Multi-Class Classifier in Parkinson’s Disease Using an Evolutionary Multi-Objective Optimization Algorithm
Abstract
:1. Introduction
2. State-of-the-Art
2.1. Diagnosis Tools
2.2. Computer Studies and Algorithms in PD Classification
3. Materials and Methods
3.1. Subjects Cohort
3.2. Pre-processing: Normalization and Segmentation
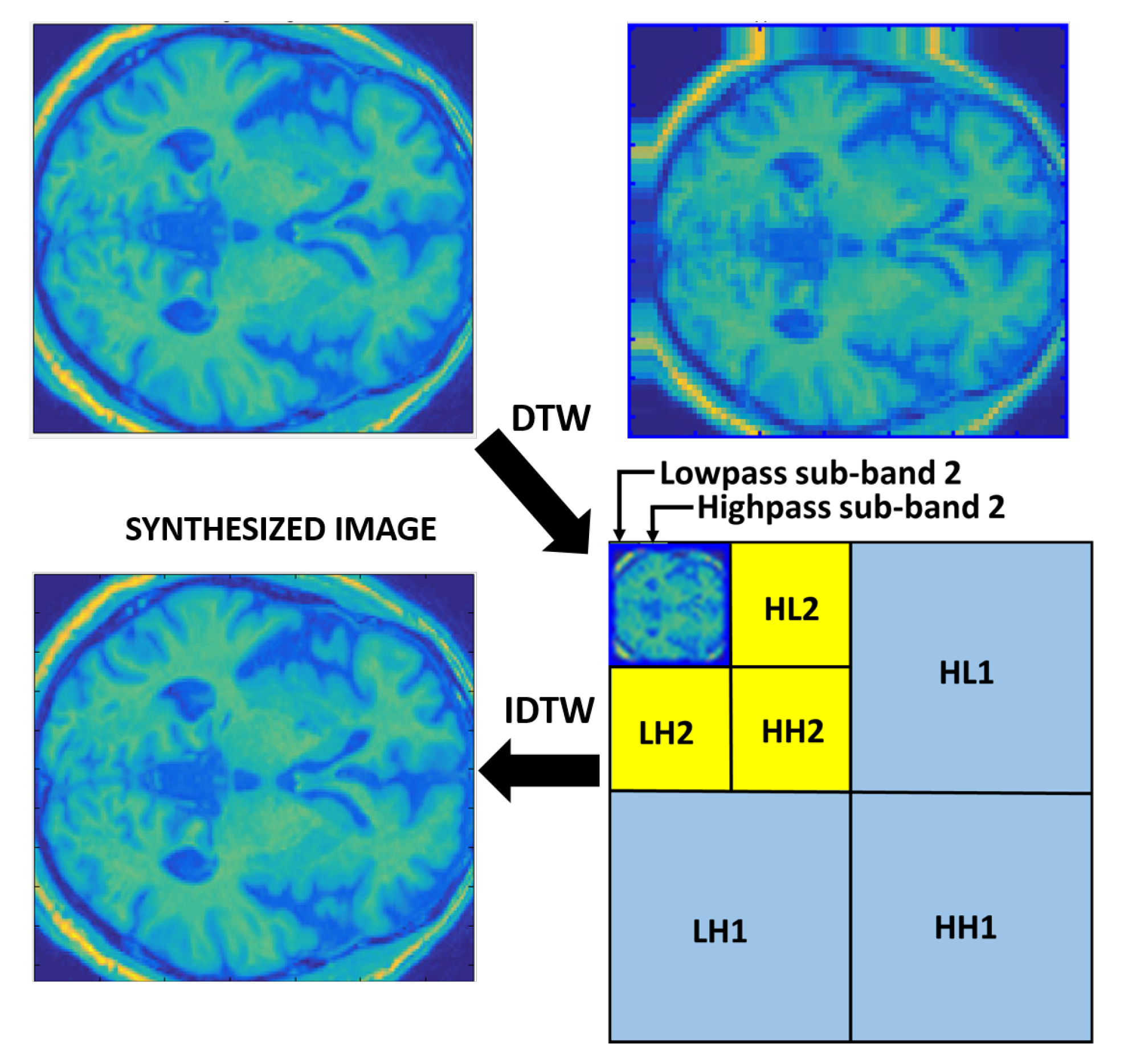
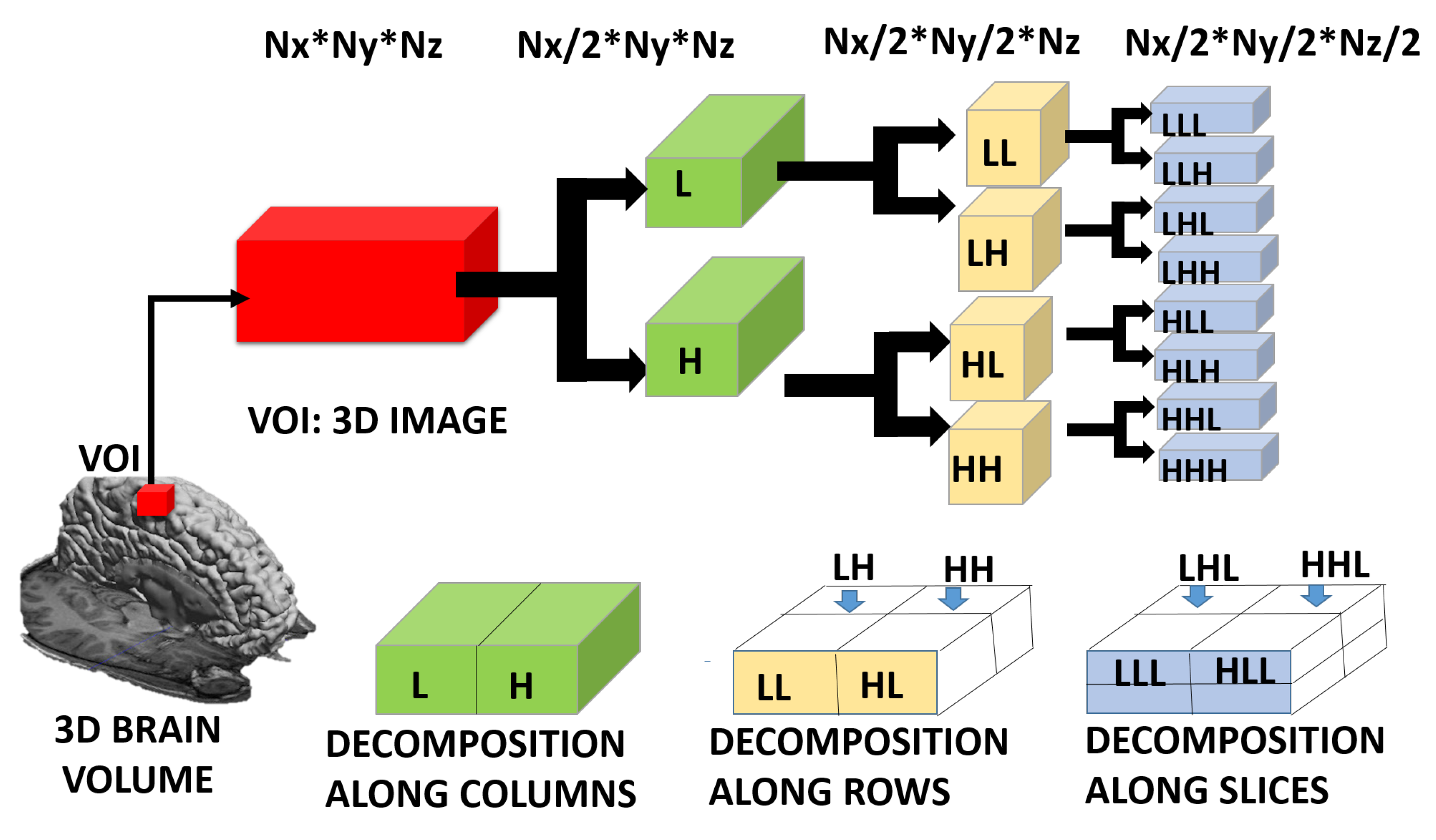
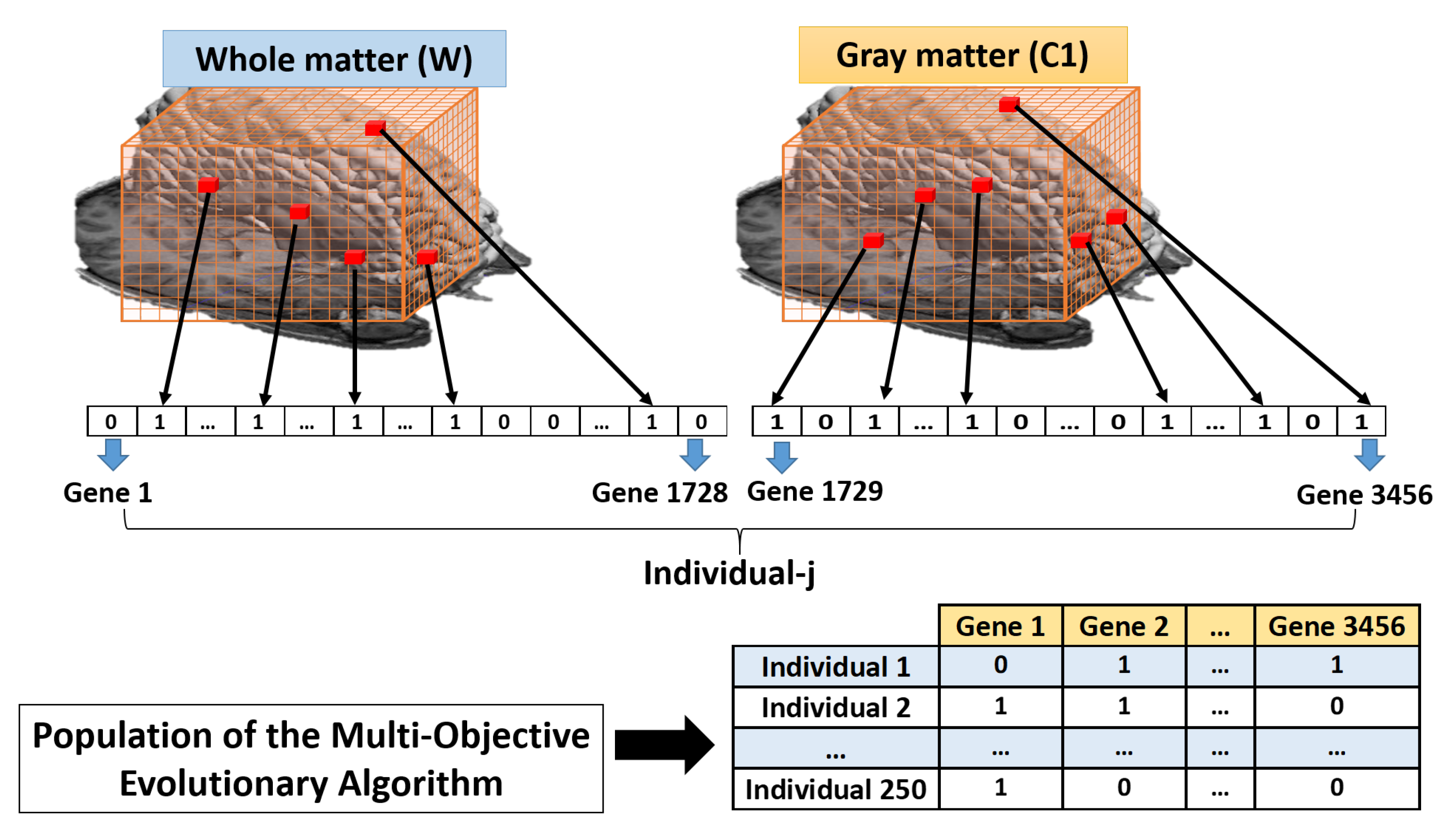
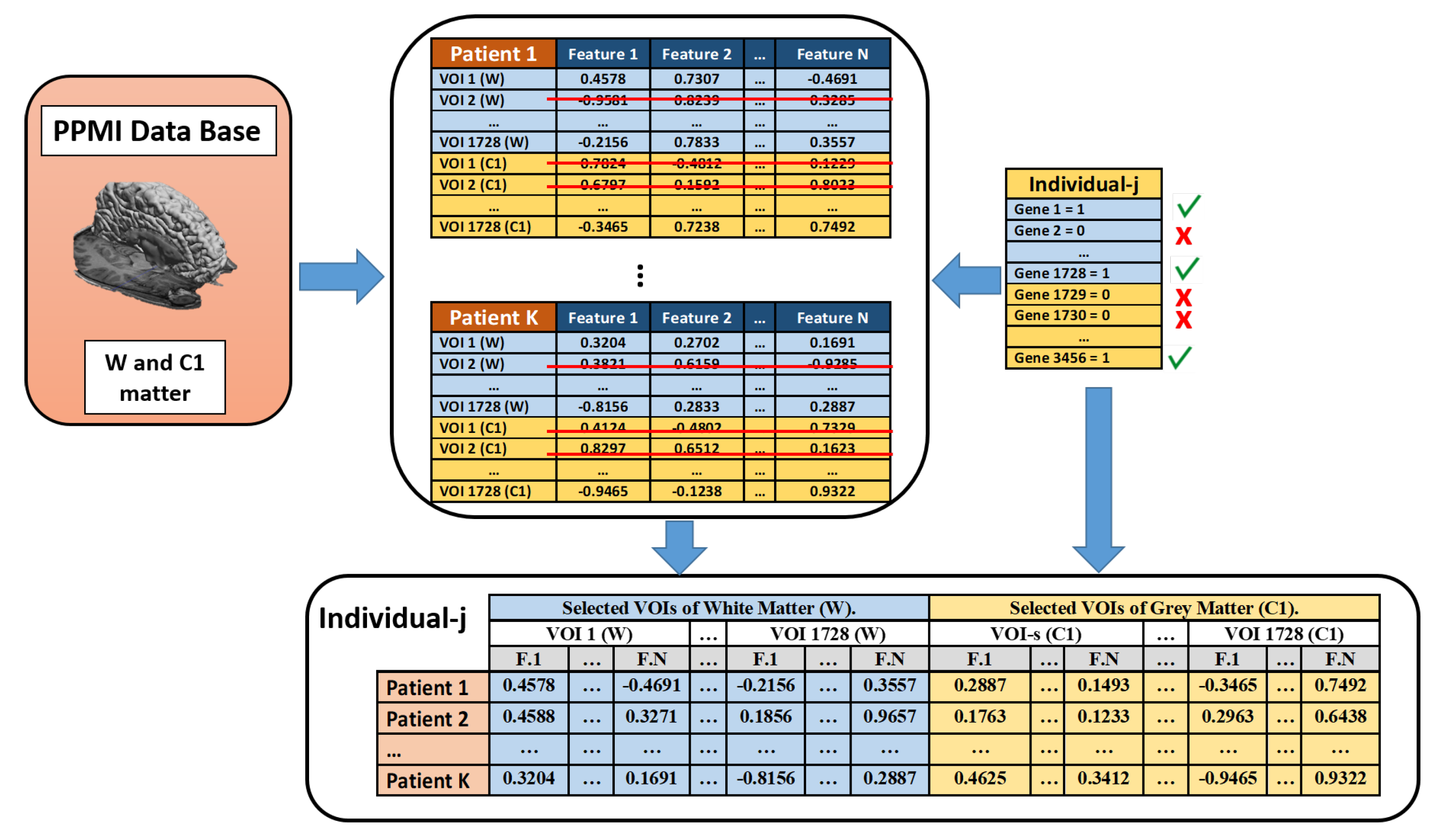
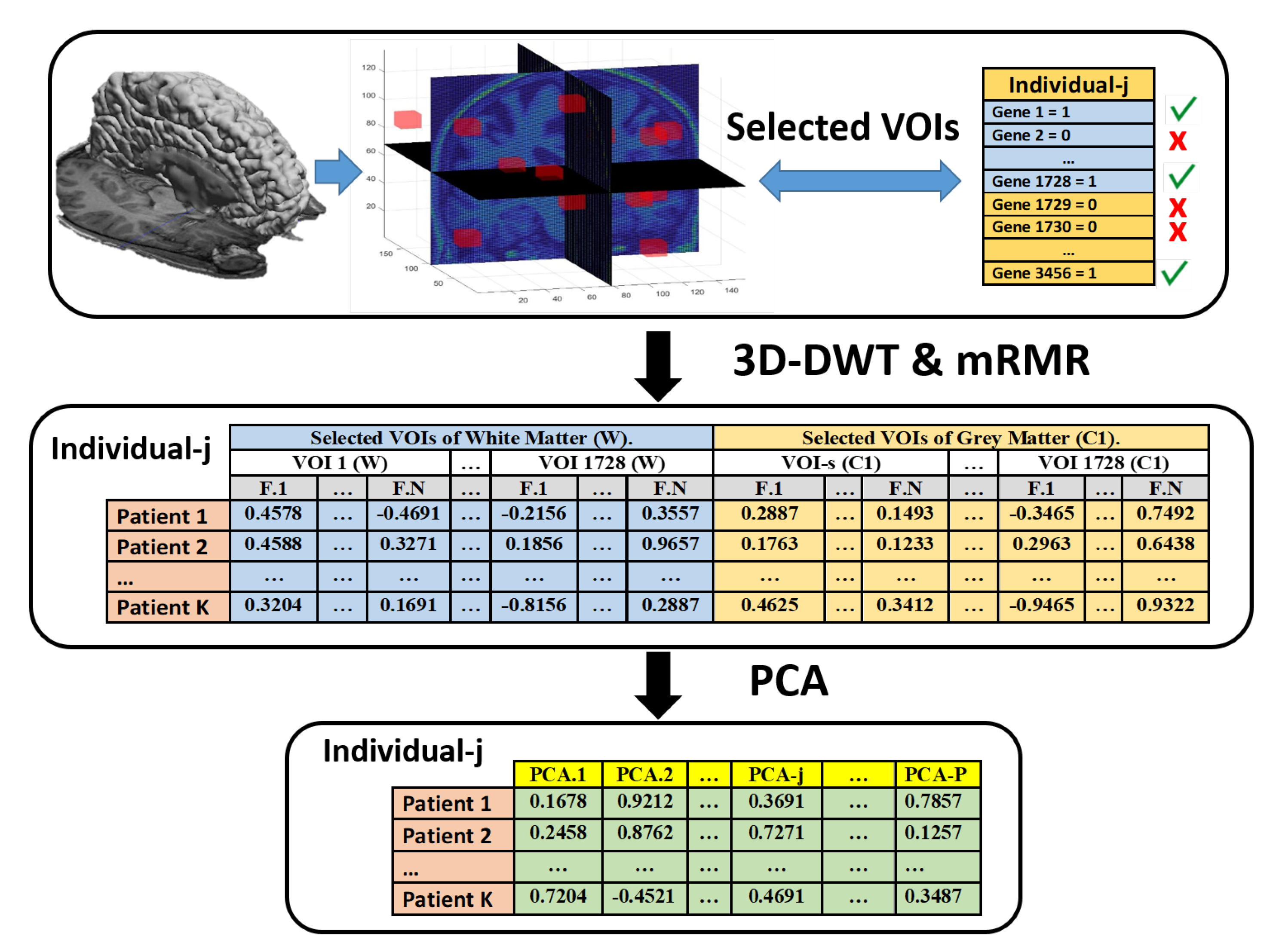
3.3. Feature Extraction
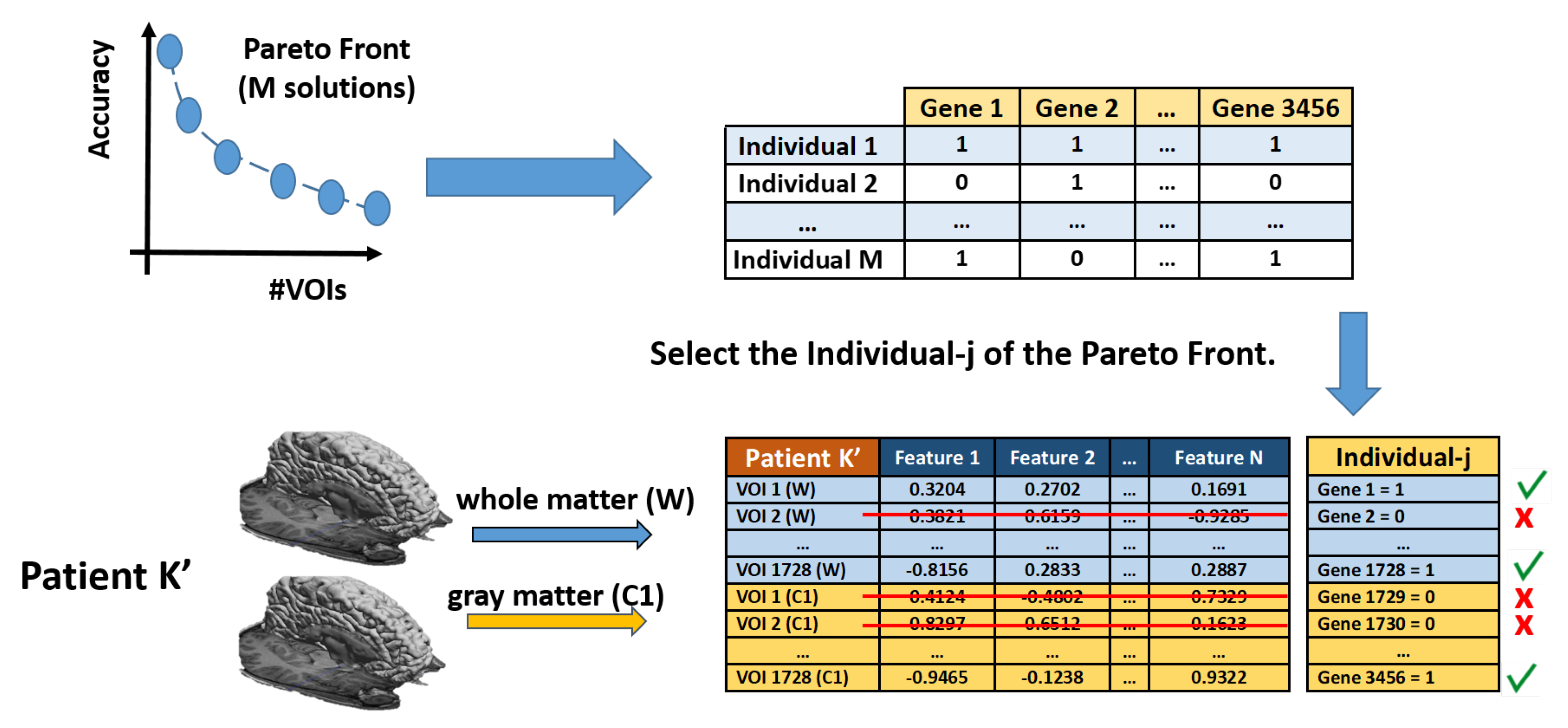
3.4. Feature Selection
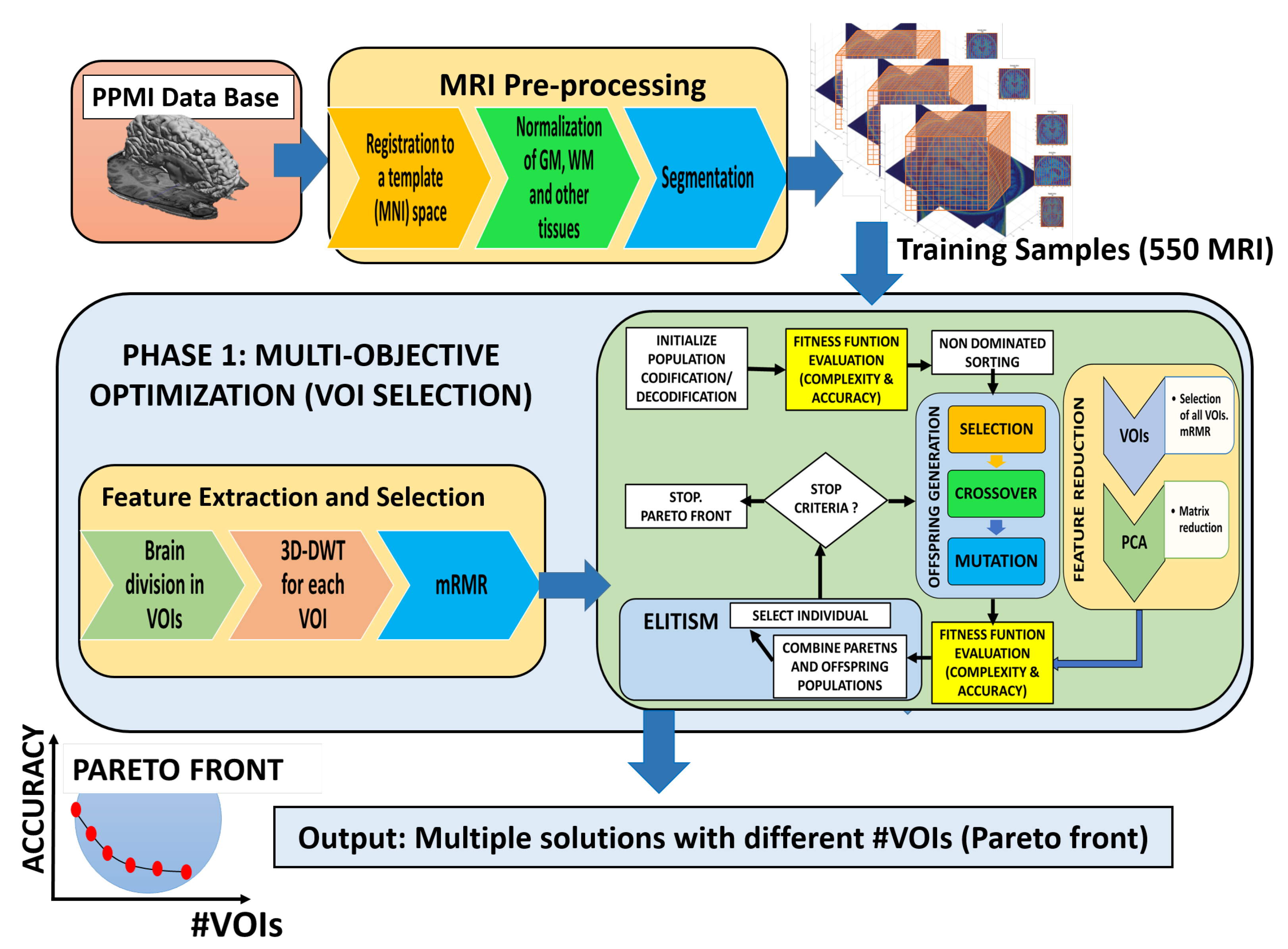
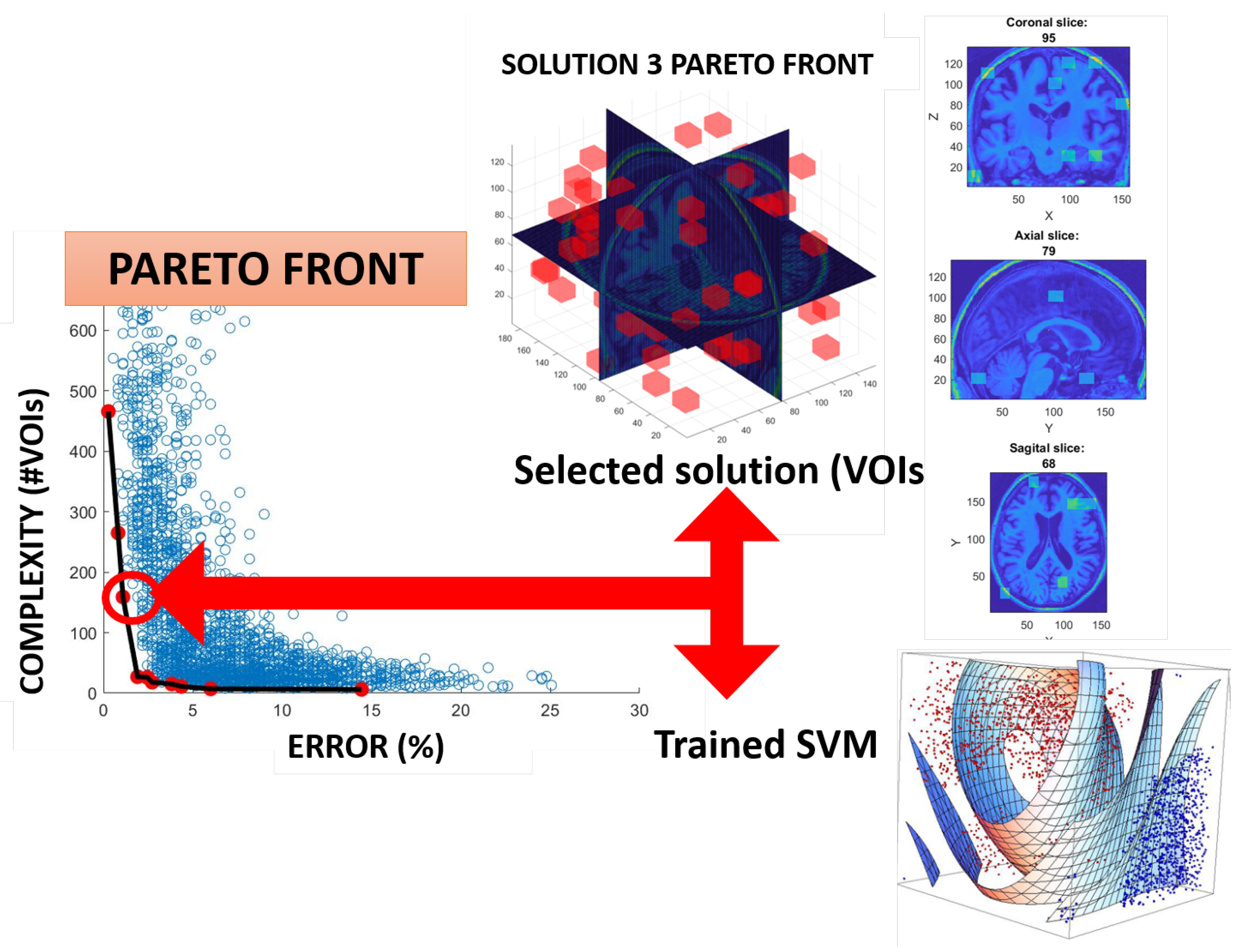
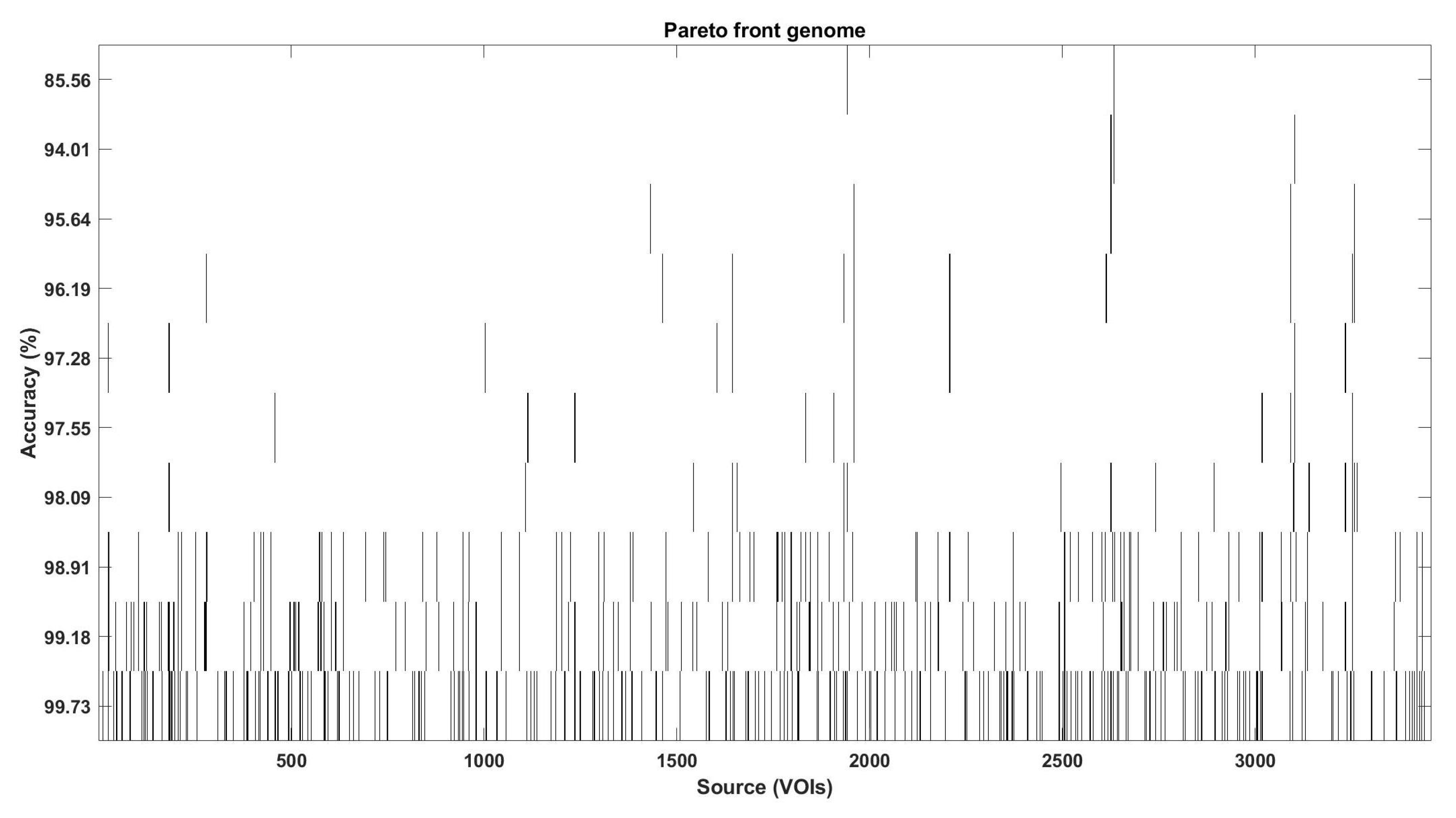
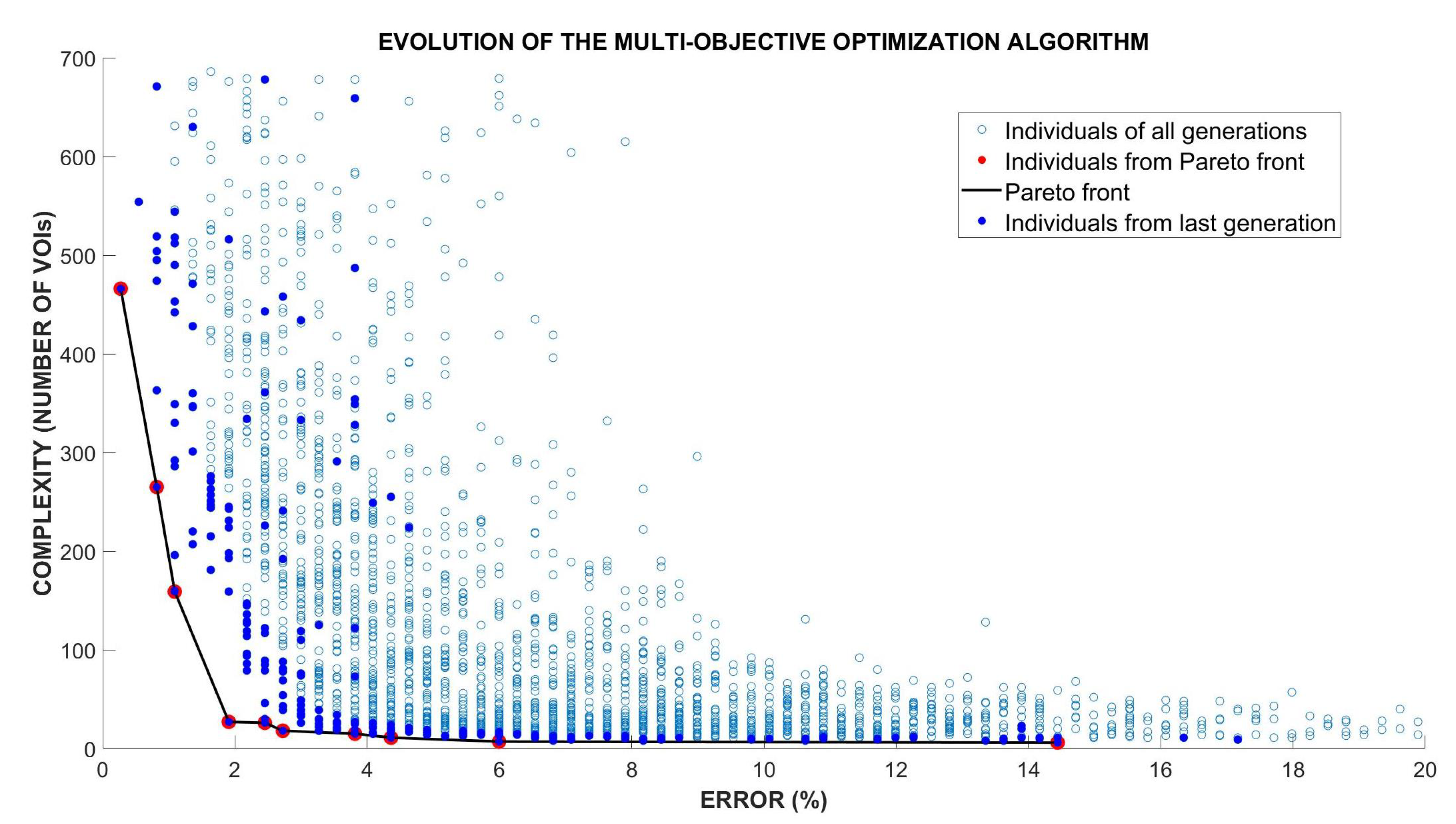
3.5. Optimization of the VOIs Selection Using Multi-Objective Optimization Evolutionary Algorithm (MOE)
- Initialization of the Multi-Objective Optimization Evolutionary Algorithm (MOE), through an initial population , and the creation of an initial (empty) set of non-dominated solutions (). The population size that was used in the simulation was 250.
- Calculate the fitness value for each individual. For this calculation, using the set of training patients, the mean precision value (10-k-fold is used) of the SVM classifier used in the MOE fitness function must be computed, together with the number of genes (complexity of the system) that are set to one.
- In the NSGA-II algorithm, there are two components that are relevant in its elitist strategy: sorting the non-dominated solutions and calculating the crowding distance. With this methodology, individuals from the population will be selected based on their fitness function.
- The evolutionary algorithm generates a new population of individuals. To achieve this, a copy of the non-dominated individuals in and is made to
- During the execution of a new generation, operators of the evolutionary algorithm are applied to the individuals of the population. These include the binary tournament selection with replacement in . As we have indicated before, there is an elitist process where the best solutions are copied from generation to generation, and MOE operators complete the population (until the 250 individuals used in this role are obtained). Binary crossover and mutation are also applied, and is set on the resulting population.
- Evaluation of fitness and Pareto functions from the construction. Our fitness function has two goals: Complexity of the obtained solution (measured as the number of VOIs selected) and accuracy of the classifier (using an SVM).
- Repeat until Stopping Condition is reached (typically, a maximum number of generations is reached).
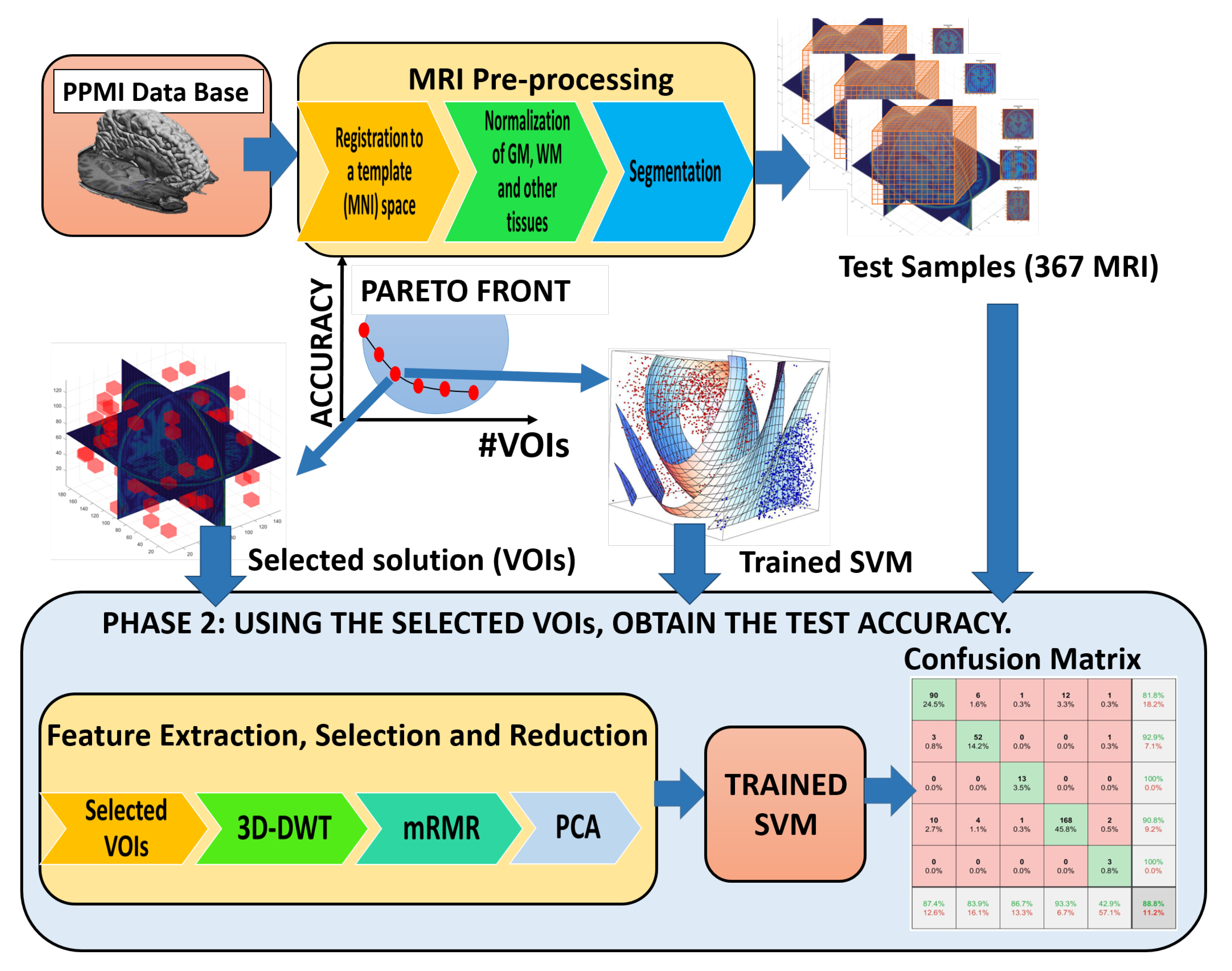
4. Pipeline
5. Results
5.1. Performance
5.2. Classification Results
5.3. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gouda, N.A.; Elkamhawy, A.; Cho, J. Emerging Therapeutic Strategies for Parkinson’s Disease and Future Prospects: A 2021 Update. Biomedicines 2022, 10, 371. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Neurological Disorders: Public Health Challenges; World Health Organization: Genebra, Switzerland, 2006; pp. 140–150. [Google Scholar]
- Olanow, C.; Schapira, A.; Obeso, J. Parkinson’s disease and other movement disorders. In Harrison’s Principles of Internal Medicine, 19th ed.; McGraw-Hill Education: New York, NY, USA, 2015; pp. 2609–2626. [Google Scholar]
- Dorsey, E.; Shererb, T.; Okunc, M.; Bloem, B. The Emerging Evidence of the Parkinson Pandemic. J. Park. Dis. 2018, 8, 140–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boshkoska, B.; Miljković, D.; Valmarska, A.; Gatsios, D.; Rigas, G.; Konitsiotis, S.; Tsiouris, K.; Fotiadis, D.; Bohanec, M. Decision Support for Medication Change of Parkinson’s Disease Patients. Comput. Methods Programs Biomed. 2020, 196, 105552. [Google Scholar] [CrossRef] [PubMed]
- Hughes, A.; Daniel, S.; Ben-Shlomo, Y.; Lees, A. The accuracy of diagnosis of parkinsonian syndromes in a specialist movement disorder service. Brain 2002, 125, 861–870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foundation, P. Stages of Parkinson’s. 2020. Available online: https://www.parkinson.org/ (accessed on 9 March 2020).
- Kalia, L.; Lang, A. Parkinson’s disease. Lancet 2015, 896–912. [Google Scholar] [CrossRef]
- Chougar, L.; Faouzi, J.; Pyatigorskaya, N.; Yahia-Cherif, L.; Gaurav, R.; Biondetti, E.; Villotte, M.; Valabregue, R.; Corvol, J.C.; Brice, A.; et al. Automated Categorization of Parkinsonian Syndromes Using Magnetic Resonance Imaging in a Clinical Setting. Mov. Disord. 2021, 36, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Biase, L.; Santo, A.; Caminiti, M.; Liso, A.; Shah, S.; Ricci, L.; Lazzaro, V. Gait Analysis in Parkinson’s Disease: An Overview of the Most Accurate Markers for Diagnosis and Symptoms Monitoring. Sensors 2020, 12, 3529. [Google Scholar] [CrossRef] [PubMed]
- Saeed, U.; Compagnone, J.; Aviv, R.E.A. Imaging biomarkers in Parkinson’s disease and Parkinsonian syndromes: Current and emerging concepts. Transl. Neurodegener. 2017, 6, 1–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salvatore, C.; Cerasa, A.; Castiglioni, I.; Gallivanone, F.; Augimeri, A.; Lopez, M.; Arabia, G.; Morelli, M.; Gilardi, M.; Quattrone, A. Machine learning on brain MRI data for differential diagnosis of Parkinson’s disease and Progressive Supranuclear Palsy. J. Neurosci. Methods 2014, 222, 230–237. [Google Scholar] [CrossRef]
- Eidelberg, D. Imaging in Parkinson’s Disease; Oxford University Press: New York, NY, USA, 2012; pp. 3–8. [Google Scholar]
- Chien, C.Y.; Hsu, S.W.; Lee, T.L.; Sung, P.S.; Lin, C.C. Using Artificial Neural Network to Discriminate Parkinson’s Disease from Other Parkinsonisms by Focusing on Putamen of Dopamine Transporter SPECT Images. Biomedicines 2020, 9, 12. [Google Scholar] [CrossRef]
- Spetsieris, P.; Eidelberg, D. Spectral guided sparse inverse covariance estimation of metabolic networks in Parkinson’s disease. Neuroimage 2021, 226, 117568. [Google Scholar] [CrossRef] [PubMed]
- Halliday, G.M.; Leverenz, J.S.; Schneider, R.; Adler, C.H. The neurobiological basis of cognitive impairment in Parkinson’s disease. Mov. Disord. 2014, 29, 634–650. [Google Scholar] [CrossRef] [Green Version]
- Simuni, T.; Uribe, L.; Cho, H.R.; Caspell-Garcia, C.; Coffey, C.S.; Siderowf, A.; Trojanowski, J.Q.; Shaw, L.M.; Seibyl, J.; Singleton, A.; et al. Clinical and dopamine transporter imaging characteristics of non-manifest LRRK2 and GBA mutation carriers in the Parkinson’s Progression Markers Initiative (PPMI): A cross-sectional study. Lancet Neurol. 2020, 19, 71–80. [Google Scholar] [CrossRef]
- Amoroso, N.; La Rocca, M.; Monaco, A.; Bellotti, R.; Tangaro, S. Complex networks reveal early MRI markers of Parkinson’s disease. Med. Image Anal. 2018, 48, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Shindea, S.; Prasadb, S.; Sabooa, Y.; Kaushicka, R.; Sainid, J.; Palb, P.; Ingalhalikara, M. Predictive markers for Parkinson’s disease using deep neural nets on neuromelanin sensitive MRI. Neuroimage Clin. 2019, 22, 101748. [Google Scholar] [CrossRef] [PubMed]
- Pang, H.; Yu, Z.; Li, R.; Yang, H.; Fan, G. MRI-Based Radiomics of Basal Nuclei in Differentiating Idiopathic Parkinson’s Disease From Parkinsonian Variants of Multiple System Atrophy: A Susceptibility-Weighted Imaging Study. Front. Aging Neurosci. 2020, 12, 379. [Google Scholar] [CrossRef]
- Olivares, R.; Munoz, R.; Soto, R.; Crawford, B.; Cárdenas, D.; Ponce, A.; Taramasco, C. An Optimized Brain-Based Algorithm for Classifying Parkinson’s Disease. Appl. Sci. 2020, 10, 1827. [Google Scholar] [CrossRef] [Green Version]
- Rana, B.; Juneja, A.; Saxena, M.; Gudwani, S.; Kumaran, S.S.; Agrawal, R.; Behari, M. Regions-of-interest based automated diagnosis of Parkinson’s disease using T1-weighted MRI. Expert Syst. Appl. 2015, 42, 4506–4516. [Google Scholar] [CrossRef]
- Long, D.; Wang, J.; Xuan, M.; Gu, Q.; Xu, X.; Kong, D.; Zhang, M. Automatic classification of early Parkinson’s disease with multimodal MR Imaging. PLoS ONE 2012, 7, e47714. [Google Scholar]
- Lei, H.; Zhao, Y.; Wen, Y.; Luo, Q.; Cai, Y.; Liu, G.; Lei, B. Sparse feature learning for multi-class Parkinson’s disease classification. Technol. Health Care 2018, 26, 193–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliveira, F.; Faria, D.; Costa, D.; Castelo-Branco, M.; Tavares, J. Extraction, selection and comparison of features for an effective automated computer-aided diagnosis of Parkinson’s disease based on [123I]FP-CIT SPECT images. Eur. Nucl. Med. Mol. Imaging 2018, 45, 1052–1062. [Google Scholar] [CrossRef] [Green Version]
- Pereira, H.; Ferreira, H. Classification of Patients with Parkinson’s Disease Using Medical Imaging and Artificial Intelligence Algorithms. In MEDICON 2019, IFMBE Proceedings 76; Springer Nature: Cham, Switzerland, 2020; pp. 2043–2056. [Google Scholar]
- Sivaranjini, S.; Sujatha, C. Deep learning based diagnosis of Parkinson’s disease using convolutional neural network. Multimed. Tools Appl. 2020, 79, 15467–15479. [Google Scholar] [CrossRef]
- Esmaeilzadeh, S.; Yang, Y.; Adeli, E. End-to-End Parkinson Disease Diagnosisusing Brain MR-Images by 3D-CNN. arXiv 2018, arXiv:1806.05233. [Google Scholar]
- Marek, K.; Jennings, D.; Lasch, S.; Siderowf, A.; Tanner, C.; Simuni, T.; Coffey, C.; Kieburtz, K.; Flagg, E.; Chowdhury, S.; et al. The parkinson progression marker initiative (PPMI). Prog. Neurobiol. 2011, 95, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Akkaoui, M.; Geoffroy, P.; Roze, E.; Degos, B.; Garcin, B. Functional Motor Symptoms in Parkinson’s Disease and Functional Parkinsonism: A Systematic Review. J. Neuropsychiatry Clin. Neurosci. 2020, 32, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Song, Y.; Kim, H.; Ku, B.; Lee, W. Patients with scans without evidence of dopaminergic deficit (SWEDD) do not have early Parkinson’s disease: Analysis of the PPMI data. PLoS ONE 2021, 16, e0246881. [Google Scholar] [CrossRef] [PubMed]
- Erro, R.; Schneider, S.; Stamelou, M. What do patients with scans without evidence of dopaminergic deficit (SWEDD) have? New evidence and continuing controversies. J. Neurol. Neurosurg. Psychiatry 2016, 87, 319–323. [Google Scholar] [CrossRef]
- Choi, H.; Ha, S.; Im, H. Refining diagnosis of Parkinson’s disease with deep learning-based interpretation of dopamine transporter imaging. NeuroImage Clin. 2017, 16, 586–594. [Google Scholar] [CrossRef]
- Schneider, S. Patients with adult-onset dystonic tremor resembling Parkinsonian tremor have scans without evidence of dopaminergic deficit (SWEDDs). Mov. Disord. 2007, 22, 2210–2215. [Google Scholar] [CrossRef]
- Nicastro, N.; Garibotto, V.; Badoud, S.; Burkhard, P. Scan without evidence of dopaminergic deficit: A 10-year retrospective study. Park. Relat. Disord. 2016, 31, 53–58. [Google Scholar] [CrossRef] [Green Version]
- Salmanpour, M.R.; Shamsaei, M.; Saberi, A.; Hajianfar, G.; Soltanian-Zadeh, H.; Rahmim, A. Robust identification of Parkinson’s disease subtypes using radiomics and hybrid machine learning. Comput. Biol. Med. 2021, 129, 104142. [Google Scholar] [CrossRef] [PubMed]
- Mahlknecht, P.; Seppi, K.; Poewe, W. The Concept of Prodromal Parkinson’s Disease. J. Parkinsons Dis. 2015, 5, 681–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Postuma, R.; Aarsland, D.; Barone, P.; Burn, D.; Hawkes, C.; Oertel, W.; Ziemssen, T. Identifying prodromal Parkinson’s disease: Pre-Motor disorders in Parkinson’s disease. Mov. Disord. Off. J. Mov. Disord. Soc. 2012, 27, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Penny, W.D.; Friston, K.J.; Ashburner, J.T.; Kiebel, S.J.; Nichols, T.E. (Eds.) Statistical Parametric Mapping: The Analysis of Functional Brain Images; Academic Press: Cambridge, MA, USA, 2006. [Google Scholar]
- Flandin, G.; Friston, K. Analysis of family-wise error rates in statistical parametric mapping using random field theory. Hum. Brain Mapp. 2017, 40, 2052–2054. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, J.; Friston, K. Unified segmentation. NeuroImage 2005, 26, 839–851. [Google Scholar] [CrossRef] [PubMed]
- Nanni, L.; Lumini, A. Wavelet decomposition tree selection for palm and face authentication. Pattern Recognit. Lett. 2008, 29, 343–353. [Google Scholar] [CrossRef]
- Challis, E.; Hurley, P.; Serra, L.; Bozzali, M.; Oliver, S.; Cercignani, M. Gaussian process classification of Alzheimer’s disease and mild cognitive impairment from resting-state fMRI. NeuroImage 2015, 112, 232–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haq, E.; Huang, J.; Kang, L.; Haq, H.; Zhan, T. Image-based state-of-the-art techniques for the identification and classification of brain diseases: A review. Med. Biol. Eng. Comput. 2020, 58, 2603–2620. [Google Scholar] [CrossRef]
- Bharati, S.; Podder, P.; Al-Masud, M. Brain Magnetic Resonance Imaging Compression Using Daubechies Biorthogonal Wavelet with the fusion of STW and SPIHT. In Proceedings of the 2018 International Conference on Advancement in Electrical and Electronic Engineering (ICAEEE), Gazipur, Bangladesh, 22–24 November 2018; pp. 1–4. [Google Scholar]
- Kovalev, V.; Kruggel, F.; Gertz, H.; Von Cramon, D.Y. Three-dimensional texture analysis of MRI brain datasets. IEEE Trans. Med. Imaging 2001, 20, 424–433. [Google Scholar] [CrossRef]
- Valenzuela, O.; Jiang, X.; Carrillo, A.; Rojas, I. Multi-Objective Genetic Algorithms to Find Most Relevant Volumes of the Brain Related to Alzheimer’s Disease and Mild Cognitive Impairment. Int. J. Neural Syst. 2018, 28, 961–972. [Google Scholar] [CrossRef]
- He, C.; Dong, J.; Zheng, Y.; Gao, Z. 3-D coefficient tree structure for 3-D wavelet video coding. IEEE Trans. Circuits Syst. Video Technol. 2003, 13, 961–972. [Google Scholar]
- Peng, H.C.; Long, F.H.; Ding, C. Feature selection based on mutual information: Criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef] [PubMed]
- Adeli, E.; Shi, F.; An, L.; Wee, C.Y.; Wu, G.; Wang, T.; Shen, D. Joint feature-sample selection and robust diagnosis of Parkinson’s disease from MRI data. NeuroImage 2016, 141, 206–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abos, A.; Baggio, H.C.; Segura, B.; Garcia-Diaz, A.I.; Compta, Y.; Martí, M.J. Discriminating cognitive status in parkinson’s disease through functional connectomics and machine learning. Sci. Rep. 2017, 7, 45347. [Google Scholar] [CrossRef] [PubMed]
- Ariz, M.; Abad, R.C.; Castellanos, G.; Martínez, M.; Muñoz-Barrutia, A.; Fernández-Seara, M.A.; Pastor, P.; Pastor, M.A.; Ortiz-de Solórzano, C. Dynamic Atlas-Based Segmentation and Quantification of Neuromelanin-Rich Brainstem Structures in Parkinson Disease. IEEE Trans. Med. Imaging 2019, 38, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhai, D.; Yang, Y.; Zhang, Y.; Wang, C. A novel semi-supervised multi-view clustering framework for screening Parkinson’s disease. Math. Biosci. Eng. 2020, 17, 3395–3411. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Lee, P.; Lee, S.; Chung, S.; Shin, N.Y. The diagnostic potential of multimodal neuroimaging measures in Parkinson’s disease and atypical parkinsonism. Brain Behav. 2020, 10, e01808. [Google Scholar] [CrossRef]
- Solana-Lavalle, G.; Rosas-Romero, R. Classification of PPMI MRI scans with voxel-based morphometry and machine learning to assist in the diagnosis of Parkinson’s disease. Comput. Methods Programs Biomed. 2021, 198, 105793. [Google Scholar] [CrossRef] [PubMed]
- Talai, A.S.; Sedlacik, J.; Boelmans, K.; Forkert, N.D. Utility of Multi-Modal MRI for Differentiating of Parkinson’s Disease and Progressive Supranuclear Palsy Using Machine Learning. Front. Neurol. 2021, 12, 546. [Google Scholar] [CrossRef]

| Measures | Normal (258) | SWEDD (154) | ||||||
|---|---|---|---|---|---|---|---|---|
| Sex (Male/Female) | 181 | 77 | 100 | 54 | ||||
| Age (years) | 61.4 | 11.7 | 59.0 | 8.2 | 64.1 | 9.7 | 57.8 | 10.7 |
| Weight (kg) | 83.0 | 10.7 | 67.9 | 17.1 | 88.6 | 12.8 | 75.8 | 12.8 |
| Mean | Std | Mean | Std | Mean | Std | Mean | Std | |
| Measures | Prodromal (38) | PD (450) | ||||||
|---|---|---|---|---|---|---|---|---|
| Sex (Male/Female) | 30 | 8 | 288 | 162 | ||||
| Age (years) | 67.1 | 3.9 | 67.8 | 3.1 | 62.4 | 9.8 | 60.2 | 8.7 |
| Weight (kg) | 88.8 | 14.5 | 68.5 | 9.7 | 90.8 | 16.1 | 66.3 | 11.9 |
| Mean | Std | Mean | Std | Mean | Std | Mean | Std | |
| Measures | GeneCohort (17) | |||
|---|---|---|---|---|
| Sex (Male/Female) | 14 | 3 | ||
| Age | 62.1 | 10.6 | 72.2 | 9.0 |
| Weight (kg) | 87.2 | 11.3 | 64.7 | 5.7 |
| Mean | Std | Mean | Std | |
| Pareto Solution | Accuracy (%) | Total VOIs | W-VOIs | C1-VOIs |
|---|---|---|---|---|
| 1 | 99.73 | 466 | 231 | 235 |
| 2 | 99.18 | 265 | 133 | 132 |
| 3 | 98.91 | 159 | 73 | 86 |
| 4 | 98.09 | 27 | 6 | 21 |
| 5 | 97.54 | 26 | 4 | 22 |
| 6 | 97.27 | 18 | 6 | 12 |
| 7 | 96.18 | 15 | 4 | 11 |
| 8 | 95.64 | 11 | 4 | 7 |
| 9 | 94.00 | 7 | 0 | 7 |
| 10 | 85.55 | 6 | 0 | 6 |
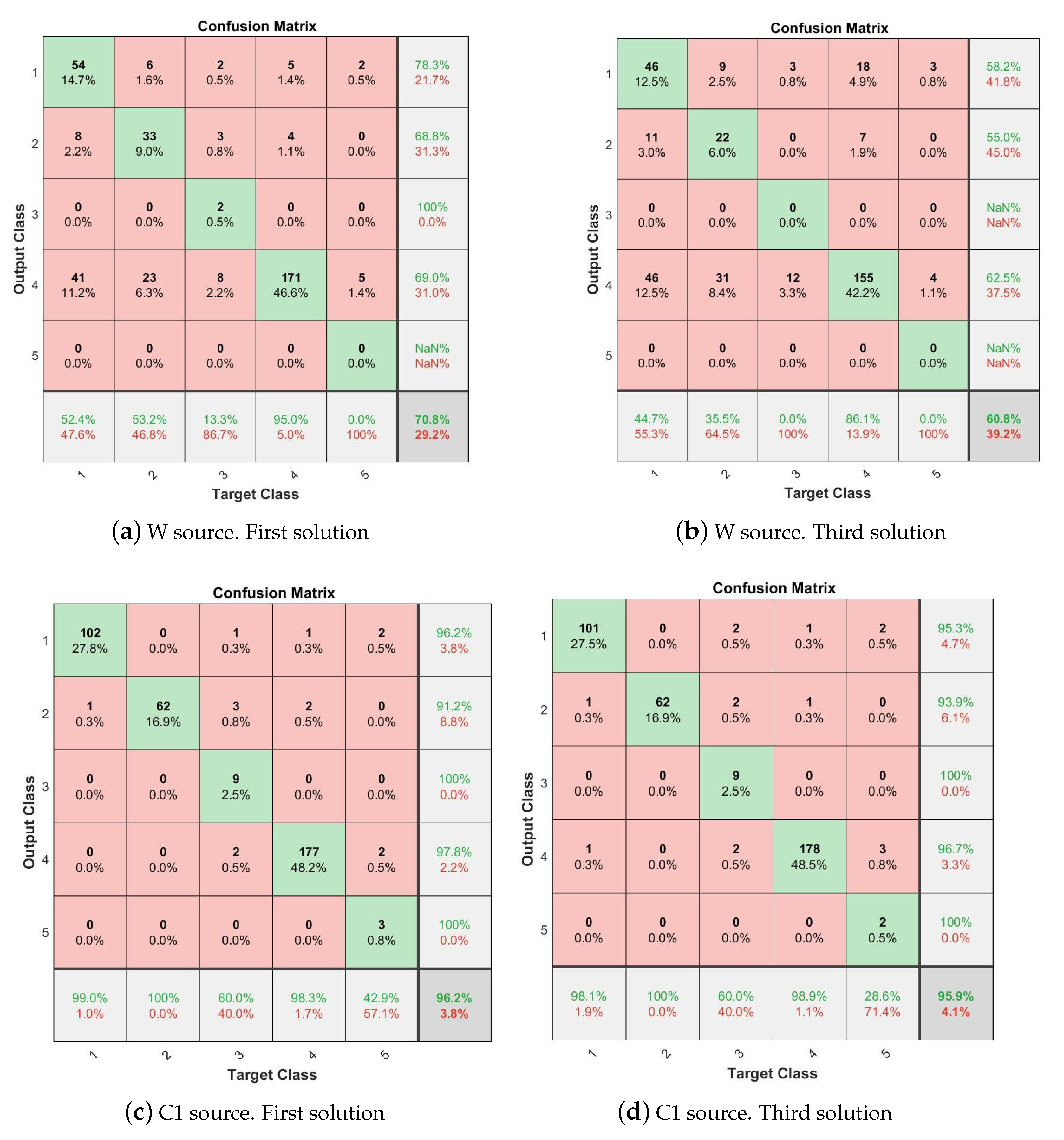
| Solution (PF) | Source | #VOIs | #Features | #PCA | Accuracy | Conf.Matrix |
|---|---|---|---|---|---|---|
| 1 | W | 231 | 6930 | 511 | 70.8 | Figure 13a |
| C1 | 235 | 7050 | 464 | 96.2 | Figure 13c | |
| W+C1 | 466 | 13,980 | 535 | 97.0 | ||
| 2 | W | 133 | 3990 | 419 | 63.8 | |
| C1 | 132 | 3960 | 416 | 95.9 | ||
| W+C1 | 265 | 7950 | 498 | 96.2 | ||
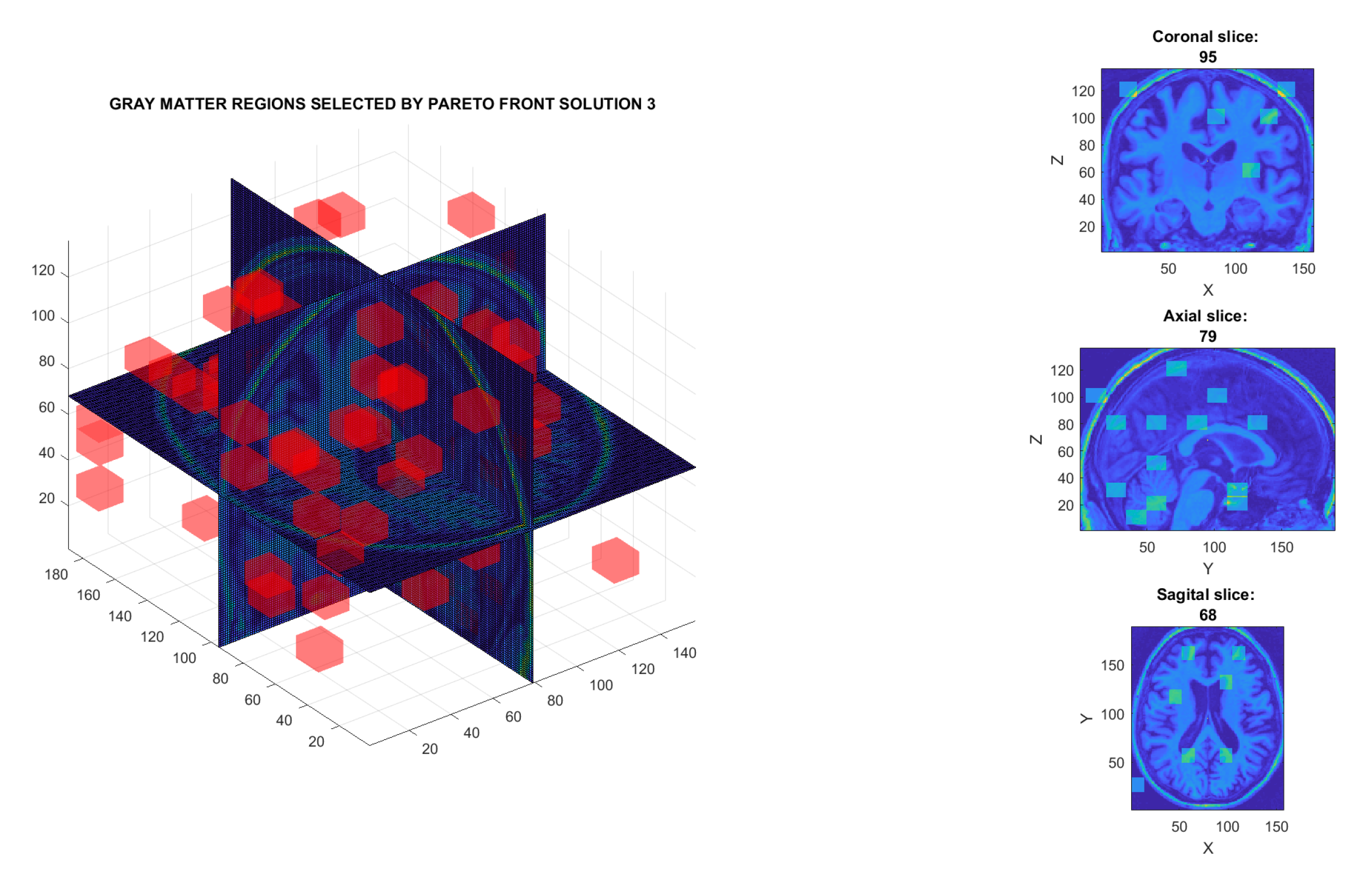
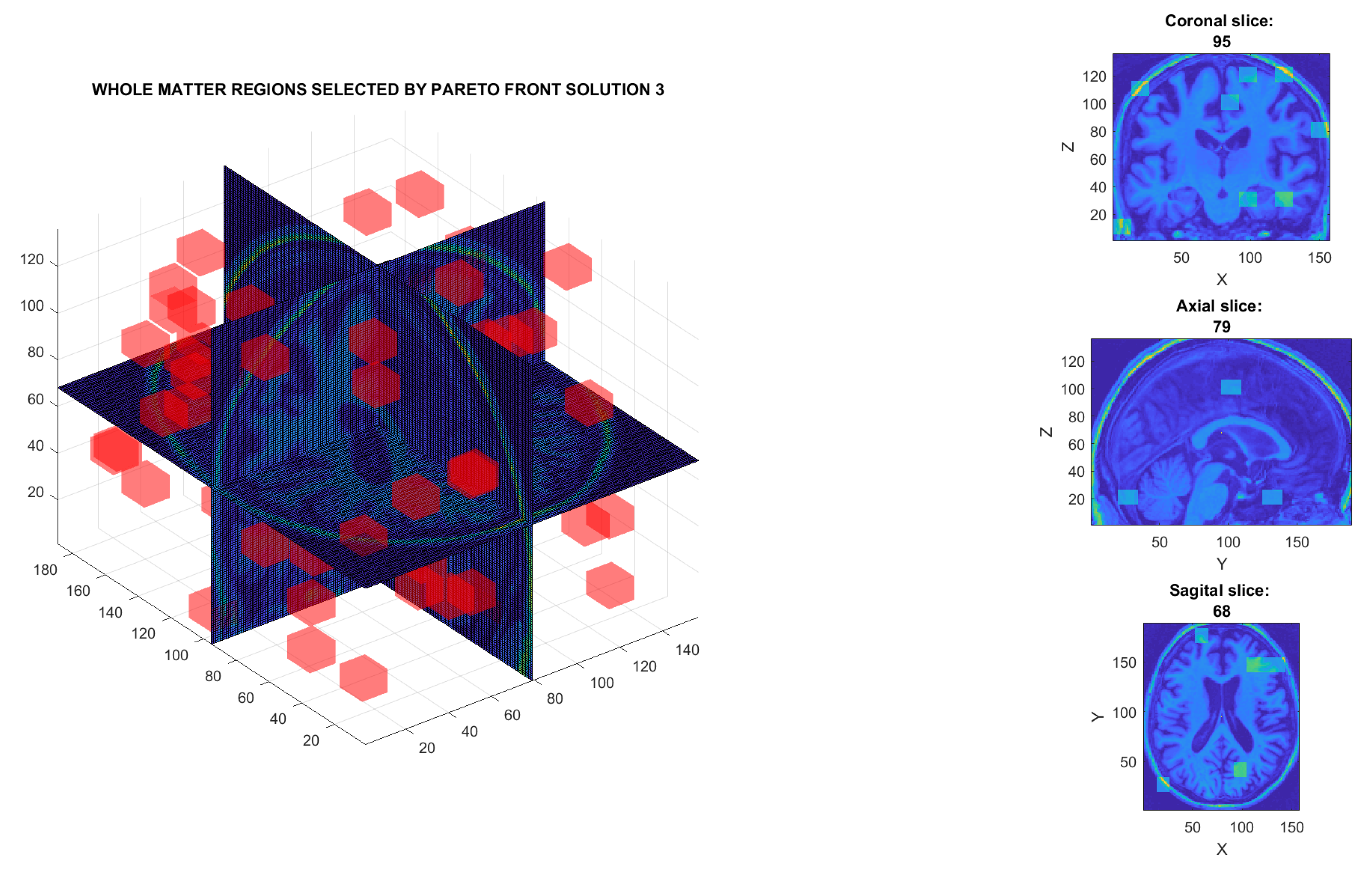
| 3 | W | 73 | 2190 | 339 | 60.8 | Figure 13b |
| C1 | 86 | 2580 | 380 | 95.9 | Figure 13d | |
| W+C1 | 159 | 4770 | 448 | 96 | ||
| 4 | W | 6 | 180 | 32 | 48.0 | |
| C1 | 21 | 630 | 224 | 93.7 | ||
| WC1 | 27 | 810 | 232 | 93.5 | ||
| 5 | W | 4 | 120 | 37 | 49.86 | |
| C1 | 22 | 660 | 221 | 92.9 | ||
| WC1 | 26 | 780 | 230 | 93.18 | ||
| 7 | W | 4 | 120 | 3 | 49.0 | |
| C1 | 11 | 330 | 136 | 88.8 | ||
| WC1 | 15 | 450 | 138 | 90.5 |
| Source | Description of VOI Locations |
|---|---|
| W | [1,13,35,49,16,26]; [1,13,35,49,36,46]; [1,13,95,109,6,16]; [1,13,110,124,86,96]; [1,13,110,124,106,116]; [1,13,125,139,76,86]; [1,13,155,169,46,56]; [14,26,20,34,6,16]; [14,26,20,34,66,76]; [14,26,50,64,46,56]; [14,26,80,94,36,46]; [14,26,80,94,116,126]; [14,26,95,109,106,116]; [14,26,125,139,116,126]; [14,26,170,184,36,46]; [14,26,170,184,56,66]; [27,39,5,19,86,96]; [27,39,140,154,76,86]; [27,39,170,184,16,26]; [27,39,170,184,76,86]; [40,52,20,34,36,46]; [40,52,170,184,86,96]; [40,52,170,184,96,106]; [53,65,5,19,36,46]; [53,65,5,19,86,96]; [53,65,35,49,36,46]; [53,65,65,79,116,126]; [53,65,110,124,6,16]; [53,65,125,139,46,56]; [53,65,140,154,96,106]; [53,65,155,169,6,16]; [53,65,170,184,66,76]; [53,65,170,184,106,116]; [66,78,20,34,76,86]; [66,78,35,49,16,26]; [66,78,80,94,86,96]; [66,78,155,169,6,16]; [66,78,155,169,16,26]; [79,91,20,34,16,26]; [79,91,95,109,96,106]; [79,91,125,139,16,26]; [92,104,35,49,66,76]; [92,104,50,64,16,26]; [92,104,65,79,6,16]; [92,104,65,79,86,96]; [92,104,95,109,26,36]; [92,104,95,109,116,126]; [92,104,125,139,46,56]; [105,117,35,49,116,126]; [105,117,50,64,106,116]; [105,117,65,79,16,26]; [105,117,80,94,116,126]; [105,117,125,139,46,56]; [105,117,140,154,66,76]; [118,130,5,19,16,26]; [118,130,5,19,46,56]; [118,130,20,34,36,46]; [118,130,80,94,86,96]; [118,130,95,109,26,36]; [118,130,95,109,116,126]; [118,130,110,124,56,66]; [118,130,125,139,86,96]; [118,130,140,154,26,36]; [118,130,140,154,66,76]; [118,130,155,169,26,36]; [131,143,35,49,76,86]; [131,143,140,154,66,76]; [131,143,170,184,96,106]; [144,156,65,79,116,126]; [144,156,95,109,76,86]; [144,156,125,139,96,106]; [144,156,140,154,76,86]; [144,156,155,169,106,116] |
| C1 | [1,13,5,19,86,96]; [1,13,20,34,66,76]; [1,13,35,49,76,86]; [1,13,35,49,96,106]; [1,13,50,64,96,106]; [1,13,65,79,36,46]; [1,13,80,94,76,86]; [1,13,80,94,96,106]; [1,13,110,124,96,106]; [1,13,125,139,96,106]; [1,13,140,154,96,106]; [1,13,170,184,26,36]; [1,13,170,184,46,56]; [1,13,170,184,56,66]; [14,26,20,34,116,126]; [14,26,50,64,6,16]; [14,26,65,79,86,96]; [14,26,80,94,16,26]; [14,26,95,109,116,126]; [27,39,5,19,106,116]; [27,39,125,139,76,86]; [27,39,125,139,86,96]; [27,39,125,139,116,126]; [40,52,20,34,56,66]; [40,52,50,64,116,126]; [40,52,80,94,6,16]; [40,52,110,124,6,16]; [40,52,110,124,66,76]; [40,52,110,124,116,126]; [40,52,155,169,6,16]; [53,65,35,49,26,36]; [53,65,50,64,66,76]; [53,65,50,64,106,116]; [53,65,80,94,76,86]; [53,65,110,124,46,56]; [53,65,125,139,46,56]; [53,65,140,154,106,116]; [53,65,155,169,66,76]; [66,78,20,34,46,56]; [66,78,20,34,106,116]; [66,78,65,79,96,106]; [66,78,80,94,116,126]; [66,78,110,124,96,106]; [66,78,155,169,96,106]; [79,91,5,19,96,106]; [79,91,20,34,26,36]; [79,91,20,34,76,86]; [79,91,35,49,6,16]; [79,91,50,64,16,26]; [79,91,50,64,46,56]; [79,91,50,64,76,86]; [79,91,65,79,116,126]; [79,91,80,94,76,86]; [79,91,95,109,96,106]; [79,91,110,124,16,26]; [79,91,110,124,26,36]; [79,91,125,139,76,86]; [92,104,50,64,66,76]; [92,104,65,79,86,96]; [92,104,80,94,116,126]; [92,104,125,139,66,76]; [92,104,140,154,86,96]; [92,104,140,154,96,106]; [105,117,65,79,26,36]; [105,117,65,79,36,46]; [105,117,65,79,86,96]; [105,117,65,79,106,116]; [105,117,80,94,46,56]; [105,117,95,109,56,66]; [105,117,155,169,66,76]; [105,117,155,169,116,126]; [105,117,170,184,56,66]; [105,117,170,184,106,116]; [118,130,50,64,76,86]; [118,130,65,79,86,96]; [118,130,80,94,76,86]; [118,130,95,109,96,106]; [118,130,140,154,36,46]; [131,143,80,94,86,96]; [131,143,95,109,116,126]; [144,156,35,49,6,16]; [144,156,65,79,36,46]; [144,156,65,79,46,56]; [144,156,80,94,36,46]; [144,156,125,139,116,126]; [144,156,155,169,16,26] |
| Author, Year | Cohort Data Base | Number of Subjects | Method Employed | Accuracy (%) |
|---|---|---|---|---|
| Long et al., 2012 [23] | Zhejiang University. | PD (n = 19) vs. Normal (n = 26) | SVM, LOOCV | 86.9 |
| Salvatore et al., 2014 [12] | Own data-base. | PD (n = 28) vs. Normal (n = 28) | PCA, SVM, VBM | 83.2 |
| Rana et al., 2015 [22] | Own data-base. | PD (n = 30) vs. Normal (n = 30) | SVM, ROI | 86.67 |
| Adeli et al., 2016 [50] | PPMI. | PD (n = 374) vs. Normal (n = 169) | Joint feature-sample selection, RLDA | 81.9 |
| Abos et al., 2017 [51] | Own data-base. | PD (n = 27) vs. Normal (n = 38) | SVM, Functional connectome | 80.0 |
| Lei et al., 2018 [24] | PPMI. | PD (n = 123) vs. SWEDD (n = 29) vs. Normal (n = 56) | SVM, GDC,CSF, DSSM | 78.37 |
| Amoroso et al., 2018 [18] | PPMI. | PD (n = 378) vs. Normal (n = 169) | Random Forest, SVM | 93 |
| Oliveira et al., 2018 [25] | PPMI. FP-CIT SPECT | PD (n = 443) vs. Normal (n = 209) | SV, LOOCV, ROI, k-NN | 97.9 |
| Ariz et al., 2019 [52] | UK Brain Bank. | PD (n = 39) vs. Normal (n = 40) | Artificial Neural Network | 79.9 |
| Shinde et al., 2019 [19] | NIMHANS, India. | PD (n = 45) vs. Normal (n = 35) | CNNs | 80.00 |
| Pang et al., 2020 [20] | China Medical University. | IPD (n = 83) vs. MSA-P (n = 102) | Seven radiomic features, SVM | 86.2 |
| Zhang et al., 2020 [53] | PPMI. | PD (n = 201) vs. Prodromal (n = 193) vs. Normal (n = 204) | Semi-supervised, LDA Multi-view learning Clustering | 74 |
| Park et al., 2020 [54] | Yonsei Univ. HealthSystem. | PD (n = 77) vs. Normal (n = 53) | SVM, Vol + ReHo + DegCen | 75.3 |
| Pereira et al., 2020 [26] | PPMI. | (a) PD (n = 378) vs. SWEDD (n = 58) (b) PD (n = 378) vs. Normal (n = 168) | CNNs | (a) 73.3 (b) 97.4 |
| Solana et al., 2021 [55] | PPMI | PD (n = 348) vs. Normal (n = 159) | Seven classifiers Haralick and Wrapper FS | 93 |
| Talai et al., 2021 [56] | U.Medical Center Hamburg. | PD (n = 45) vs. PSP-RS (n = 20) vs. Normal (n = 159) | Regional morphology features quantitative T2 values diffusion-tensor | 95.1 |
| Proposed method Solution 1. C1 matter Figure 13c | PPMI. | PD (n = 450) vs. Normal (n = 258) vs. SWEDD (n = 154) vs. Prodromal (n = 38) vs. GeneCohort (n = 17) | 3D-DWT, MOE, SVM | 96.2 |
| Proposed method Solution 3. C1 matter Figure 13d | PPMI. | PD (n = 450) vs. Normal (n = 258) vs. vs. SWEDD (n = 154) vs. Prodromal (n = 38) vs. GeneCohort (n = 17) | 3D-DWT, MOE, SVM | 95.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojas-Valenzuela, I.; Valenzuela, O.; Delgado-Marquez, E.; Rojas, F. Multi-Class Classifier in Parkinson’s Disease Using an Evolutionary Multi-Objective Optimization Algorithm. Appl. Sci. 2022, 12, 3048. https://doi.org/10.3390/app12063048
Rojas-Valenzuela I, Valenzuela O, Delgado-Marquez E, Rojas F. Multi-Class Classifier in Parkinson’s Disease Using an Evolutionary Multi-Objective Optimization Algorithm. Applied Sciences. 2022; 12(6):3048. https://doi.org/10.3390/app12063048
Chicago/Turabian StyleRojas-Valenzuela, Ignacio, Olga Valenzuela, Elvira Delgado-Marquez, and Fernando Rojas. 2022. "Multi-Class Classifier in Parkinson’s Disease Using an Evolutionary Multi-Objective Optimization Algorithm" Applied Sciences 12, no. 6: 3048. https://doi.org/10.3390/app12063048
APA StyleRojas-Valenzuela, I., Valenzuela, O., Delgado-Marquez, E., & Rojas, F. (2022). Multi-Class Classifier in Parkinson’s Disease Using an Evolutionary Multi-Objective Optimization Algorithm. Applied Sciences, 12(6), 3048. https://doi.org/10.3390/app12063048






