Cardiac Magnetic Resonance Left Ventricle Segmentation and Function Evaluation Using a Trained Deep-Learning Model
Abstract
:1. Introduction
2. Methods
2.1. Cardiac MRI Datasets
2.2. Algorithm Workflow
- A naive method (Naive): The trained U-net was used to segment the 90 ACDC test subjects directly.
- A combined method (Combined) that integrated MCD, spatial augmentation, and style-intensity augmentation method. We explored the effects of MCD, spatial augmentation, and advanced style-intensity augmentation for U-net training; the optimal combination of the three components constitutes the combined method. A recent study [17] proposed style-intensity augmentation during network training to tackle the domain shift issue and demonstrated state-of-the-art performance in breast segmentation in MRI datasets from a different domain. Style-intensity augmentation comprises style transfer and intensity remapping, which produce non-realistic looking MR scans while preserving the image shapes. The style transfer procedure uses features extracted from style images to augment the training images, randomizing the color, texture and contrast but preserving the geometry [29]. The intensity remapping technique generates a random mapping function to map the original image signal intensities to new values. This method is based on the assumption that by considerably changing the appearance of training images, the network will focus on non-domain specific features, e.g., the geometric shape of breast that is preserved in different breast MR datasets [17]. The optimized combined method was applied to the ACDC test dataset.
- DeepLab: DeepLabV3+ [30], a top performing neural network in several medical image segmentation challenges, was trained on the LVSC dataset and tested on the ACDC test dataset.
2.3. Evaluation Methods
2.4. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| DSC ([0, 1]) | ASSD (mm) | |||||
|---|---|---|---|---|---|---|
| MCD | Spa. Aug. | Sty.-Int. Aug. | LVM | LVC | LVM | LVC |
| ine ✗ | ✗ | ✗ | 0.33 ± 0.22 | 0.46 ± 0.30 | 16.85 ± 21.52 | 15.28 ± 18.02 |
| ✗ | ✗ | ✓ | 0.49 ± 0.18 | 0.68 ± 0.22 | 8.38 ± 7.33 | 8.55 ± 8.69 |
| ✗ | ✓ | ✗ | 0.73 ± 0.12 | 0.85 ± 0.14 | 2.92 ± 2.86 | 3.34 ± 3.48 |
| ✗ | ✓ | ✓ | 0.77 ± 0.07 | 0.87 ± 0.11 | 2.39 ± 2.05 | 2.80 ± 2.49 |
| ✓ | ✗ | ✗ | 0.34 ± 0.21 | 0.49 ± 0.29 | 11.30 ± 16.93 | 9.97 ± 15.36 |
| ✓ | ✗ | ✓ | 0.55 ± 0.17 | 0.71 ± 0.21 | 7.47 ± 7.00 | 7.37 ± 8.22 |
| ✓ | ✓ | ✗ | 0.75 ± 0.11 | 0.87 ± 0.12 | 2.30 ± 1.64 | 2.39 ± 1.85 |
| ✓ | ✓ | ✓ | 0.78 ± 0.08 | 0.87 ± 0.12 | 2.71 ± 2.50 | 2.87 ± 2.61 |
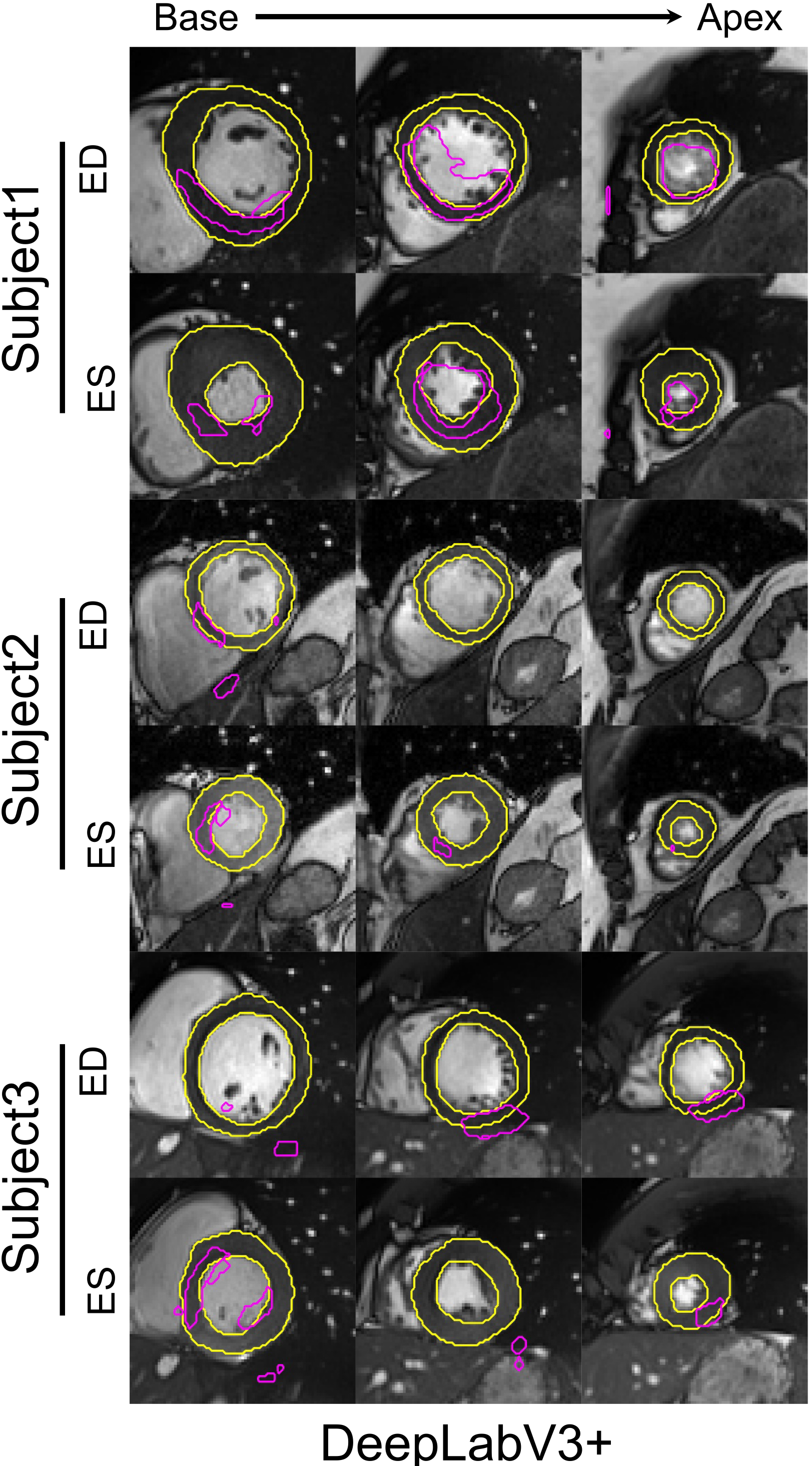
References
- Flachskampf, F.A.; Biering-Sørensen, T.; Solomon, S.D.; Duvernoy, O.; Bjerner, T.; Smiseth, O.A. Cardiac imaging to evaluate left ventricular diastolic function. JACC Cardiovasc. Imaging 2015, 8, 1071–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Members, W.C.; Hundley, W.G.; Bluemke, D.A.; Finn, J.P.; Flamm, S.D.; Fogel, M.A.; Friedrich, M.G.; Ho, V.B.; Jerosch-Herold, M.; Kramer, C.M.; et al. ACCF/ACR/AHA/NASCI/SCMR 2010 expert consensus document on cardiovascular magnetic resonance: A report of the American College of Cardiology Foundation Task Force on Expert Consensus Documents. Circulation 2010, 121, 2462–2508. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Ng, M.; Goubran, M.; Petersen, S.E.; Piechnik, S.K.; Neubauer, S.; Wright, G. Improving cardiac MRI convolutional neural network segmentation on small training datasets and dataset shift: A continuous kernel cut approach. Med. Image Anal. 2020, 61, 101636. [Google Scholar] [CrossRef] [PubMed]
- Petitjean, C.; Dacher, J.N. A review of segmentation methods in short axis cardiac MR images. Med. Image Anal. 2011, 15, 169–184. [Google Scholar] [CrossRef] [Green Version]
- Peng, P.; Lekadir, K.; Gooya, A.; Shao, L.; Petersen, S.E.; Frangi, A.F. A review of heart chamber segmentation for structural and functional analysis using cardiac magnetic resonance imaging. Magn. Reson. Mater. Phys. Biol. Med. 2016, 29, 155–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef] [Green Version]
- Leiner, T.; Rueckert, D.; Suinesiaputra, A.; Baeßler, B.; Nezafat, R.; Išgum, I.; Young, A.A. Machine learning in cardiovascular magnetic resonance: Basic concepts and applications. J. Cardiovasc. Magn. Reson. 2019, 21, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Bai, W.; Sinclair, M.; Tarroni, G.; Oktay, O.; Rajchl, M.; Vaillant, G.; Lee, A.M.; Aung, N.; Lukaschuk, E.; Sanghvi, M.M.; et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J. Cardiovasc. Magn. Reson. 2018, 20, 65. [Google Scholar] [CrossRef] [Green Version]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G.; et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Chen, C.; Qin, C.; Qiu, H.; Tarroni, G.; Duan, J.; Bai, W.; Rueckert, D. Deep Learning for Cardiac Image Segmentation: A Review. Front. Cardiovasc. Med. 2020, 7, 25. [Google Scholar] [CrossRef]
- Yan, W.; Wang, Y.; Gu, S.; Huang, L.; Yan, F.; Xia, L.; Tao, Q. The Domain Shift Problem of Medical Image Segmentation and Vendor-Adaptation by Unet-GAN. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2019; pp. 623–631. [Google Scholar]
- Tzeng, E.; Hoffman, J.; Saenko, K.; Darrell, T. Adversarial discriminative domain adaptation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 7167–7176. [Google Scholar]
- Zhu, Y.; Fahmy, A.S.; Duan, C.; Nakamori, S.; Nezafat, R. Automated Myocardial T2 and Extracellular Volume Quantification in Cardiac MRI Using Transfer Learning—Based Myocardium Segmentation. Radiol. Artif. Intell. 2020, 2, e190034. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.; Xu, Z.; Moon, H.; Bao, S.; Assad, A.; Moyo, T.K.; Savona, M.R.; Abramson, R.G.; Landman, B.A. Synseg-net: Synthetic segmentation without target modality ground truth. IEEE Trans. Med. Imaging 2018, 38, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.A. Unsupervised bidirectional cross-modality adaptation via deeply synergistic image and feature alignment for medical image segmentation. IEEE Trans. Med. Imaging 2020, 39, 2494–2505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Hesse, L.S.; Kuling, G.; Veta, M.; Martel, A. Intensity augmentation to improve generalizability of breast segmentation across different MRI scan protocols. IEEE Trans. Biomed. Eng. 2020, 68, 759–770. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, X.; Yang, D.; Sanford, T.; Harmon, S.; Turkbey, B.; Wood, B.J.; Roth, H.; Myronenko, A.; Xu, D.; et al. Generalizing deep learning for medical image segmentation to unseen domains via deep stacked transformation. IEEE Trans. Med. Imaging 2020, 39, 2531–2540. [Google Scholar] [CrossRef]
- Guo, F.; Ng, M.; Roifman, I.; Wright, G. Cardiac MRI Left Ventricular Segmentation and Function Quantification Using Pre-trained Neural Networks. In International Conference on Functional Imaging and Modeling of the Heart; Springer: Cham, Switzerland, 2021; pp. 46–54. [Google Scholar]
- Radau, P.; Lu, Y.; Connelly, K.; Paul, G.; Dick, A.; Wright, G. Evaluation framework for algorithms segmenting short axis cardiac MRI. MIDAS J.-Card. MR Left Ventricle Segmentation Chall. 2009, 49. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; pp. 234–241. [Google Scholar]
- Gal, Y.; Ghahramani, Z. Dropout as a bayesian approximation: Representing model uncertainty in deep learning. In Proceedings of the 33rd International Conference on Machine Learning, New York, NY, USA, 20–22 June 2016; pp. 1050–1059. [Google Scholar]
- Guo, F.; Capaldi, D.P.; McCormack, D.G.; Fenster, A.; Parraga, G. Ultra-short Echo-time Magnetic Resonance Imaging Lung Segmentation with Under-Annotations and Domain Shift. Med. Image Anal. 2021, 72, 102107. [Google Scholar] [CrossRef]
- Guo, F.; Ng, M.; Wright, G. Cardiac cine MRI left ventricle segmentation combining deep learning and graphical models. In Medical Imaging 2020: Image Processing; International Society for Optics and Photonics: Bellingham, WA, USA, 2020; Volume 11313. [Google Scholar]
- Guo, F.; Krahn, P.R.; Escartin, T.; Roifman, I.; Wright, G. Cine and late gadolinium enhancement MRI registration and automated myocardial infarct heterogeneity quantification. Magn. Reson. Med. 2021, 85, 2842–2855. [Google Scholar] [CrossRef]
- Tang, M.; Ben Ayed, I.; Marin, D.; Boykov, Y. Secrets of grabcut and kernel k-means. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015; pp. 1555–1563. [Google Scholar]
- Yuan, J.; Bae, E.; Tai, X.C. A study on continuous max-flow and min-cut approaches. In Proceedings of the 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Francisco, CA, USA, 13–18 June 2010; pp. 2217–2224. [Google Scholar]
- Guo, F.; Yuan, J.; Rajchl, M.; Svenningsen, S.; Capaldi, D.P.; Sheikh, K.; Fenster, A.; Parraga, G. Globally optimal co-segmentation of three-dimensional pulmonary 1H and hyperpolarized 3He MRI with spatial consistence prior. Med. Image Anal. 2015, 23, 43–55. [Google Scholar] [CrossRef]
- Jackson, P.T.; Abarghouei, A.A.; Bonner, S.; Breckon, T.P.; Obara, B. Style augmentation: Data augmentation via style randomization. In Proceedings of the CVPR Workshops, Long Beach, CA, USA, 16–21 June 2019; pp. 83–92. [Google Scholar]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European conference on computer vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Guo, F.; Ng, M.; Wright, G. Cardiac MRI left ventricle segmentation and quantification: A framework combining U-Net and continuous max-flow. In International Workshop on Statistical Atlases and Computational Models of the Heart; Springer: Cham, Switzerland, 2018; pp. 450–458. [Google Scholar]
- Nai, Y.H.; Teo, B.W.; Tan, N.L.; O’Doherty, S.; Stephenson, M.C.; Thian, Y.L.; Chiong, E.; Reilhac, A. Comparison of metrics for the evaluation of medical segmentations using prostate MRI dataset. Comput. Biol. Med. 2021, 134, 104497. [Google Scholar] [CrossRef]
- Maier-Hein, L.; Eisenmann, M.; Reinke, A.; Onogur, S.; Stankovic, M.; Scholz, P.; Arbel, T.; Bogunovic, H.; Bradley, A.P.; Carass, A.; et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grothues, F.; Smith, G.C.; Moon, J.C.; Bellenger, N.G.; Collins, P.; Klein, H.U.; Pennell, D.J. Comparison of interstudy reproducibility of cardiovascular magnetic resonance with two-dimensional echocardiography in normal subjects and in patients with heart failure or left ventricular hypertrophy. Am. J. Cardiol. 2002, 90, 29–34. [Google Scholar] [CrossRef]
- Kirby, M.; Svenningsen, S.; Owrangi, A.; Wheatley, A.; Farag, A.; Ouriadov, A.; Santyr, G.E.; Etemad-Rezai, R.; Coxson, H.O.; McCormack, D.G.; et al. Hyperpolarized 3He and 129Xe MR imaging in healthy volunteers and patients with chronic obstructive pulmonary disease. Radiology 2012, 265, 600–610. [Google Scholar] [CrossRef]
- Damen, F.W.; Newton, D.T.; Lin, G.; Goergen, C.J. Machine Learning Driven Contouring of High-Frequency Four-Dimensional Cardiac Ultrasound Data. Appl. Sci. 2021, 11, 1690. [Google Scholar] [CrossRef]
- Lee, H.; Yoon, T.; Yeo, C.; Oh, H.; Ji, Y.; Sim, S.; Kang, D. Cardiac Arrhythmia Classification Based on One-Dimensional Morphological Features. Appl. Sci. 2021, 11, 9460. [Google Scholar] [CrossRef]
- Komatsu, M.; Sakai, A.; Komatsu, R.; Matsuoka, R.; Yasutomi, S.; Shozu, K.; Dozen, A.; Machino, H.; Hidaka, H.; Arakaki, T.; et al. Detection of cardiac structural abnormalities in fetal ultrasound videos using deep learning. Appl. Sci. 2021, 11, 371. [Google Scholar] [CrossRef]
- Tao, Q.; Yan, W.; Wang, Y.; Paiman, E.H.; Shamonin, D.P.; Garg, P.; Plein, S.; Huang, L.; Xia, L.; Sramko, M.; et al. Deep learning–based method for fully automatic quantification of left ventricle function from cine MR images: A multivendor, multicenter study. Radiology 2019, 290, 81–88. [Google Scholar] [CrossRef] [Green Version]
- Wong, K.C.; Moradi, M.; Tang, H.; Syeda-Mahmood, T. 3D segmentation with exponential logarithmic loss for highly unbalanced object sizes. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2018; pp. 612–619. [Google Scholar]
- Wang, Y.; Zhang, Y.; Xuan, W.; Kao, E.; Cao, P.; Tian, B.; Ordovas, K.; Saloner, D.; Liu, J. Fully automatic segmentation of 4D MRI for cardiac functional measurements. Med. Phys. 2019, 46, 180–189. [Google Scholar] [CrossRef] [Green Version]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef] [Green Version]

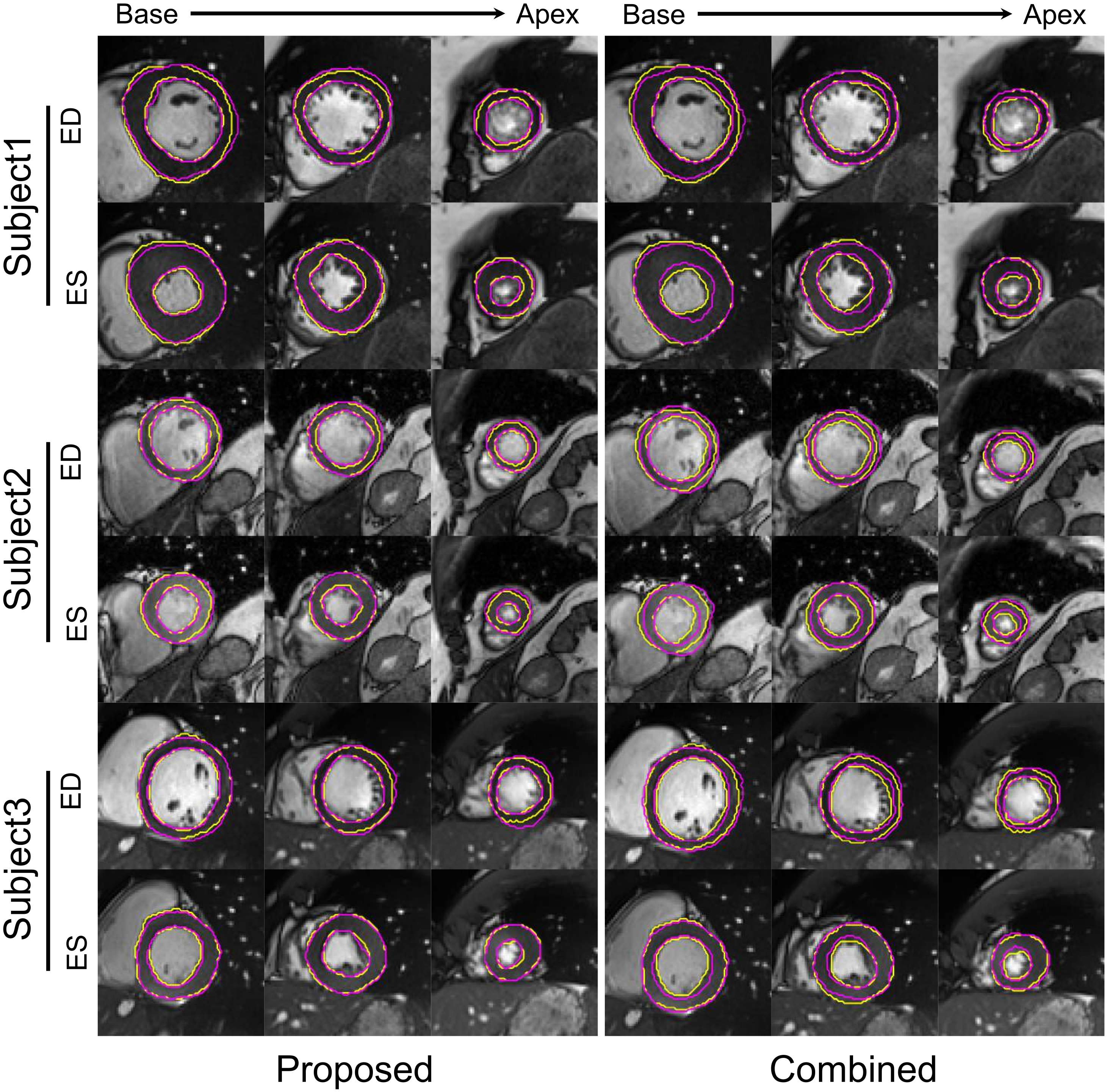
| DSC ([0, 1]) | ASSD (mm) | |||
|---|---|---|---|---|
| Methods | LVM | LVV | LVM | LVC |
| Proposed | 0.81 ± 0.09 | 0.90 ± 0.09 | 2.04 ± 1.77 | 1.82 ± 2.18 |
| Naive | 0.74 ± 0.12 * | 0.87 ± 0.12 * | 2.43 ± 2.16 * | 2.40 ± 2.58 * |
| Combined | 0.78 ± 0.08 * | 0.87 ± 0.12 * | 2.71 ± 2.50 * | 2.87 ± 2.61 * |
| DeepLab | 0.26 ± 0.18 * | 0.32 ± 0.27 * | 18.60 ± 17.48 * | 17.33 ± 12.37 * |
| Manual | Proposed | Naive | Combined | DeepLab | |
|---|---|---|---|---|---|
| LVMM (g) ¥ | 138.1 ± 54.3 | 129.3 ± 49.8 | 110.8 ± 48.2 | 154.4 ± 83.6 | 46.4 ± 34.7 |
| LVEDV (mL) | 163.8 ± 75.2 | 162.9 ± 72.0 | 174.6 ± 74.5 | 175.8 ± 72.8 | 71.8 ± 69.3 |
| LVESV (mL) | 99.4 ± 80.4 | 99.2 ± 76.7 | 108.2 ± 80.0 | 118.4 ± 76.2 | 58.3 ± 62.1 |
| LVSV (mL) | 64.4 ± 24.6 | 63.7 ± 25.8 | 66.5 ± 31.5 | 57.4 ± 25.9 | 13.5 ± 24.2 |
| LVEF (%) | 46.2 ± 20.4 | 45.5 ± 20.5 | 43.0 ± 23.6 | 36.7 ± 18.4 | −4.4 ± 142.2 |
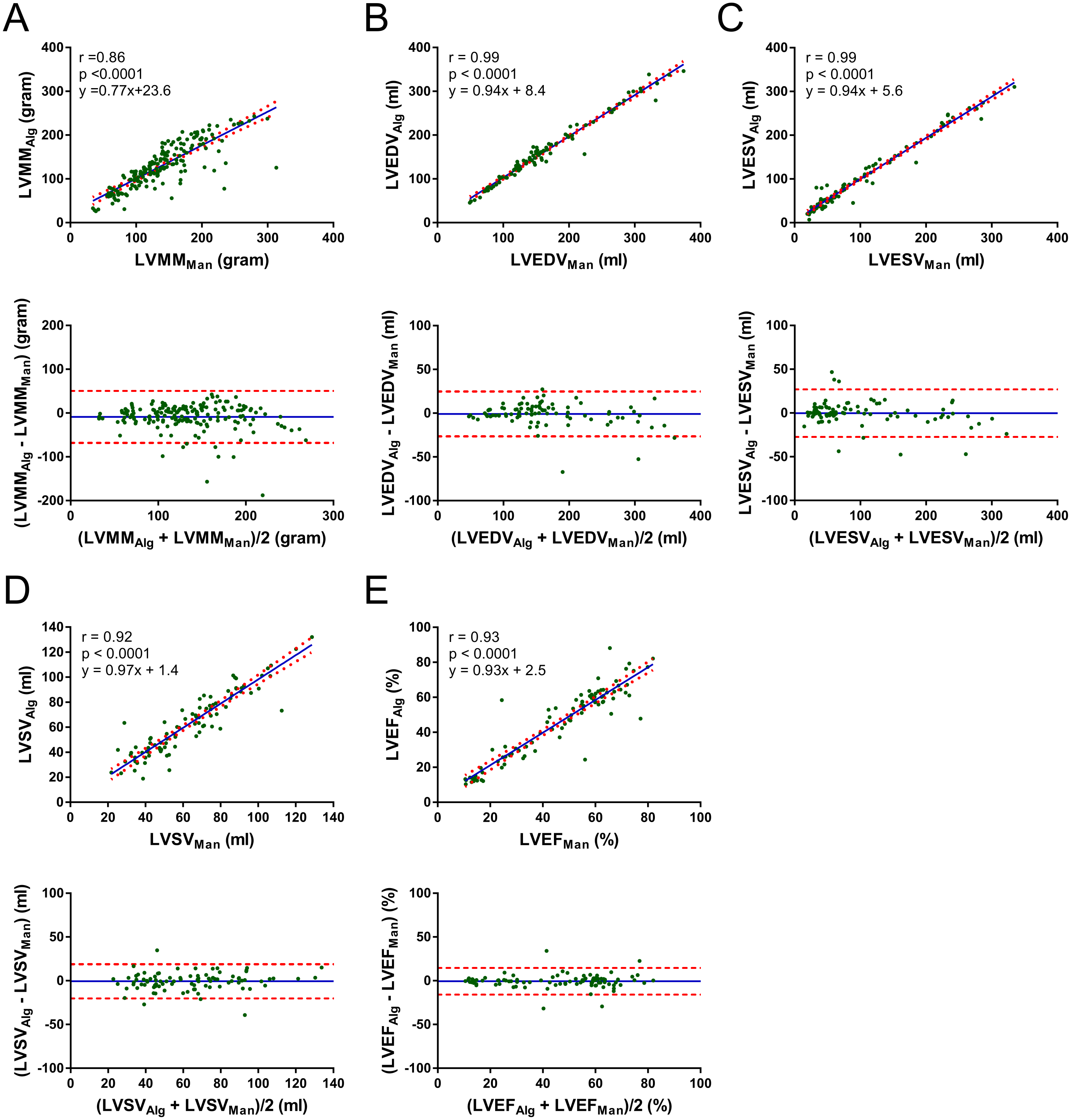
| Pearson (r, 95% CI) | Proposed vs. Manual | Naive vs. Manual | Combined. vs. Manual | DeepLab vs. Manual |
|---|---|---|---|---|
| ine LVMM (g) ¥ | 0.86 ([0.80, 0.90]) | 0.79 ([0.73, 0.84]) | 0.41 ([0.28, 0.52]) | 0.47 ([0.35, 0.58]) |
| LVEDV (mL) | 0.99 ([0.98, 0.99]) | 0.98 ([0.97, 0.99]) | 0.99 ([0.99, 0.99]) | 0.57 ([0.41, 0.69]) |
| LVESV (mL) | 0.99 ([0.98, 0.99]) | 0.98 ([0.97, 0.99]) | 0.97 ([0.96, 0.98]) | 0.65 ([0.51, 0.75]) |
| LVSV (mL) | 0.92 ([0.88, 0.95]) | 0.84 ([0.76, 0.89]) | 0.83 ([0.75, 0.89]) | 0.13 ([−0.08, 0.33]) |
| LVEF (%) | 0.93 ([0.89, 0.95]) | 0.75 ([0.65, 0.83]) | 0.76 ([0.65, 0.83]) | 0.08 ([−0.13, 0.28]) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, F.; Ng, M.; Roifman, I.; Wright, G. Cardiac Magnetic Resonance Left Ventricle Segmentation and Function Evaluation Using a Trained Deep-Learning Model. Appl. Sci. 2022, 12, 2627. https://doi.org/10.3390/app12052627
Guo F, Ng M, Roifman I, Wright G. Cardiac Magnetic Resonance Left Ventricle Segmentation and Function Evaluation Using a Trained Deep-Learning Model. Applied Sciences. 2022; 12(5):2627. https://doi.org/10.3390/app12052627
Chicago/Turabian StyleGuo, Fumin, Matthew Ng, Idan Roifman, and Graham Wright. 2022. "Cardiac Magnetic Resonance Left Ventricle Segmentation and Function Evaluation Using a Trained Deep-Learning Model" Applied Sciences 12, no. 5: 2627. https://doi.org/10.3390/app12052627
APA StyleGuo, F., Ng, M., Roifman, I., & Wright, G. (2022). Cardiac Magnetic Resonance Left Ventricle Segmentation and Function Evaluation Using a Trained Deep-Learning Model. Applied Sciences, 12(5), 2627. https://doi.org/10.3390/app12052627






