Deep Transformers for Computing and Predicting ALCOA+Data Integrity Compliance in the Pharmaceutical Industry
Abstract
1. Introduction
- 1.
- Attributable: This means that all collected data should include information about the individuals who collected the data, the individuals who took action, and the time when the action was carried out.
- 2.
- Legible: This refers to the requirement for data to be accurate and comprehensible. However, legibility encompasses more than just reading the written content, it also involves understanding the surrounding context of the information.
- 3.
- Contemporaneous: This addresses the issue of recording data promptly and accurately for both individuals and systems. When dealing with electronic data, it is customary to timestamp the activities or actions to ensure a chronological order.
- 4.
- Original: This refers to keeping records in their original form rather than using duplicates or transcriptions, especially when it comes to manual record-keeping. The initial recording of the data, whether it is on paper or in a digital system, should serve as the primary record. In the case of digitally recorded data, it is crucial to have technical and procedural measures set to prevent alterations to the original recording.
- 5.
- Accurate: This ensures that all records accurately depict the actual events without any mistakes. Moreover, it is crucial to refrain from altering the original information in a manner that causes its loss (e.g., compression, rounding of numerical values, acronyms, etc.).
- 6.
- Complete: It is necessary for all captured information to possess a comprehensive audit trail to demonstrate the absence of deletions or losses. This requirement encompasses not only the primary data recording but also includes metadata, retest data, analysis data, and other related elements. Additionally, audit trails must be in place to track any modifications made to the data.
- 7.
- Consistent: This refers to the need for data timestamping, that must be sequential in an ascending order. Timestamps should also be added to indicate modifications made to the original data recording.
- 8.
- Enduring: This refers to the long-term storage of data that must be able to be read and understood for many years after their creation.
- 9.
- Available: This refers to the ability of data to be accessible to every interested party and at any time during its life cycle.
- it describes an approach to assess compliance with ALCOA+ principles;
- it describes a model to predict compliance with ALOCA+ principles;
- it evaluates the AI models on a real dataset consisting of data stemming from two different pharmaceuticals production lines.
2. Related Work
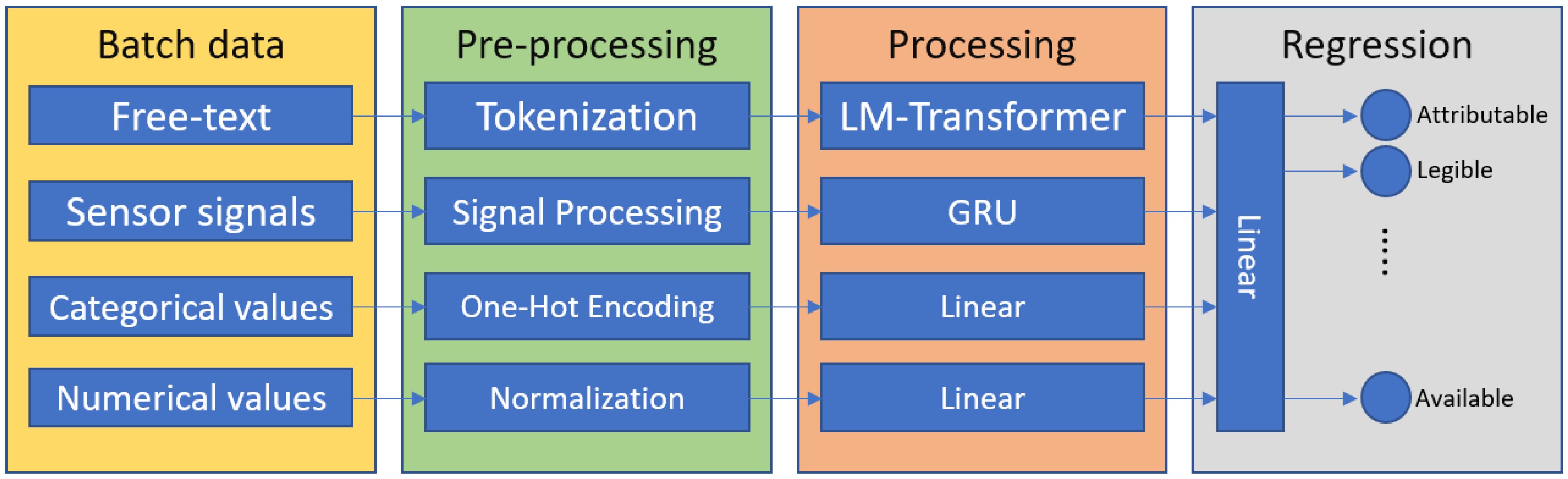
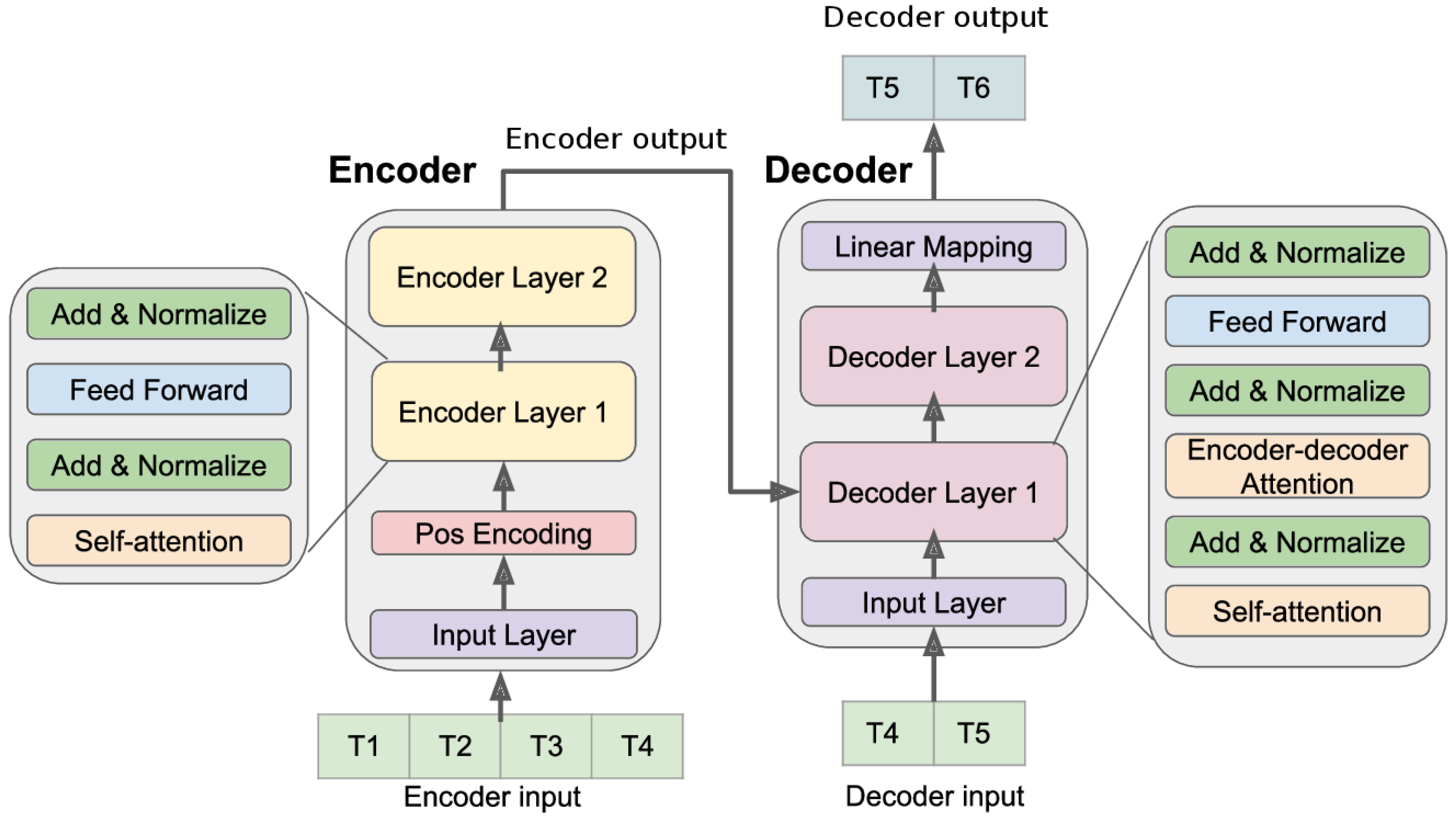
3. Description of the ALCOAi Model
- An ALCOA+ regression model
- An ALCOA+ forecasting model
4. Performance Evaluation
4.1. Dataset Description and Hyperparameters
- 1.
- Gather each input data sample and associate a set of the corresponding ALCOA+ scores calculated as in [5].
- 2.
- Normalize and encode each input sample as described in the previous section.
- 3.
- Feed each data sample to the network and gather its output.
- 4.
- Compare the output of the network to the corresponding effective ALCOA+ values and calculate the mean squared error loss (Formula (1)).
- 5.
- Back-propagate the error.
4.2. Network Configuration Selection
4.3. Results and Discussion
4.3.1. ALCOA+ Value Regression
4.3.2. ALCOA+ Value Prediction
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hole, G.; Hole, A.S.; McFalone-Shaw, I. Digitalization in pharmaceutical industry: What to focus on under the digital implementation process? Int. J. Pharm. X 2021, 3, 100095. [Google Scholar] [PubMed]
- Alosert, H.; Savery, J.; Rheaume, J.; Cheeks, M.; Turner, R.; Spencer, C.; Farid, S.S.; Goldrick, S. Data integrity within the biopharmaceutical sector in the era of Industry 4.0. Biotechnol. J. 2022, 17, 2100609. [Google Scholar] [CrossRef]
- Fisher, A.C.; Liu, W.; Schick, A.; Ramanadham, M.; Chatterjee, S.; Brykman, R.; Lee, S.L.; Kozlowski, S.; Boam, A.B.; Tsinontides, S.C.; et al. An audit of pharmaceutical continuous manufacturing regulatory submissions and outcomes in the US. Int. J. Pharm. 2022, 622, 121778. [Google Scholar]
- McDermott, O.; Antony, J.; Sony, M.; Daly, S. Barriers and enablers for continuous improvement methodologies within the Irish pharmaceutical industry. Processes 2022, 10, 73. [Google Scholar] [CrossRef]
- Durá, M.; Sánchez-García, Á.; Sáez, C.; Leal, F.; Chis, A.E.; González-Vélez, H.; García-Gómez, J.M. Towards a computational approach for the assessment of compliance of ALCOA+ Principles in pharma industry. Stud. Health Technol. Inform. 2022, 294, 755–759. [Google Scholar]
- Vignesh, M.; Ganesh, G. Current status, challenges and preventive strategies to overcome data integrity issues in the pharmaceutical industry. Int. J. Appl. Pharm. 2020, 12, 19–23. [Google Scholar]
- Rattan, A.K. Data integrity: History, issues, and remediation of issues. PDA J. Pharm. Sci. Technol. 2018, 72, 105–116. [Google Scholar] [CrossRef]
- Barenji, R.V.; Akdag, Y.; Yet, B.; Oner, L. Cyber-physical-based PAT (CPbPAT) framework for Pharma 4.0. Int. J. Pharm. 2019, 567, 118445. [Google Scholar]
- Wolf, T.; Debut, L.; Sanh, V.; Chaumond, J.; Delangue, C.; Moi, A.; Cistac, P.; Rault, T.; Louf, R.; Funtowicz, M.; et al. Transformers: State-of-the-art natural language processing. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing: System Demonstrations, Online, 16–20 November 2020; pp. 38–45. [Google Scholar]
- Zhou, H.; Zhang, S.; Peng, J.; Zhang, S.; Li, J.; Xiong, H.; Zhang, W. Informer: Beyond efficient transformer for long sequence time-series forecasting. In Proceedings of the AAAI Conference on Artificial Intelligence, Virtually, 2–9 February 2021; Volume 35, pp. 11106–11115. [Google Scholar]
- Meng, T.; Jing, X.; Yan, Z.; Pedrycz, W. A survey on machine learning for data fusion. Inf. Fusion 2020, 57, 115–129. [Google Scholar]
- Qiu, J.; Wu, Q.; Ding, G.; Xu, Y.; Feng, S. A survey of machine learning for big data processing. EURASIP J. Adv. Signal Process. 2016, 2016, 1–16. [Google Scholar]
- Zhang, Q.; Yang, L.T.; Chen, Z.; Li, P. A survey on deep learning for big data. Inf. Fusion 2018, 42, 146–157. [Google Scholar]
- Pires, I.M.; Garcia, N.M.; Pombo, N.; Flórez-Revuelta, F. From data acquisition to data fusion: A comprehensive review and a roadmap for the identification of activities of daily living using mobile devices. Sensors 2016, 16, 184. [Google Scholar] [PubMed]
- Ding, W.; Jing, X.; Yan, Z.; Yang, L.T. A survey on data fusion in internet of things: Towards secure and privacy-preserving fusion. Inf. Fusion 2019, 51, 129–144. [Google Scholar]
- Alam, F.; Mehmood, R.; Katib, I.; Albogami, N.N.; Albeshri, A. Data fusion and IoT for smart ubiquitous environments: A survey. IEEE Access 2017, 5, 9533–9554. [Google Scholar]
- Salkuti, S.R. A survey of big data and machine learning. Int. J. Electr. Comput. Eng. (2088–8708) 2020, 10, 575–580. [Google Scholar] [CrossRef]
- Jauro, F.; Chiroma, H.; Gital, A.Y.; Almutairi, M.; Shafi’i, M.A.; Abawajy, J.H. Deep learning architectures in emerging cloud computing architectures: Recent development, challenges and next research trend. Appl. Soft Comput. 2020, 96, 106582. [Google Scholar]
- Sengupta, S.; Basak, S.; Saikia, P.; Paul, S.; Tsalavoutis, V.; Atiah, F.; Ravi, V.; Peters, A. A review of deep learning with special emphasis on architectures, applications and recent trends. Knowl.-Based Syst. 2020, 194, 105596. [Google Scholar]
- Elbadawi, M.; Gaisford, S.; Basit, A.W. Advanced machine-learning techniques in drug discovery. Drug Discov. Today 2021, 26, 769–777. [Google Scholar]
- Patel, L.; Shukla, T.; Huang, X.; Ussery, D.W.; Wang, S. Machine learning methods in drug discovery. Molecules 2020, 25, 5277. [Google Scholar]
- Lavecchia, A. Machine-learning approaches in drug discovery: Methods and applications. Drug Discov. Today 2015, 20, 318–331. [Google Scholar]
- Dara, S.; Dhamercherla, S.; Jadav, S.S.; Babu, C.M.; Ahsan, M.J. Machine learning in drug discovery: A review. Artif. Intell. Rev. 2022, 55, 1947–1999. [Google Scholar] [PubMed]
- Réda, C.; Kaufmann, E.; Delahaye-Duriez, A. Machine learning applications in drug development. Comput. Struct. Biotechnol. J. 2020, 18, 241–252. [Google Scholar] [PubMed]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Dey, R.; Salem, F.M. Gate-variants of gated recurrent unit (GRU) neural networks. In Proceedings of the 2017 IEEE 60th international midwest symposium on circuits and systems (MWSCAS), Boston, MA, USA, 6–9 August 2017; pp. 1597–1600. [Google Scholar]
- Li, S.; Jin, X.; Xuan, Y.; Zhou, X.; Chen, W.; Wang, Y.X.; Yan, X. Enhancing the locality and breaking the memory bottleneck of transformer on time series forecasting. Adv. Neural Inf. Process. Syst. 2019, 32. [Google Scholar]
- Wu, N.; Green, B.; Ben, X.; O’Banion, S. Deep transformer models for time series forecasting: The influenza prevalence case. arXiv 2020, arXiv:2001.08317. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Cho, K.; Van Merriënboer, B.; Gulcehre, C.; Bahdanau, D.; Bougares, F.; Schwenk, H.; Bengio, Y. Learning phrase representations using RNN encoder-decoder for statistical machine translation. arXiv 2014, arXiv:1406.1078. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Rojat, T.; Puget, R.; Filliat, D.; Del Ser, J.; Gelin, R.; Díaz-Rodríguez, N. Explainable artificial intelligence (xai) on timeseries data: A survey. arXiv 2021, arXiv:2104.00950. [Google Scholar]


| Hyperparameter Name | Value |
|---|---|
| Learning rate | 0.0001 |
| Batch size | 32 |
| Optimizer | Adam [29] |
| 0.9 | |
| 2 | 0.999 |
| Max. epochs | 200 |
| LSTM | GRU | ALCOAi | |
|---|---|---|---|
| Number of layers | 2 | 2 | - |
| Number of hidden states | 256 | 256 | - |
| Number of encoder layers | - | - | 4 |
| Number of decoder layers | - | - | 4 |
| Number of attention heads | - | - | 16 |
| Dropout | 0.3 | 0.3 | 0.5 |
| ALCOA+ Principle | I600 | I1000 | ||||||
|---|---|---|---|---|---|---|---|---|
| Min | Max | Mean | Std | Min | Max | Mean | Std | |
| Attributable | 25 | 50 | 48.17 | 2.13 | 0 | 50 | 47.8 | 3.34 |
| Legible | 75 | 94 | 83.1 | 3.34 | 79 | 91 | 84.66 | 1.71 |
| Contemporaneous | 62 | 95 | 87.65 | 6.31 | 50 | 94 | 87.45 | 4.62 |
| Original * | 100 | 100 | 100 | 0 | 100 | 100 | 100 | 0 |
| Accurate | 0 | 50 | 37.22 | 6.43 | 0 | 100 | 35.41 | 5.74 |
| Complete | 84 | 98 | 96.76 | 2.85 | 83 | 100 | 97.2 | 1.82 |
| Consistent * | 100 | 100 | 100 | 0 | 100 | 100 | 100 | 0 |
| Available * | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Enduring * | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ALCOA+ Principle | I600 | I1000 | ||||
|---|---|---|---|---|---|---|
| ALCOAi | GRU | LSTM | ALCOAi | GRU | LSTM | |
| Attributable | 1.24 | 1.88 | 1.96 | 1.44 | 2.05 | 1.68 |
| Legible | 2.37 | 2.66 | 2.64 | 4.61 | 4.73 | 5.30 |
| Contemporaneous | 2.38 | 3.79 | 4.00 | 3.46 | 4.88 | 5.73 |
| Accurate | 2.83 | 3.07 | 3.07 | 3.04 | 3.69 | 3.61 |
| Complete | 1.99 | 2.22 | 2.12 | 2.40 | 2.48 | 2.54 |
| Average | 2.16 | 2.72 | 2.76 | 2.99 | 3.57 | 3.77 |
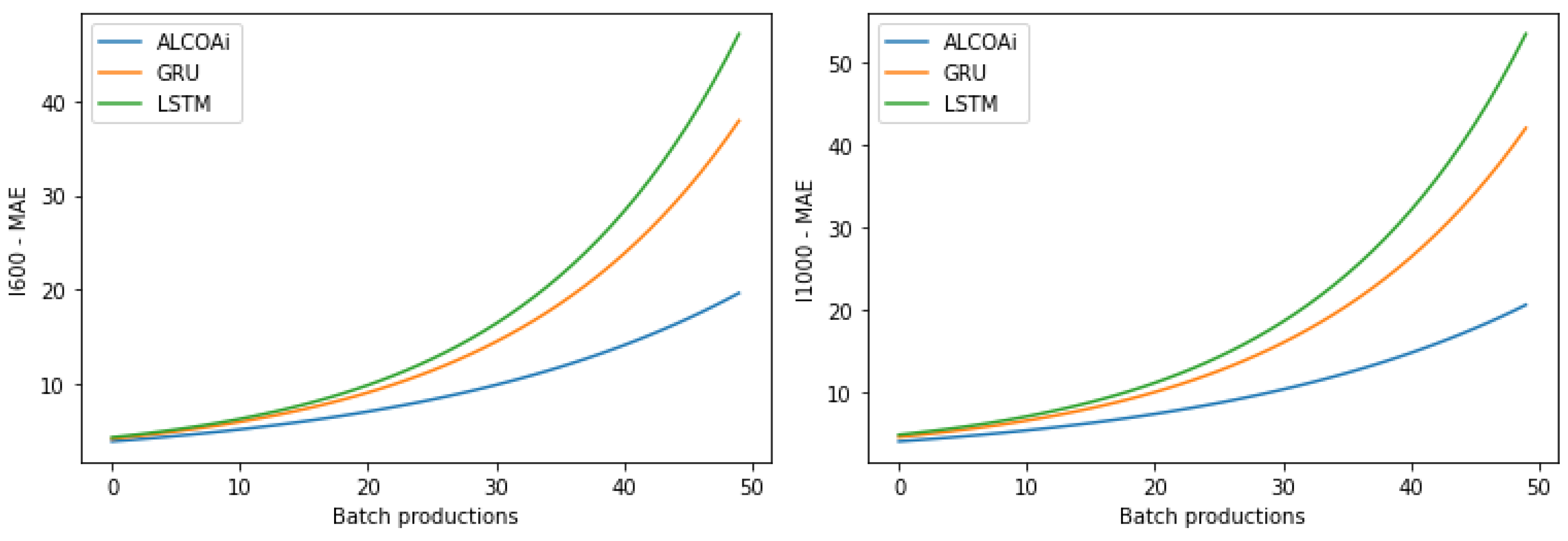
| Model | I600 Dataset | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Batch | 5 | 10 | 15 | 20 | 25 | 30 | 35 | 40 | 45 | 50 |
| ALCOAi | 2.34 | 3.87 | 6.16 | 7.99 | 9.24 | 10.05 | 10.84 | 13.37 | 14.74 | 20.41 |
| GRU | 2.81 | 3.99 | 6.45 | 9.11 | 11.36 | 15.19 | 17.74 | 23.58 | 31.39 | 38.27 |
| Difference | 20.1 | 3.1 | 4.76 | 14.02 | 22.94 | 51.14 | 63.65 | 76.37 | 112.96 | 87.51 |
| LSTM | 2.82 | 4.21 | 7.00 | 9.88 | 13.25 | 17.09 | 22.33 | 30.01 | 39.43 | 49.99 |
| Difference | 20.51 | 8.79 | 13.64 | 23.65 | 43.40 | 70.05 | 106 | 124.46 | 167.5 | 144.93 |
| Model | I1000 Dataset | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Batch | 5 | 10 | 15 | 20 | 25 | 30 | 35 | 40 | 45 | 50 |
| ALCOAi | 3.15 | 4.83 | 6.31 | 8.22 | 9.78 | 10.44 | 13.65 | 15.72 | 18.51 | 22.38 |
| GRU | 4.01 | 6.91 | 7.81 | 9.63 | 11.12 | 15.79 | 19.98 | 24.75 | 32.34 | 43.10 |
| Difference | 27.3 | 43.06 | 23.77 | 17.15 | 13.7 | 51.24 | 46.37 | 57.44 | 74.72 | 92.58 |
| LSTM | 4.49 | 7.88 | 8.42 | 10.71 | 13.97 | 17.82 | 22.94 | 29.99 | 43.65 | 59.22 |
| Difference | 42.54 | 63.15 | 33.44 | 30.29 | 42.84 | 70.69 | 68.06 | 90.78 | 135.82 | 164.61 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kavasidis, I.; Lallas, E.; Leligkou, H.C.; Oikonomidis, G.; Karydas, D.; Gerogiannis, V.C.; Karageorgos, A. Deep Transformers for Computing and Predicting ALCOA+Data Integrity Compliance in the Pharmaceutical Industry. Appl. Sci. 2023, 13, 7616. https://doi.org/10.3390/app13137616
Kavasidis I, Lallas E, Leligkou HC, Oikonomidis G, Karydas D, Gerogiannis VC, Karageorgos A. Deep Transformers for Computing and Predicting ALCOA+Data Integrity Compliance in the Pharmaceutical Industry. Applied Sciences. 2023; 13(13):7616. https://doi.org/10.3390/app13137616
Chicago/Turabian StyleKavasidis, Isaak, Efthimios Lallas, Helen C. Leligkou, Georgios Oikonomidis, Dimitrios Karydas, Vassilis C. Gerogiannis, and Anthony Karageorgos. 2023. "Deep Transformers for Computing and Predicting ALCOA+Data Integrity Compliance in the Pharmaceutical Industry" Applied Sciences 13, no. 13: 7616. https://doi.org/10.3390/app13137616
APA StyleKavasidis, I., Lallas, E., Leligkou, H. C., Oikonomidis, G., Karydas, D., Gerogiannis, V. C., & Karageorgos, A. (2023). Deep Transformers for Computing and Predicting ALCOA+Data Integrity Compliance in the Pharmaceutical Industry. Applied Sciences, 13(13), 7616. https://doi.org/10.3390/app13137616







