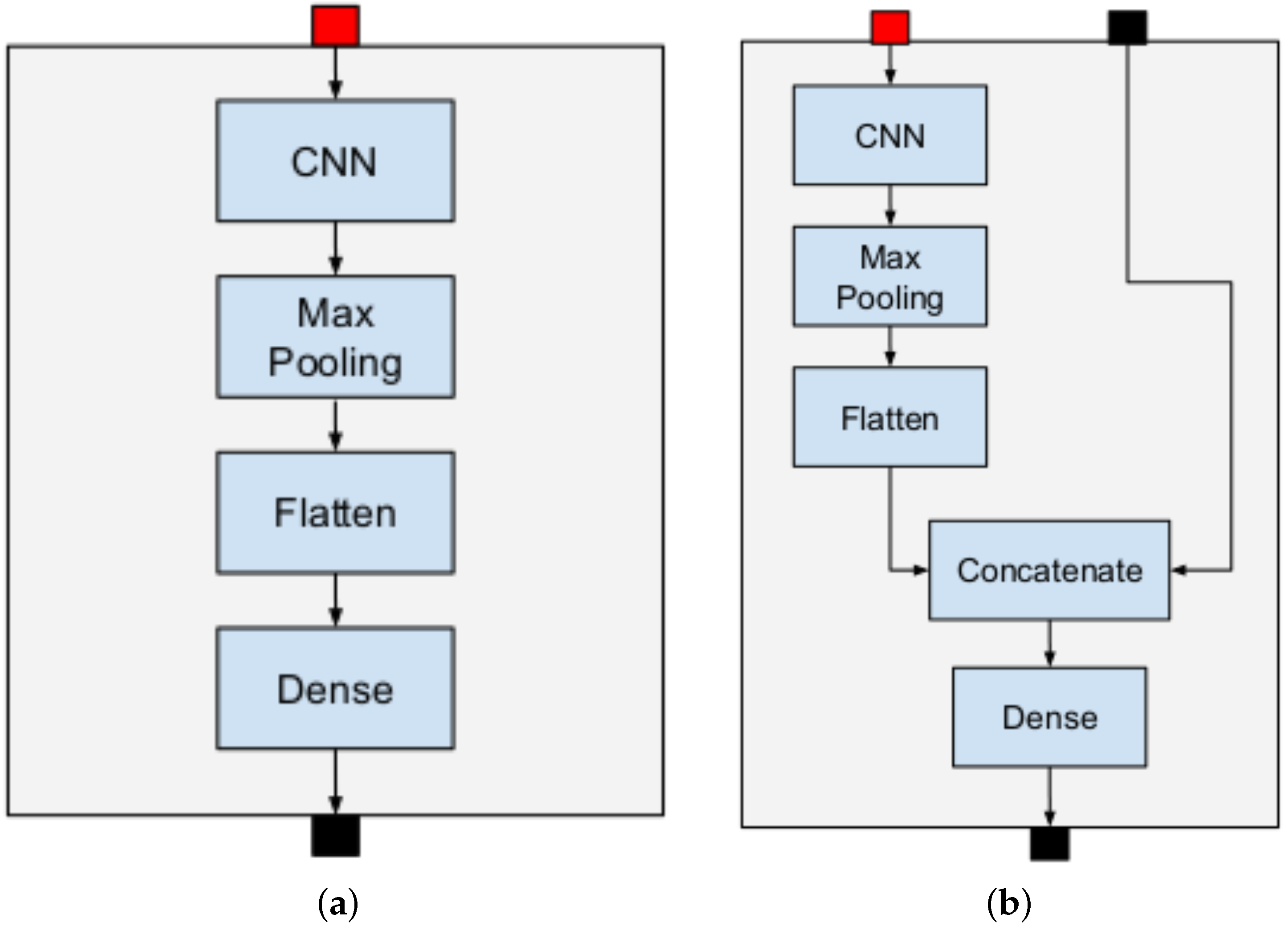
3.1. Hierarchical Deep Neural Network
- Preprocessing node: Where textual data in the input are converted into a word-embeddings-based representation (the green ones in the upper left part of the figure);
- Internal node: A DNN basic block whose inputs are word-embeddings-based and the classification result is obtained as the output of its parent node. In the same way, its output is connected in input to the corresponding children node;
- Root Node: A special internal node that corresponds to the root of the hierarchy. It differs from the other internal nodes because its input is directly and only connected to the output of the preprocessing node.
| Algorithm 1 Pseudocode for HDDN generation |
|
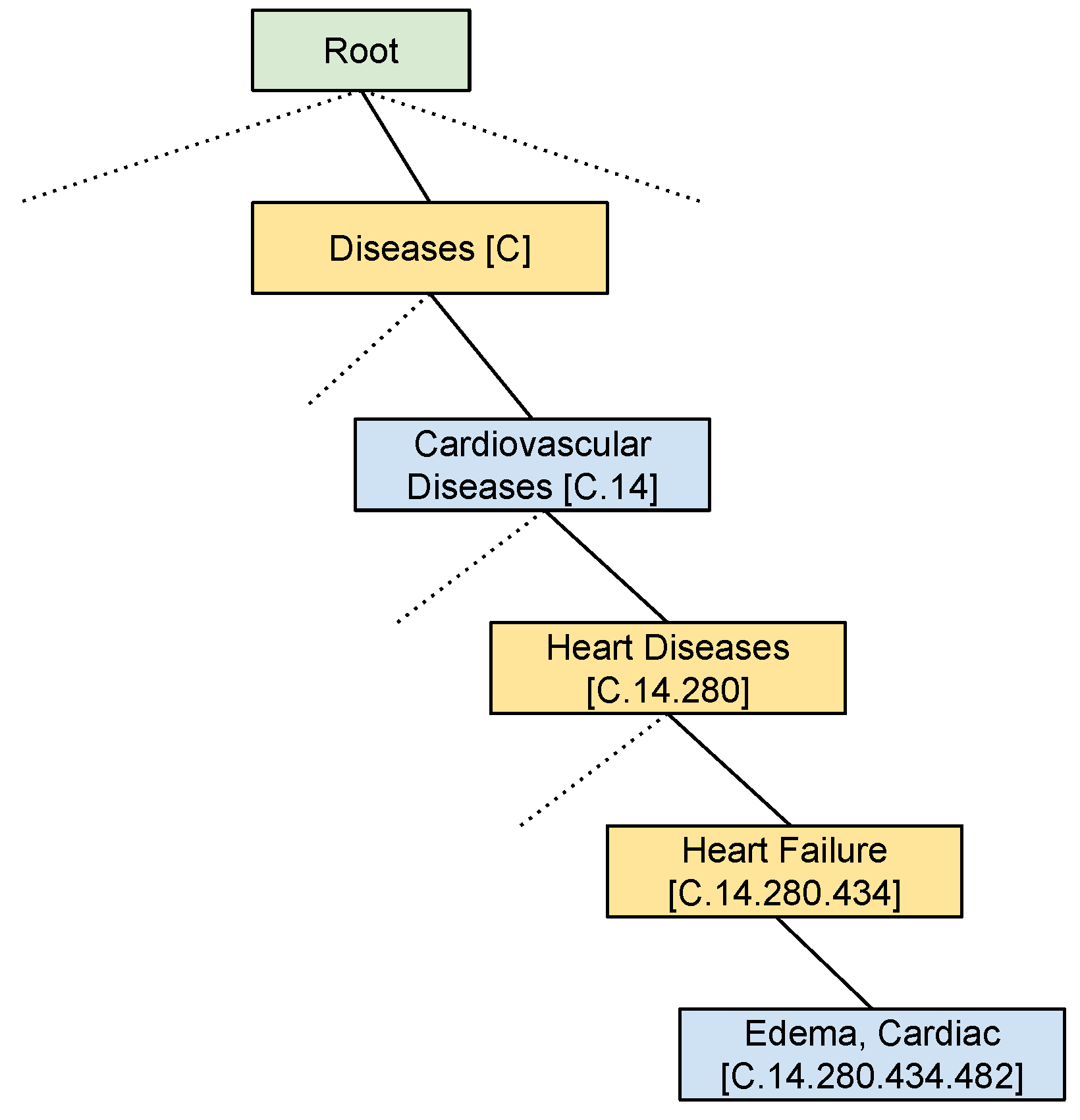
3.2. Hierarchical Label Set Expansion
4. Experimental Assessment and Discussion
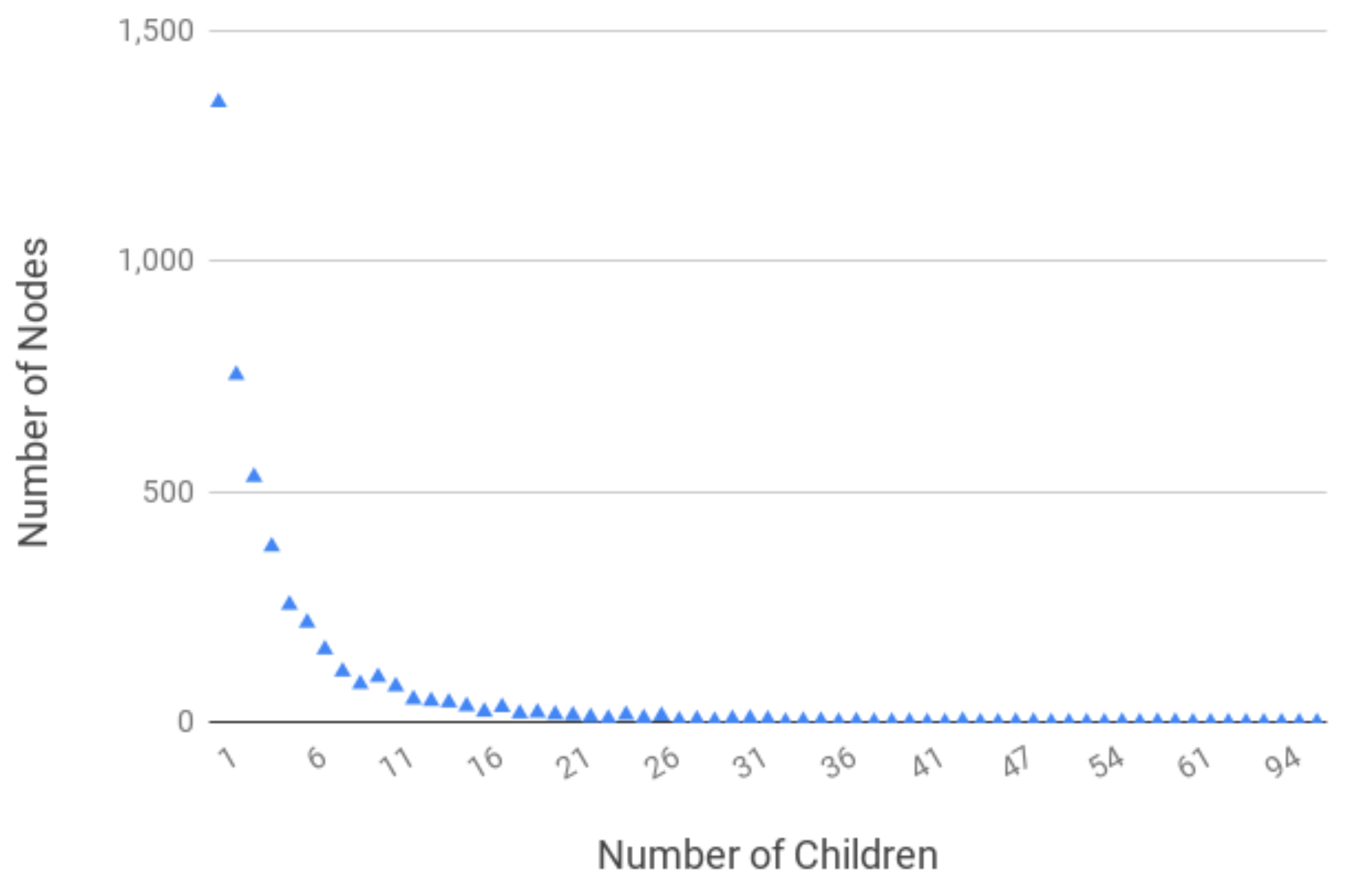
4.1. Dataset
4.2. Word Embedding Models
4.3. Tools and Hardware
4.4. Evaluation Metrics
4.5. Results and Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, L.; Polosukhin, I. Attention is All you Need. In Proceedings of the Advances in Neural Information Processing Systems 30: Annual Conference on Neural Information Processing Systems 2017, Long Beach, CA, USA, 4–9 December 2017; pp. 5998–6008. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Minneapolis, MN, USA, 2–7 June 2019; ACL: Minneapolis, MN, USA, 2019; Volume 1, pp. 4171–4186. [Google Scholar] [CrossRef]
- Shamshad, F.; Khan, S.; Zamir, S.W.; Khan, M.H.; Hayat, M.; Khan, F.S.; Fu, H. Transformers in medical imaging: A survey. Med. Image Anal. 2023, 88, 102802. [Google Scholar] [CrossRef]
- Yang, X.; Bian, J.; Hogan, W.R.; Wu, Y. Clinical concept extraction using transformers. J. Am. Med. Inform. Assoc. 2020, 27, 1935–1942. [Google Scholar] [CrossRef]
- Xiao, H.; Li, L.; Liu, Q.; Zhu, X.; Zhang, Q. Transformers in medical image segmentation: A review. Biomed. Signal Process. Control 2023, 84, 104791. [Google Scholar] [CrossRef]
- Stylianou, N.; Vlahavas, I. TransforMED: End-to-End transformers for evidence-based medicine and argument mining in medical literature. J. Biomed. Inform. 2021, 117, 103767. [Google Scholar] [CrossRef]
- Alicante, A.; Benerecetti, M.; Corazza, A.; Silvestri, S. A distributed architecture to integrate ontological knowledge into information extraction. Int. J. Grid Util. Comput. 2016, 7, 245–256. [Google Scholar] [CrossRef]
- Yin, Y.; Lai, S.; Song, L.; Zhou, C.; Han, X.; Yao, J.; Su, J. An External Knowledge Enhanced Graph-based Neural Network for Sentence Ordering. J. Artif. Intell. Res. 2021, 70, 545–566. [Google Scholar] [CrossRef]
- Gu, T.; Zhao, H.; He, Z.; Li, M.; Ying, D. Integrating external knowledge into aspect-based sentiment analysis using graph neural network. Knowl.-Based Syst. 2023, 259, 110025. [Google Scholar] [CrossRef]
- Marcus, G. Deep Learning: A Critical Appraisal. arXiv 2018, arXiv:abs/1801.00631. [Google Scholar]
- Liu, J.; Chang, W.; Wu, Y.; Yang, Y. Deep Learning for Extreme Multi-label Text Classification. In Proceedings of the 40th International ACM SIGIR Conference on Research and Development in Information Retrieval, Tokyo, Japan, 7–11 August 2017; ACM: Tokyo, Japan, 2017; pp. 115–124. [Google Scholar] [CrossRef]
- Gargiulo, F.; Silvestri, S.; Ciampi, M.; De Pietro, G. Deep neural network for hierarchical extreme multi-label text classification. Appl. Soft Comput. 2019, 79, 125–138. [Google Scholar] [CrossRef]
- Nentidis, A.; Bougiatiotis, K.; Krithara, A.; Paliouras, G.; Kakadiaris, I. Results of the fifth edition of the BioASQ Challenge. In Proceedings of the BioNLP 2017 Workshop, Vancouver, BC, Canada, 4 August 2017; ACL: Vancouver, BC, Canada, 2017; pp. 48–57. [Google Scholar]
- Nentidis, A.; Krithara, A.; Bougiatiotis, K.; Paliouras, G. Overview of BioASQ 8a and 8b: Results of the Eighth Edition of the BioASQ Tasks a and b. In Proceedings of the Working Notes of CLEF 2020—Conference and Labs of the Evaluation Forum, Thessaloniki, Greece, 22–25 September 2020; CEUR Workshop Proceedings. Cappellato, L., Eickhoff, C., Ferro, N., Névéol, A., Eds.; CEUR-WS.org: Aachen, Germany, 2020; Volume 2696. [Google Scholar]
- Nentidis, A.; Katsimpras, G.; Vandorou, E.; Krithara, A.; Paliouras, G. Overview of BioASQ Tasks 9a, 9b and Synergy in CLEF2021. In Proceedings of the Working Notes of CLEF 2021—Conference and Labs of the Evaluation Forum, Bucharest, Romania, 21–24 September 2021; CEUR Workshop Proceedings. Faggioli, G., Ferro, N., Joly, A., Maistro, M., Pirpi, F., Eds.; CEUR-WS.org: Aachen, Germany, 2021; Volume 2936. [Google Scholar]
- Nentidis, A.; Krithara, A.; Paliouras, G.; Gasco, L.; Krallinger, M. BioASQ at CLEF2022: The Tenth Edition of the Large-scale Biomedical Semantic Indexing and Question Answering Challenge. In Advances in Information Retrieval; Hagen, M., Verberne, S., Macdonald, C., Seifert, C., Balog, K., Norvaag, K., Setty, V., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 429–435. [Google Scholar]
- Peng, S.; You, R.; Wang, H.; Zhai, C.; Mamitsuka, H.; Zhu, S. DeepMeSH: Deep semantic representation for improving large-scale MeSH indexing. Bioinformatics 2016, 32, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Yoon, W.; Kim, S.; Kim, D.; Kim, S.; So, C.H.; Kang, J. BioBERT: A pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 2019, 36, 1234–1240. [Google Scholar] [CrossRef] [PubMed]
- You, R.; Liu, Y.; Mamitsuka, H.; Zhu, S. BERTMeSH: Deep contextual representation learning for large-scale high-performance MeSH indexing with full text. Bioinformatics 2020, 37, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Tinn, R.; Cheng, H.; Lucas, M.; Usuyama, N.; Liu, X.; Naumann, T.; Gao, J.; Poon, H. Domain-Specific Language Model Pretraining for Biomedical Natural Language Processing. ACM Trans. Comput. Health 2022, 3, 1–23. [Google Scholar] [CrossRef]
- Mustafa, F.E.; Boutalbi, R.; Iurshina, A. Annotating PubMed Abstracts with MeSH Headings using Graph Neural Network. In Proceedings of the Fourth Workshop on Insights from Negative Results in NLP, Dubrovnik, Croatia, 5 May 2023; Association for Computational Linguistics: Dubrovnik, Croatia, 2023; pp. 75–81. [Google Scholar] [CrossRef]
- Gargiulo, F.; Silvestri, S.; Ciampi, M. Exploit Hierarchical Label Knowledge for Deep Learning. In Proceedings of the 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS), Córdoba, Spain, 5–7 June 2019; pp. 539–542. [Google Scholar] [CrossRef]
- Brust, C.A.; Denzler, J. Integrating Domain Knowledge: Using Hierarchies to Improve Deep Classifiers. In Pattern Recognition; Palaiahnakote, S., Sanniti di Baja, G., Wang, L., Yan, W.Q., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 3–16. [Google Scholar]
- Chen, F.; Yin, G.; Dong, Y.; Li, G.; Zhang, W. KHGCN: Knowledge-Enhanced Recommendation with Hierarchical Graph Capsule Network. Entropy 2023, 25, 697. [Google Scholar] [CrossRef] [PubMed]
- Kowsari, K.; Brown, D.E.; Heidarysafa, M.; Meimandi, K.J.; Gerber, M.S.; Barnes, L.E. HDLTex: Hierarchical Deep Learning for Text Classification. In Proceedings of the 2017 16th IEEE International Conference on Machine Learning and Applications (ICMLA), Cancun, Mexico, 18–21 December 2017; pp. 364–371. [Google Scholar] [CrossRef]
- Mukherjee, A.; Garg, I.; Roy, K. Encoding Hierarchical Information in Neural Networks Helps in Subpopulation Shift. IEEE Trans. Artif. Intell. 2023, 1–2. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, Y. Topic-aware hierarchical multi-attention network for text classification. Int. J. Mach. Learn. Cybern. 2023, 14, 1863–1875. [Google Scholar] [CrossRef]
- Aminizadeh, S.; Heidari, A.; Toumaj, S.; Darbandi, M.; Navimipour, N.J.; Rezaei, M.; Talebi, S.; Azad, P.; Unal, M. The applications of machine learning techniques in medical data processing based on distributed computing and the Internet of Things. Comput. Methods Programs Biomed. 2023, 241, 107745. [Google Scholar] [CrossRef]
- Woźniak, M.; Wieczorek, M.; Siłka, J. BiLSTM deep neural network model for imbalanced medical data of IoT systems. Future Gener. Comput. Syst. 2023, 141, 489–499. [Google Scholar] [CrossRef]
- Joloudari, J.H.; Marefat, A.; Nematollahi, M.A.; Oyelere, S.S.; Hussain, S. Effective Class-Imbalance Learning Based on SMOTE and Convolutional Neural Networks. Appl. Sci. 2023, 13, 4006. [Google Scholar] [CrossRef]
- Xiong, J.; Yu, L.; Niu, X.; Leng, Y. XRR: Extreme multi-label text classification with candidate retrieving and deep ranking. Inf. Sci. 2023, 622, 115–132. [Google Scholar] [CrossRef]
- Ye, X.; Xiao, M.; Ning, Z.; Dai, W.; Cui, W.; Du, Y.; Zhou, Y. NEEDED: Introducing Hierarchical Transformer to Eye Diseases Diagnosis. In Proceedings of the 2023 SIAM International Conference on Data Mining (SDM), Minneapolis, MN, USA, 27–29 April 2023; SIAM: Philadelphia, PA, USA, 2023; pp. 667–675. [Google Scholar] [CrossRef]
- Mikolov, T.; Chen, K.; Corrado, G.; Dean, J. Efficient Estimation of Word Representations in Vector Space. In Proceedings of the 1st International Conference on Learning Representations, ICLR 2013, Scottsdale, AZ, USA, 2–4 May 2013; Workshop Track Proceedings. Bengio, Y., LeCun, Y., Eds.; ICLR: Scottsdale, AZ, USA, 2013. [Google Scholar]
- Silvestri, S.; Gargiulo, F.; Ciampi, M. Improving Biomedical Information Extraction with Word Embeddings Trained on Closed-Domain Corpora. In Proceedings of the 2019 IEEE Symposium on Computers and Communications (ISCC), Barcelona, Spain, 29 June–3 July 2019; pp. 1129–1134. [Google Scholar] [CrossRef]
- Gargiulo, F.; Silvestri, S.; Ciampi, M. A Big Data architecture for knowledge discovery in PubMed articles. In Proceedings of the 2017 IEEE Symposium on Computers and Communications, ISCC 2017, Heraklion, Greece, 3–6 July 2017; IEEE: Heraklion, Greece, 2017; pp. 82–87. [Google Scholar] [CrossRef]
- Řehůřek, R.; Sojka, P. Software Framework for Topic Modelling with Large Corpora. In Proceedings of the LREC 2010 Workshop on New Challenges for NLP Frameworks, La Valleta, Malta, 22 May 2010; ELRA: Luxemburg, 2010; pp. 45–50. [Google Scholar] [CrossRef]
- Tsoumakas, G.; Katakis, I.; Vlahavas, I. Mining multi-label data. In Data Mining and Knowledge Discovery Handbook, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 667–685. [Google Scholar] [CrossRef]
- Manning, C.D.; Raghavan, P.; Schütze, H. Chetion to Information Retrieval; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar] [CrossRef]
- Aho, A.V.; Hopcroft, J.E.; Ullman, J.D. On finding lowest common ancestors in trees. SIAM J. Comput. 1976, 5, 115–132. [Google Scholar] [CrossRef]
- Gargiulo, F.; Silvestri, S.; Ciampi, M. Deep Convolution Neural Network for Extreme Multi-label Text Classification. In Proceedings of the 11th International Joint Conference on Biomedical Engineering Systems and Technologies (BIOSTEC 2018)—Volume 5: HEALTHINF, Funchal, Madeira, Portugal, 19–21 January 2018; SciTePress: Madeira, Portugal, 2018; pp. 641–650. [Google Scholar] [CrossRef][Green Version]
- Zaman, S.; Moon, T.; Benson, T.; Jacobs, S.A.; Chiu, K.; Van Essen, B. Parallelizing Graph Neural Networks via Matrix Compaction for Edge-Conditioned Networks. In Proceedings of the 2022 22nd IEEE International Symposium on Cluster, Cloud and Internet Computing (CCGrid), Taormina, Italy, 16–19 May 2022; pp. 386–395. [Google Scholar] [CrossRef]
- Petit, Q.R.; Li, C.; Emad, N. Distributed and Parallel Sparse Computing for Very Large Graph Neural Networks. In Proceedings of the 2022 IEEE International Conference on Big Data (Big Data), Osaka, Japan, 17–20 December 2022; pp. 6796–6798. [Google Scholar] [CrossRef]
- Fu, Q.; Ji, Y.; Huang, H.H. TLPGNN: A Lightweight Two-Level Parallelism Paradigm for Graph Neural Network Computation on GPU. In Proceedings of the 31st International Symposium on High-Performance Parallel and Distributed Computing, Minneapolis, MN, USA, 27 June–1 July 2022; Association for Computing Machinery: New York, NY, USA, 2022; pp. 122–134. [Google Scholar] [CrossRef]
- Buonaiuto, G.; Gargiulo, F.; De Pietro, G.; Esposito, M.; Pota, M. Best practices for portfolio optimization by quantum computing, experimented on real quantum devices. Sci. Rep. 2023, in press. [CrossRef] [PubMed]
| L | L* | ||
|---|---|---|---|
| 5 | 23,255 | 18,740.05 | |
| All | 57,859 | 19,614.76 |
| Vector Size | Training Algorithm | Window Size | Training Set Size | Vocabulary Size |
|---|---|---|---|---|
| 200 | CBOW | 5 | 10,876,004 | 1,701,632 |
| BERT Model | Training Corpus Word Count | Biomedical Domain Corpus Word Count |
|---|---|---|
| BERT-large-cased | ≈21.3 Billion words | ≈18 Billion words |
| Test | FLAT Measures | ||||||
|---|---|---|---|---|---|---|---|
| Acc | EBP | EBR | EBF | MiP | MiR | MiF | |
| CNN WE | 0.1353 | 0.7528 | 0.1398 | 0.2324 | 0.7522 | 0.1336 | 0.2268 |
| CNN BioBERT | 0.2344 | 0.7828 | 0.1619 | 0.2684 | 0.7810 | 0.1592 | 0.2644 |
| HDNN WE | 0.1719 | 0.7578 | 0.1828 | 0.2894 | 0.7578 | 0.1739 | 0.2829 |
| HDNN BioBERT | 0.3015 | 0.8213 | 0.2187 | 0.3453 | 0.8191 | 0.2041 | 0.3270 |
| Test | Hierarchical Measures | |||||
|---|---|---|---|---|---|---|
| HiP | HiR | HiF | LCa-P | LCa-R | LCa-F | |
| CNN WE | 0.7839 | 0.1635 | 0.2665 | 0.5530 | 0.0738 | 0.1273 |
| CNN BioBERT | 0.8117 | 0.1775 | 0.2899 | 0.5824 | 0.1113 | 0.1819 |
| HDNN WE | 0.7820 | 0.2052 | 0.3194 | 0.6350 | 0.1239 | 0.2020 |
| HDNN BioBERT | 0.8229 | 0.2573 | 0.3941 | 0.7177 | 0.1944 | 0.3005 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silvestri, S.; Gargiulo, F.; Ciampi, M. Integrating PubMed Label Hierarchy Knowledge into a Complex Hierarchical Deep Neural Network. Appl. Sci. 2023, 13, 13117. https://doi.org/10.3390/app132413117
Silvestri S, Gargiulo F, Ciampi M. Integrating PubMed Label Hierarchy Knowledge into a Complex Hierarchical Deep Neural Network. Applied Sciences. 2023; 13(24):13117. https://doi.org/10.3390/app132413117
Chicago/Turabian StyleSilvestri, Stefano, Francesco Gargiulo, and Mario Ciampi. 2023. "Integrating PubMed Label Hierarchy Knowledge into a Complex Hierarchical Deep Neural Network" Applied Sciences 13, no. 24: 13117. https://doi.org/10.3390/app132413117
APA StyleSilvestri, S., Gargiulo, F., & Ciampi, M. (2023). Integrating PubMed Label Hierarchy Knowledge into a Complex Hierarchical Deep Neural Network. Applied Sciences, 13(24), 13117. https://doi.org/10.3390/app132413117








