Featured Application
This paper presents a prediction method for the biochemical recurrence of prostate cancer following radical prostatectomy using partial correlation and a neural network.
Abstract
Biochemical recurrence (BCR) of prostate cancer occurs when the PSA level increases after treatment. BCR prediction is necessary for successful prostate cancer treatment. We propose a model to predict the BCR of prostate cancer using a partial correlation neural network (PCNN). Our study used data from 1021 patients with prostate cancer who underwent radical prostatectomy at a tertiary hospital. There were nine input variables with BCR as the outcome variable. Feature-sensitive and partial correlation analyses were performed to develop the PCNN. The PCNN provides an NN architecture that is optimized for BCR prediction. The proposed PCNN achieved higher performance in BCR prediction than other machine learning methodologies, with accuracy, sensitivity, and specificity values of 87.16%, 90.80%, and 85.62%, respectively. The enhanced performance of the PCNN is owing to the reduction in unnecessary predictive factors through the correlation between the variables that are used. The PCNN can be used in the clinical treatment stage following prostate treatment. It is expected to be used as a clinical decision-making system in clinical follow-ups for prostate cancer.
1. Introduction
Prostate cancer (PCa) is the second most common cancer in men worldwide, and continuous efforts have been made toward its treatment and prevention. Various PCa treatment options exist, including radical prostatectomy (RP), radiation therapy, and androgen-deprivation therapy (ADT) [1]. However, PCa may recur following treatment. Continuous follow-up management is currently used to identify the risk of recurrence. Recurrence is determined by prostate-specific antigen (PSA) values or cancer stages. Biochemical recurrence (BCR) occurs when the PSA level increases following treatment [2,3,4].
BCR prediction is necessary for successful PCa treatment [5]. Many studies have been conducted on the prediction of BCR, and research on prediction methods using machine learning (ML) technology is currently in progress. Prediction using ML has been reported to be excellent for diagnosis [6,7,8,9,10]. At present, the healthcare field is attempting to apply digital twins, and ML can be viewed as a technology for implementing digital twins [11,12,13,14,15]. Among the ML technologies, the most widely used, i.e., neural network (NN), can contribute to BCR prediction. The NN is good at analyzing data without domain knowledge of the BCR. Furthermore, an NN can analyze complex data to discover new patterns and information relating to BCR [16,17,18,19]. However, the “black box” characteristic of NN does not indicate the importance of learning variables [20,21,22]. eXplainable artificial intelligence (XAI) is a method for determining the internal calculation process after learning [23,24]. However, XAI does not know the correlations between variables before learning. Moreover, NN architectures with many connections between unnecessary nodes reduce the learning costs and performance. Therefore, an NN technique that can reduce the costs and improve the accuracy based on a few variables is required. Several optimizations are available, and we use a pruning NN to improve the architecture [25,26].
We propose a partial correlation-based NN for predicting BCR for PCa. The proposed method first analyzes the significance of the variables and outcomes through NN-based sensitivity, learning of the variables that are used. Second, based on the learned sensitivity model, the partial correlation between each variable is analyzed. Finally, we provide a partial correlation neural network (PCNN) architecture that can predict BCR using the analyzed results.
2. Materials and Methods
2.1. Study Design
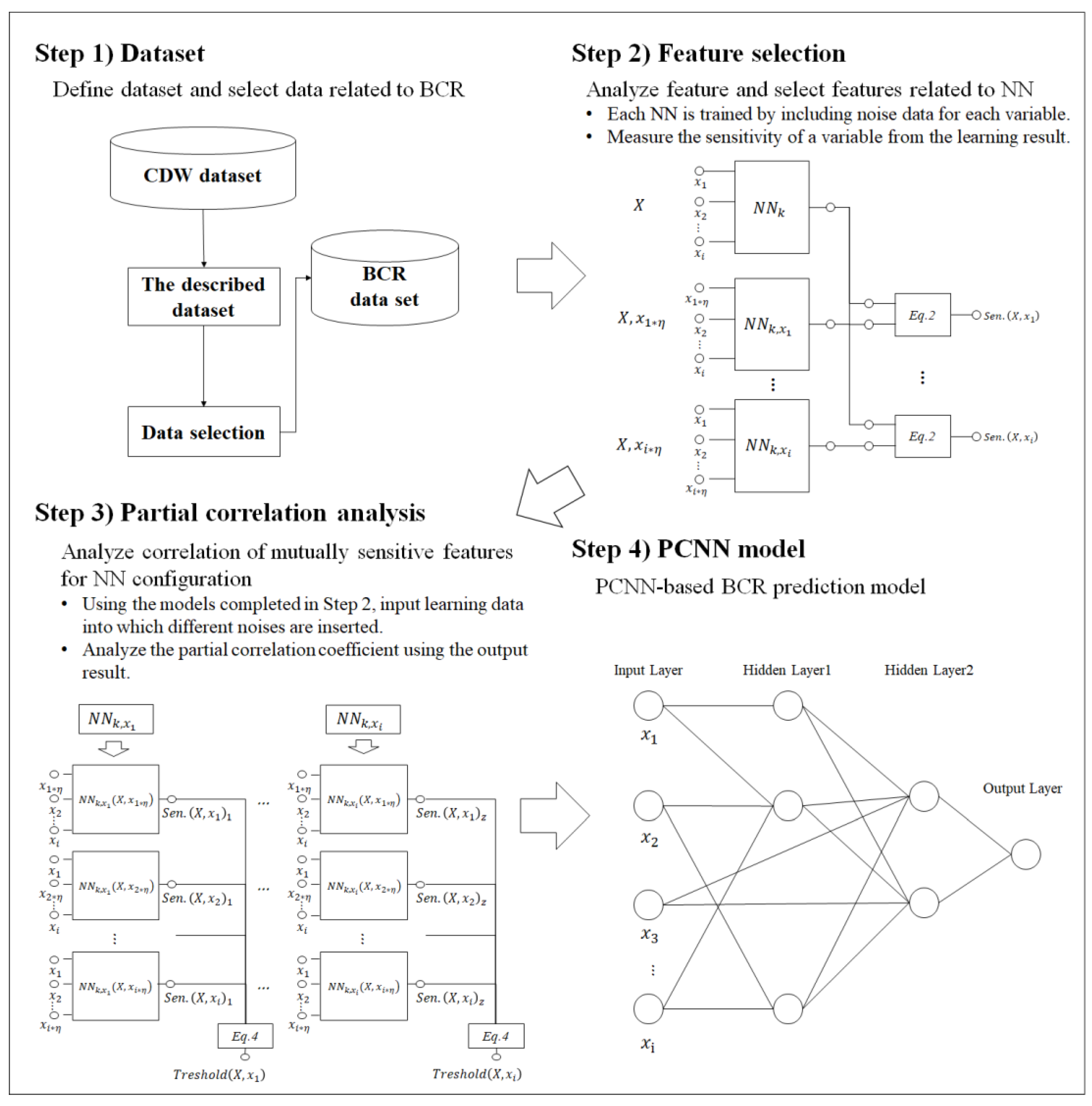
Our study was conducted according to the process shown in Figure 1. First, we describe the dataset that was used for predicting BCR. This is the process of composing the BCR dataset from data that were acquired from the Clinical Data Warehouse (CDW). Second, sensitivity learning was performed using noise values for each variable. Sensitivity training was performed for the BCR prediction. The NN was used to measure the importance of the variables. Third, the partial correlations were analyzed by substituting each variable into the learned sensitivity model. A partial correlation relationship was established by selecting a variable that had a large effect depending on the noise value. Subsequently, the final PCNN model was developed using the analyzed partial correlation results. Finally, we measured the performance of the PCNN model for BCR prediction.
Figure 1.
Study design. CDW, Clinical Data Warehouse; BCR, biochemical recurrence; NN, neural network; PCNN, partial correlation neural network.
2.2. Dataset
We used CDW data from 3046 patients who underwent robot-assisted radical prostatectomy treatment at tertiary care institutions involved in PCa. Data were obtained for the period October 2011 to October 2021. In total, 1021 patient data related to BCR were used in the study. We used follow-up data of more than 60 months from surgery. Therefore, we predicted BCR likelihood at 5 years after surgery. A total of 283 patients with BCR and 738 patients with non-BCR for a follow-up period of more than 60 months were included. The remaining 2025 patients did not have a full follow-up period of at least 60 months and did not have a robot-assisted prostatectomy treatment, and missing critical data were excluded. Our study defines BCR as when the PSA nadir value exceeds 0.2 ng/mL in a PCa patient who had completed RP treatment [27]. We used a total of 10 input variables to predict the BCR: age at diagnosis, BMI, initial PSA value, Gleason score (sum) group, pathology T stage group, extracapsular extension (ECE), seminal vesicle (SV), perineural invasion (PNI), lymph node metastasis (LNM), and surgical margin (SM).
2.3. Feature-Sensitive Analysis
The “black box” problem means that the extent to which the input variable affects the learned NN model is not known. Feature-sensitivity analysis is a method for determining whether the input variables affect the ML classification task [28,29,30]. To measure the sensitivity of the input variable, the NN was trained using “noisy” data for the target variable. This method could calculate the extent to which the target variable affected the BCR prediction. The NN used backpropagation, and the Tangent function was used as the activation function. The feature-sensitivity analysis using noisy data is shown in Equation (1):
Here, NN is a neural network learning algorithm, and X is the original dataset. The NN model that is learned through data X is defined as , where η is the noise, and is the value that is obtained by applying noise to the input variable. We define the data with noise applied to the variable as . For the value of η, a continuous variable used a random value within a specific range [0, 0.01], and for a categorical variable, all data were set to 0. The NN model in which the noise value is substituted is defined as . The learning model was defined as described above, and the sensitivity of each variable was determined. Equation (2) presents the sensitivity calculation for each variable.
Here, and are the results of substituting X for each model. The in this formula is defined as the sensitivity value. A larger sensitivity value indicates a greater effect on the BCR prediction.
2.4. Partial Correlation Analysis
Partial correlation analysis is a method for measuring the intrinsic relationship between only two variables, with cross-correlation among three or more variables [31,32]. In this study, the partial correlation analysis was the process of analyzing the partial correlation of two variables using NN-based reasoning [33,34]. The previously trained model was used to analyze the partial correlation for each variable. When the noise values of the other variables are substituted in , a larger deviation indicates greater correlation. That is, when the data of (X, ) excluding ) are input into a specific model, a larger influence increases the partial correlation. Two procedures were performed for the partial correlation analysis. The first procedure calculated the sensitivity of each variable for the models for a specific variable. The sensitivity calculation is shown in Equation (3):
where z represents the noisy data for each variable. Thus, z Sensitivity(X,) was constructed. Next, to construct a partial correlation with a different variable according to the sensitivity, the threshold value of the corresponding variable was determined. The threshold was the same as that in Equation (4).
The threshold is the average of all sensitivity values that are associated with the variable. When the threshold is lower than the mean, the two variables exhibit a low partial correlation. As indicated in Equation (5), if the sensitive data are higher than the threshold, they become a candidate for partial correlation.
where 0 is not a candidate, and 1 is a candidate. Equation (6) establishes a partial correlation relationship.
Two partial correlation candidates must exist for a specific variable. Thus, when learning is performed in the NN, variables with partial correlations can be defined.
2.5. Partial Correlation Neural Network
The NN architecture was constructed using the partial correlation results. Variables with partial correlations were configured as coupled; otherwise, they were configured as uncoupled. Variables with partial correlations are connected to the same hidden layer, which yields similar results for the partially correlated variables.
The PCNN improves the accuracy because a variable with partial correlation performs learning and inference, which proceed through a second independent variable.
3. Results
3.1. Characteristics
Table 1 presents the characteristics of the patient data (non-BCR: 738; BCR, 283). The Pearson correlation coefficient of each variable was analyzed to determine whether there was a significant correlation with the outcome variable [35,36].

Table 1.
Characteristics of patient data. BMI, body mass index; PSA, prostate-specific antigen; ECE, extracapsular extension; SV, seminal vesicle; PNI, perineural invasion; LVM, lymph node metastasis, SM, surgical margin.
In 1025 patients, the average PSA level was 9.979; in non-BCR, it was 4.765 (standard deviation: SD 7.859), while in BCR, it was 23.576 (SD 109.274). The Gleason score (sum) was 5–7 in 817 patients and 8–9 in 204 individuals. The pathological T stage was T1~T2b in 136 patients and T2c~T3 in 885. ECE and SV were present in 669 and 850 patients, respectively. PNI, LNM, and SM were present in 297, 821, and 659 individuals, respectively. Age and BMI were not suitable for BCR classification because the Pearson correlation coefficient was close to 0.05. The Gleason score, pathological T stage, and SV were >0.3, which was suitable for BCR classification. The Pearson correlation coefficient analysis was performed using the Statistical Package of IBM SPSS Modeler 14.2 software (IBM, Armonk, NY, USA).
3.2. PCNN
The results of the analysis of implementing the PCNN model are also described. The NN was composed of ten input nodes, two hidden nodes, and one output node, and Tangent is used as the activation function. The hidden node bias was 0.1, and the learning rate was 0.3. The learning algorithm used was backpropagation.
The results of the feature-sensitivity analysis are presented in Table 2. The most sensitive variable for the classification of BCR was SV (9.01), followed by SM (4.75), PNI (3.79), and LNM (3.32). Age (0 years) and BMI (0.46) were the lowest. Thus, the sensitivity analysis yielded similar results to those of the Pearson correlation coefficient. In the Pearson correlation, the Gleason score and pathology T stage were effective, but in the feature sensitivity analysis, SV, SM, PNI, and LNM were effective. All variables in the ranking can be considered as useful for predicting the BCR. The parameters of the NN were as follows: learning rate = 0.1, activation function = sigmoid, learning algorithm = backpropagation, batch size = 10, and epochs = 1000.

Table 2.
Results of feature sensitivity analysis.
The results of the partial correlation analysis are presented in Table 3. A total of 10 variables were determined based on their mutual influence. That is, the correlated variables affected the sensitivity change owing to their mutual influence. For example, age (+η) affected the value of age as the candidate groups LNM and SM changed. Thus, LNM and SM were amplified with age. As both LNM and SM had age in the candidate group, they had a partial correlation. There was no partial correlation in the PSA levels because SM was in the candidate group, but other variables were not in the candidate group. Partial correlations were constructed for the remaining features in the same manner.

Table 3.
Results of partial correlation analysis. PSA, prostate-specific antigen; BMI, body mass index; GS, Gleason score (sum); PT, pathology T stage; ECE, extracapsular extension; SV, seminal vesicle; PNI, perineural invasion; LNM, lymph node metastasis; SM, surgical margin.
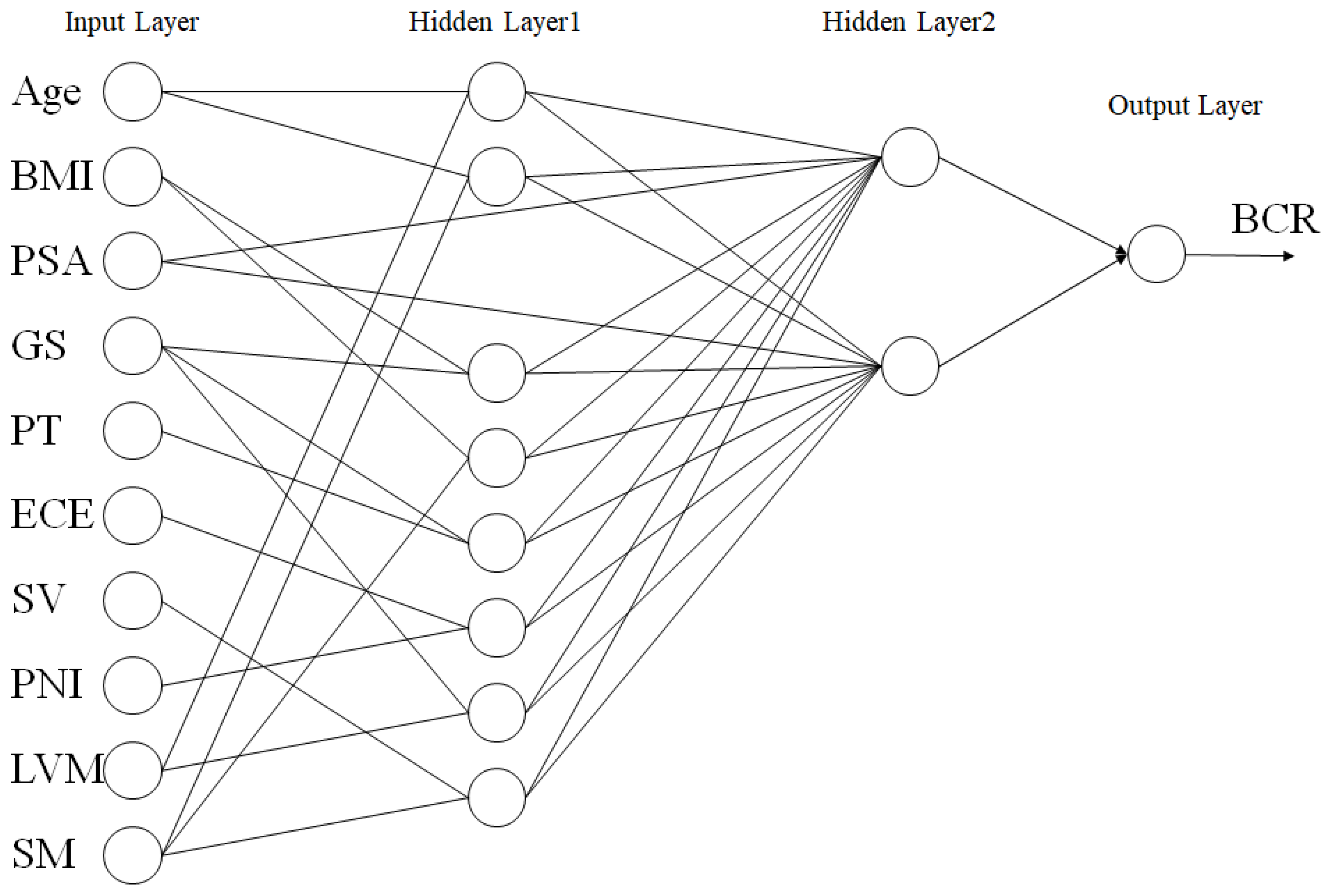
The PCNN, which connects the hidden layers based on the correlation of variables, was modeled as depicted in Figure 2. The PCNN consists of an input layer composed of ten nodes, hidden layer 1 composed of eight nodes, hidden layer 2 composed of two nodes, and an output layer composed of one node. Learning and inference are performed between variables with partial correlations in the eight layers. The PSA level is the most significant variable for PCa and may be effective for inference because it is unaffected by other factors. Hidden layer 2 was designed to solve the “XOR problem” of the perceptron.
Figure 2.
PCNN. PSA, prostate-specific antigen; BMI, body mass index; GS, Gleason score (sum); PT, pathology T stage; ECE, extracapsular extension; SV, seminal vesicle; PNI, perineural invasion; LNM, lymph node metastasis, SM, surgical margin.
3.3. Performance Measures
We measured the accuracy, sensitivity, and specificity using a confusion matrix to evaluate the proposed PCNN. The experimental models for predicting the BCR were as follows: support vector machine (SVM), Bayesian network (BN), random forest (RF), XGBoost (LR), XGBoost (Tree), neural network (NN), recurrent neural network (RNN), long short-term memory (LSTM), and the proposed PCNN. The compared models were implemented using SPSS IBM Modeler 14.2 (IBM, Armonk, New York, USA), and default model parameters were used. The parameters of the PCNN were as follows: learning rate = 0.1, activation function = sigmoid, learning algorithm = backpropagation, batch size = 10, and epochs = 1000.
As the training and testing sets had a low class imbalance and used small amounts of data, we conducted experiments using 66:33 sets (674:347 patients). Table 4 presents the experimental results of the accuracy, sensitivity, and specificity. In the training set, the tree-based algorithms XGBoost (Tree) and RF had the highest accuracy (94.60%) and specificity (100%). The proposed PCNN achieved better performance (93.6%, 97.60%, and 92.28%) than other algorithms such as SVM and NN. The performance of XGBoost (Tree) and RF in the testing set was low (72.14% and 72.05%, respectively). As the boosting technique is used in these methods, they are not suitable for datasets that have not been trained, owing to overfitting problems. The proposed model achieved exceptionally high performance (74.70%, 77.57%, and 72.15%) and was the most effective.

Table 4.
Performance measures (training, testing = 66:33). SVM, support vector machine; BN, Bayesian network; RF, random forest; NN, neural network; RNN, recurrent neural network; LSTM, long short-term memory; PCNN, partial correlation neural network.
Subsequently, the performance was measured using 10-fold cross-validation, which is a suitable method for measuring small datasets. The accuracy, sensitivity, and specificity were measured, and the measurement results are displayed in Table 5. The proposed PCNN model achieved the highest performance (87.16%, 90.80%, and 85.62%). XGBoost (Tree) achieved high performance in the sensitivity (86.60%, 91.43%, and 84.94%).

Table 5.
Performance measures (10-fold cross-validation). SVM, support vector machine; BN, Bayesian network; RF, random forest; NN, neural network; RNN, recurrent neural network; LSTM, long short-term memory; PCNN, partial correlation neural network.
4. Discussion
AI has been used to build models in the field of urology in recent years. Several studies have reported the effectiveness of applying AI to PCa. We have proposed an NN-based BCR prediction model and pruned the model to remove unnecessary node connections in the NN. The PCNN was constructed by performing feature-sensitivity analysis and partial correlation analysis for optimized BCR prediction. This study is the first to optimize an NN model for developing a BCR-prediction model. The PCNN achieved good performance (accuracy: 87.16%) in BCR prediction.
XGBoost had a higher sensitivity than the PCNN but a lower accuracy and specificity. Overall, the PCNN achieved higher performance, and the value of the specificity consisting of a small number of classes for predicting BCR was higher. Moreover, the PCNN is more effective than XGBoost because it consumes a lower cost and time. The tree algorithms (XGBoost (Tree) and RF) are generally suitable for these data because more categorical variables results in better performance. Furthermore, the performance was high when the boosting technique was used. Deep learning models generally achieve high performance when they have numerous data and features. Therefore, the performance of RNN and LSTM was low in the data used in this study. The PCNN analyzes the partial correlation of input variables and configures the most suitable NN architecture for BCR prediction; thus, the experimental performance was high. The proposed PCNN improved the performance by approximately 4% compared to other algorithms and is suitable for BCR prediction. The performance is expected to be higher for large amounts of data.
Ekşi [9] predicted the accuracy of BCR using Cox regression, RF, KNN, and LR after robot-assisted prostatectomy. When using the same control RF, the same accuracy was achieved at 86.6%. Thus, the data used in this study can be considered as significant for predicting BCR. Momenzadeh [37] predicted the accuracy using LR, SVM, and XGBoost to predict whether death due to PCa occurred. When the same XGBoost was used in the study of Momenzadeh [37] and our study, the performance was 94.68% and 86.16%, respectively, but the results may differ depending on the number of samples in the dataset used. The data used in this study consisted of a small dataset, and the performance may be low in such cases. As the proposed PCNN achieved higher performance than XGBoost, the algorithm of this study is superior. Most studies to date have used the vanilla model as a predictive model or a combination of other models. This study optimized NN differently, and the proposed PCNN model outperformed the models of previous studies. Therefore, the proposed PCNN can provide various treatment options for clinical follow-up. Given the excellent performance of BCR prediction following surgery, it can be used for clinically relevant purposes. The limitations of this study are as follows: First, there is a limitation in building a model that uses only a small amount of data. The use of data from multiple centers can provide a more accurate model. Second, because the criterion for BCR can be set to 0.2 or 0.4, it is different for each researcher. Methods based on different definitions are required in future research.
5. Conclusions
We developed a model to predict BCR in patients with PCa. A partial correlation analysis of variables relating to BCR was performed, and the results were used to construct an NN architecture. Our method is novel in that we constructed the NN architecture by considering the correlation of BCR variables rather than a simple ML model. Therefore, the performance is good because it overcomes the design disadvantages of the NN. Utilizing our research, medical engineering can raise the level of AI relating to urology, reduce medical costs and overtreatment, and contribute to improving quality of life. The proposed PCNN achieves higher performance than other prediction models and is suitable for predicting BCR. The PCNN is expected to be used as part of a digital twin study for predicting the prognosis of virtual PCa and can be applied to clinical decision-support systems in the future.
Author Contributions
Conceptualization, J.-K.K. and I.-Y.C.; methodology, J.-K.K.; software, J.-K.K.; validation, J.-K.K.; formal analysis, J.-K.K.; investigation, J.-K.K., S.-H.H., and I.-Y.C.; resources, I.-Y.C.; data curation, S.-H.H.; writing—original draft preparation, J.-K.K.; writing—review and editing, J.-K.K.; visualization, J.-K.K.; supervision, I.-Y.C.; project administration, I.-Y.C.; funding acquisition, I.-Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Research Foundation of Korea (NRF-2020R1A2C2012284). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Institutional Review Board Statement
Our study protocol was approved by the Institutional Review Board of the Catholic University of Korea (IRB No. KC21WNSE0887).
Informed Consent Statement
This study protocol was approved by the institutional review board of the Catholic Medical Centre. This research was a retrospective study of CDW data, and all data were de-identified and involved no more than minimal risk to subjects. The requirement for written informed consent was waived by the Research Ethics Committee of the Catholic Medical Centre. All methods were performed in accordance with the relevant guidelines and regulations.
Data Availability Statement
The datasets of the current study are not publicly available. Due to Catholic Medical Centre policies and reasonable privacy and security concerns, the underlying CDW data are not easily redistributable to researchers from other centers but are available upon reasonable request from the corresponding authors and with permission of the Catholic Medical Centre.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ko, Y.H.; Park, S.W.; Ha, U.S.; Joung, J.Y.; Jeong, S.H.; Byun, S.S.; Jeon, S.S.; Kwak, C. A comparison of the survival outcomes of robotic-assisted radical prostatectomy and radiation therapy in patients over 75 years old with non-metastatic prostate cancer: A Korean multicenter study. Investig. Clin. Urol. 2021, 62, 535–544. [Google Scholar] [CrossRef] [PubMed]
- Briganti, A.; Karnes, R.J.; Joniau, S.; Boorjian, S.A.; Cozzarini, C.; Gandaglia, G.; Hinkelbein, W.; Haustermans, K.; Tombal, B.; Shariat, S.; et al. Prediction of outcome following early salvage radiotherapy among patients with biochemical recurrence after radical prostatectomy. Eur. Urol. 2014, 66, 479–486. [Google Scholar] [CrossRef] [PubMed]
- Van den Broeck, T.; van den Bergh, R.C.N.; Arfi, N.; Gross, T.; Moris, L.; Briers, E.; Cumberbatch, M.; De Santis, M.; Tilki, D.; Fanti, S.; et al. Prognostic value of biochemical recurrence following treatment with curative intent for prostate cancer: A systematic review. Eur. Urol. 2019, 75, 967–987. [Google Scholar] [CrossRef] [PubMed]
- Punnen, S.; Cooperberg, M.R.; D’Amico, A.V.; Karakiewicz, P.I.; Moul, J.W.; Scher, H.I.; Schlomm, T.; Freedland, S.J. Management of biochemical recurrence after primary treatment of prostate cancer: A systematic review of the literature. Eur. Urol. 2013, 64, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Remmers, S.; Verbeek, J.F.M.; Nieboer, D.; van der Kwast, T.; Roobol, M.J. Predicting biochemical recurrence and prostate cancer-specific mortality after radical prostatectomy: Comparison of six prediction models in a cohort of patients with screening- and clinically detected prostate cancer. BJU Int. 2019, 124, 635–642. [Google Scholar] [CrossRef]
- Qiao, P.; Zhang, D.; Zeng, S.; Wang, Y.; Wang, B.; Hu, X. Using machine learning method to identify MYLK as a novel marker to predict biochemical recurrence in prostate cancer. Biomark Med. 2021, 15, 29–41. [Google Scholar] [CrossRef]
- Vittrant, B.; Leclercq, M.; Martin-Magniette, M.L.; Collins, C.; Bergeron, A.; Fradet, Y.; Droit, A. Identification of a transcriptomic prognostic signature by machine learning using a combination of small cohorts of prostate cancer. Front Genet. 2020, 11, 550894. [Google Scholar] [CrossRef]
- Lee, S.J.; Yu, S.H.; Kim, Y.; Kim, J.K.; Hong, J.H.; Kim, C.; Seo, S.I.; Byun, S.S.; Jeong, C.W.; Lee, J.Y.; et al. Prediction system for prostate cancer recurrence using machine learning. Appl. Sci. 2020, 10, 1333. [Google Scholar] [CrossRef]
- Ekşi, M.; Evren, İ.; Akkaş, F.; Arıkan, Y.; Özdemir, O.; Özlü, D.N.; Ayten, A.; Sahin, S.; Tuğcu, V.; Taşçı, A.İ. Machine learning algorithms can more efficiently predict biochemical recurrence after robot-assisted radical prostatectomy. Prostate 2021, 81, 913–920. [Google Scholar] [CrossRef]
- Tan, Y.G.; Fang, A.H.S.; Lim, J.K.S.; Khalid, F.; Chen, K.; Ho, H.S.S.; Yuen, J.S.; Huang, H.H.; Tay, K.J. Incorporating artificial intelligence in urology: Supervised machine learning algorithms demonstrate comparative advantage over nomograms in predicting biochemical recurrence after prostatectomy. Prostate 2022, 82, 298–305. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, L.; Yang, Y.; Zhou, L.; Ren, L.; Wang, F.; Liu, R.; Pang, Z.; Deen, M.J. A novel cloud-based framework for the elderly healthcare services using digital twin. IEEE Access. 2019, 7, 49088–49101. [Google Scholar] [CrossRef]
- Elayan, H.; Aloqaily, M.; Guizani, M. Digital twin for intelligent context-aware IoT healthcare systems. IEEE Internet Things J. 2021, 8, 16749–16757. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, X.; Zhao, J. Create the individualized digital twin for noninvasive precise pulmonary healthcare. Significances Bioeng Biosci. 2018, 1, 2. [Google Scholar] [CrossRef]
- Zhang, J.; Li, L.; Lin, G.; Fang, D.; Tai, Y.; Huang, J. Cyber resilience in healthcare digital twin on lung cancer. IEEE Access. 2020, 8, 201900–201913. [Google Scholar] [CrossRef]
- Patrone, C.; Galli, G.; Revetria, R. A state of the art of digital twin and simulation supported by data mining in the healthcare sector. Advancing Technology Industrialization Through Intelligent Software Methodologies, Tools and Techniques. IOS Press 2019, 318, 605–615. [Google Scholar]
- Sargos, P.; Leduc, N.; Giraud, N.; Gandaglia, G.; Roumiguié, M.; Ploussard, G.; Rozet, F.; Soulié, M.; Mathieu, R.; Artus, P.M.; et al. Deep neural networks outperform the CAPRA score in predicting biochemical recurrence after prostatectomy. Front Oncol. 2021, 10, 3237. [Google Scholar] [CrossRef]
- Hu, X.H.; Cammann, H.; Meyer, H.A.; Jung, K.; Lu, H.B.; Leva, N.; Magheli, A.; Stephan, C.; Busch, J. Risk prediction models for biochemical recurrence after radical prostatectomy using prostate-specific antigen and Gleason score. Asian J. Androl. 2014, 16, 897–901. [Google Scholar]
- Peterson, L.E.; Ozen, M.; Erdem, H.; Amini, A.; Gomez, L.; Nelson, C.C.; Ittmann, M. Artificial neural network analysis of DNA microarray-based prostate cancer recurrence. In Proceedings of the 2005 IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology, San Diego, CA, USA, 14–15 November 2005. [Google Scholar]
- Yan, Y.; Shao, L.; Liu, Z.; He, W.; Yang, G.; Liu, J.; Xia, H.; Zhang, Y.; Chen, H.; Liu, C.; et al. Deep learning with quantitative features of magnetic resonance images to predict biochemical recurrence of radical prostatectomy: A multi-center study. Cancers 2021, 13, 3098. [Google Scholar] [CrossRef]
- Olden, J.D.; Jackson, D.A. Illuminating the “black box”: A randomization approach for understanding variable contributions in artificial neural networks. Ecol Modell. 2002, 154, 135–150. [Google Scholar] [CrossRef]
- Samek, W.; Wiegand, T.; Müller, K.-R. Explainable artificial intelligence: Understanding, visualizing and interpreting deep learning models. arXiv 2017, arXiv:1708.08296. [Google Scholar]
- Dayhoff, J.E.; DeLeo, J.M. Artificial neural networks: Opening the black box. Cancer 2001, 91 (Supp. Sl), 1615–1635. [Google Scholar] [CrossRef] [PubMed]
- Zednik, C. Solving the black box problem: A normative framework for explainable artificial intelligence. Philos Technol. 2021, 34, 265–288. [Google Scholar] [CrossRef]
- Buhrmester, V.; Münch, D.; Arens, M. Analysis of explainers of black box deep neural networks for computer vision: A survey. Mach Learn Knowl Extr. 2021, 3, 966–989. [Google Scholar] [CrossRef]
- Yeom, S.K.; Seegerer, P.; Lapuschkin, S.; Binder, A.; Wiedemann, S.; Müller, K.R.; Samek, W. Pruning by explaining: A novel criterion for deep neural network pruning. Pattern Recognit. 2021, 115, 107899. [Google Scholar] [CrossRef]
- Liang, T.; Glossner, J.; Wang, L.; Shi, S.; Zhang, X. Pruning and quantization for deep neural network acceleration: A survey. Neurocomputing 2021, 461, 370–403. [Google Scholar] [CrossRef]
- Freedland, S.J.; Sutter, M.E.; Dorey, F.; Aronson, W.J. Defining the ideal cutpoint for determining PSA recurrence after radical prostatectomy. Prostate-specific antigen. Urology 2003, 61, 365–369. [Google Scholar] [CrossRef]
- Naik, D.L. A novel sensitivity-based method for feature selection. J. Big Data 2021, 8, 1. [Google Scholar] [CrossRef]
- Laroza Silva, D.; Marcelo De Jesus, K.L. Backpropagation Neural Network with Feature Sensitivity Analysis: Pothole Prediction Model for Flexible Pavements using Traffic and Climate Associated Factors. In Proceedings of the 2020 the 3rd International Conference on Computing and Big Data, Taichung Taiwan, 5–7 August 2020. [Google Scholar]
- Abaker, A.A.; Saeed, F.A. Towards transparent machine learning models using feature sensitivity algorithm. J. Inform. 2020, 14, 15–22. [Google Scholar] [CrossRef]
- Yang, J.; Li, L.; Wang, A. A partial correlation-based Bayesian network structure learning algorithm under linear SEM. Knowl Based Syst. 2011, 24, 963–976. [Google Scholar] [CrossRef]
- Agastinose Ronicko, J.F.A.; Thomas, J.; Thangavel, P.; Koneru, V.; Langs, G.; Dauwels, J. Diagnostic classification of autism using resting-state fMRI data improves with full correlation functional brain connectivity compared to partial correlation. J. Neurosci Methods 2020, 345, 108884. [Google Scholar] [CrossRef]
- Epskamp, S.; Fried, E.I. A tutorial on regularized partial correlation networks. Psychol. Methods 2018, 23, 617–634. [Google Scholar] [CrossRef]
- Kim, J.K.; Kang, S. Neural network-based coronary heart disease risk prediction using feature correlation analysis. J. Healthc. Eng. 2017, 2017, 2780501. [Google Scholar] [CrossRef]
- Sajjad, U.; Hussain, I.; Imran, M.; Sultan, M.; Wang, C.C.; Alsubaie, A.S.; Mahmoud, K.H. Boiling heat transfer evaluation in nanoporous surface coatings. Nanomaterials 2021, 11, 3383. [Google Scholar] [CrossRef]
- Sajjad, U.; Hussain, I.; Raza, W.; Sultan, M.; Alarifi, I.M.; Wang, C.C. On the critical heat flux assessment of micro- and nanoscale roughened surfaces. Nanomaterials 2022, 12, 3256. [Google Scholar] [CrossRef]
- Momenzadeh, N.; Hafezalseheh, H.; Nayebpour, M.R.; Fathian, M.; Noorossana, R. A hybrid machine learning approach for predicting survival of patients with prostate cancer: A SEER-based population study. Inform. Med. Unlocked 2021, 27, 100763. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).