Genomic Insights into Two Endophytic Strains: Stenotrophomonas geniculata NWUBe21 and Pseudomonas carnis NWUBe30 from Cowpea with Plant Growth-Stimulating Attributes
Abstract
1. Introduction
2. Materials and Methods
- The isolation, morphogenomic authentication, and plant growth-stimulating screening of cowpea endophytic bacterial isolates.
- Isolation protocol.
- In vitro PGP screening assay.
- Genomic DNA extraction, whole-genome sequencing, and annotation.
3. Results
3.1. Genomic Features of the Endophytic Pseudomonas carnis Strain NWUBe30 and Stenotrophomonas geniculata Strain NWUBe21
3.2. Abundance of Plant Growth-Improving Genes in the Genomes of the Endophytic Bacteria Strains
4. Discussion
- Authentication of the endophytic bacteria strains as plant growth enhancers.
- Genetic components that are involved in plant growth enhancement.
- Iron acquisition and metabolism.
- Motility and Chemotaxis
- Nitrogen Metabolism
- Phosphorus Metabolism.
- Plant hormone-auxin biosynthesis.
- Stress Response.
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumawat, K.C.; Razdan, N.; Saharan, K. Rhizospheric microbiome: Bio-based emerging strategies for sustainable agriculture development and future perspectives. Microbiol. Res. 2022, 254, 126901. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.; Pieterse, C.M. Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Morella, N.M.; Weng, F.C.-H.; Joubert, P.M.; Metcalf, C.J.E.; Lindow, S.; Koskella, B. Successive passaging of a plant-associated microbiome reveals robust habitat and host genotype-dependent selection. Proc. Natl. Acad. Sci. USA 2020, 117, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Kumawat, K.C.; Sharma, P.; Nagpal, S.; Gupta, R.; Sirari, A.; Nair, R.M.; Bindumadhava, H.; Singh, S. Dual microbial inoculation, a game changer?—Bacterial biostimulants with multifunctional growth promoting traits to mitigate salinity stress in Spring Mungbean. Front. Microbiol. 2021, 11, 3491. [Google Scholar] [CrossRef]
- Barea, J. Future challenges and perspectives for applying microbial biotechnology in sustainable agriculture based on a better understanding of plant-microbiome interactions. J. Soil Sci. Plant Nutr. 2015, 15, 261–282. [Google Scholar] [CrossRef]
- Omomowo, O.I.; Babalola, O.O. Constraints and prospects of improving cowpea productivity to ensure food, nutritional security and environmental sustainability. Front. Plant Sci. 2021, 12, 751731. [Google Scholar] [CrossRef]
- Ayalew, T.; Samago, T.; Petra, H.; Cadisch, G. Yield response of field-grown cowpea (Vigna unguiculata (L.) Walp.) varieties to Bradyrhizobium inoculation. Agron. J. 2021, 113, 3258–3268. [Google Scholar] [CrossRef]
- Cardona-Ayala, C.; Cardona-Villadiego, C.; Peñate-Pacheco, C.; Araméndiz-Tatis, H.; Espitia-Camacho, M.M. Growth, biomass distribution, gas exchange and chlorophyll fluorescence in cowpea (Vigna unguiculata (L.) Walp.) under drought conditions. Austr. J. Crop Sci. 2020, 14, 371–381. [Google Scholar] [CrossRef]
- Omomowo, O.I.; Babalola, O.O. Bacterial and fungal endophytes: Tiny giants with immense beneficial potential for plant growth and sustainable agricultural productivity. Microorganisms 2019, 7, 481. [Google Scholar] [CrossRef]
- Compant, S.; Cambon, M.C.; Vacher, C.; Mitter, B.; Samad, A.; Sessitsch, A. The plant endosphere world–bacterial life within plants. Environ. Microbiol. 2021, 23, 1812–1829. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, B.S.; Ayangbenro, A.; Babalola, O.O. Effect of endophytic bacterium, Stenotrophomonas maltophilia JVB5 on sunflowers. Plant Prot. Sci. 2022, 58, 185–198. [Google Scholar] [CrossRef]
- Kaewkla, O.; Sukpanoa, S.; Suriyachadkun, C.; Chamroensaksi, N.; Chumroenphat, T.; Franco, C.M.M. Streptomyces spinosus sp. nov. and Streptomyces shenzhenensis subsp. oryzicola subsp. nov. endophytic actinobacteria isolated from Jasmine rice and their genome mining for potential as antibiotic producers and plant growth promoters. Antonie Van Leeuwenhoek 2022, 115, 871–888. [Google Scholar] [CrossRef] [PubMed]
- Del Carmen Orozco-Mosqueda, M.; Santoyo, G. Plant-microbial endophytes interactions: Scrutinizing their beneficial mechanisms from genomic explorations. Curr. Plant Biol. 2021, 25, 100189. [Google Scholar] [CrossRef]
- Ahmad, T.; Bashir, A.; Farooq, S.; Riyaz-Ul-Hassan, S. Burkholderia gladioli E39CS3, an endophyte of Crocus sativus Linn., induces host resistance against corm-rot caused by Fusarium oxysporum. J. Appl. Microbiol. 2022, 132, 495–508. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Yang, C.; Wang, Y.; Ma, T.; Cai, F.; Wei, L.; Jin, M.; Osei, R.; Zhang, J.; Tang, M. Potential of an endophytic bacteria Bacillus amyloliquefaciens 3–5 as biocontrol agent against potato scab. Microb. Pathog. 2022, 163, 105382. [Google Scholar] [CrossRef]
- Kaewkla, O.; Franco, C.M. Rational approaches to improving the isolation of endophytic actinobacteria from Australian native trees. Microb. Ecol. 2013, 65, 384–393. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef]
- Matsuda, R.; Handayani, M.L.; Sasaki, H.; Takechi, K.; Takano, H.; Takio, S. Production of indoleacetic acid by strains of the epiphytic bacteria Neptunomonas spp. isolated from the red alga Pyropia yezoensis and the seagrass Zostera marina. Arch. Microbiol. 2018, 200, 255–265. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Glick, B.R.; Karaturovíc, D.M.; Newell, P.C. A novel procedure for rapid isolation of plant growth promoting pseudomonads. Can. J. Microbiol. 1995, 41, 533–536. [Google Scholar] [CrossRef]
- Cappucino, J.; Sherman, N. Nitrogen Cycle. Microbiology: A Laboratory Manual; Benjamin/Cumming Pub. Co.: New York, NY, USA, 1992. [Google Scholar]
- Khan, N.; Bano, A. Exopolysaccharide producing rhizobacteria and their impact on growth and drought tolerance of wheat grown under rainfed conditions. PLoS ONE 2019, 14, e0222302. [Google Scholar] [CrossRef] [PubMed]
- Dinesh, R.; Anandaraj, M.; Kumar, A.; Bini, Y.K.; Subila, K.P.; Aravind, R. Isolation, characterization, and evaluation of multi-trait plant growth promoting rhizobacteria for their growth promoting and disease suppressing effects on ginger. Microbiol. Res. 2015, 173, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yang, F.; Zhang, L.; Wang, J. Organic acid secretion and phosphate solubilizing efficiency of Pseudomonas sp. PSB12: Effects of phosphorus forms and carbon sources. Geomicrobiol. J. 2016, 33, 870–877. [Google Scholar] [CrossRef]
- Fukuda, K.; Ogawa, M.; Taniguchi, H.; Saito, M. Molecular approaches to studying microbial communities: Targeting the 16S ribosomal RNA gene. J. UOEH 2016, 38, 223–232. [Google Scholar] [CrossRef]
- Raddadi, N.; Cherif, A.; Boudabous, A.; Daffonchio, D. Screening of plant growth promoting traits of Bacillus thuringiensis. Ann. Microbiol. 2008, 58, 47–52. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Cottingham, R. The DOE systems biology knowledgebase (KBase). In Proceedings of the 5th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics, Newport Beach, CA, USA, 20–23 September 2014. [Google Scholar]
- Arkin, A.P.; Stevens, R.L.; Cottingham, R.W.; Maslov, S.; Henry, C.S.; Dehal, P.; Ware, D.; Perez, F.; Harris, N.L.; Canon, S. The DOE systems biology knowledgebase (KBase). bioRxiv 2016, 096354. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Quintas-Nunes, F.; Rossi, M.J.; Nascimento, F.X. Genomic insights into the plant-associated lifestyle of Kosakonia radicincitans MUSA4, a diazotrophic plant-growth-promoting bacterium. Syst. Appl. Microbiol. 2022, 45, 126303. [Google Scholar] [CrossRef]
- Di, Y.N.; Kui, L.; Singh, P.; Liu, L.F.; Xie, L.Y.; He, L.L.; Li, F.S. Identification and characterization of Bacillus subtilis B9: A diazotrophic plant growth-promoting endophytic bacterium isolated from sugarcane root. J. Plant Growth Regul. 2022, 1–18. [Google Scholar] [CrossRef]
- Tran, T.; French, E.; Iyer-Pascuzzi, A.S. In vitro functional characterization predicts the impact of bacterial root endophytes on plant growth. J. Exp. Bot. 2022. [Google Scholar] [CrossRef]
- Wang, S.; Wang, J.; Zhou, Y.; Huang, Y.; Tang, X. Prospecting the plant growth–promoting activities of endophytic bacteria Cronobacter sp. YSD YN2 isolated from Cyperus esculentus L. var. sativus leaves. Ann. Microbiol. 2022, 72, 1. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Babalola, O.O. Elucidating mechanisms of endophytes used in plant protection and other bioactivities with multifunctional prospects. Front. Bioeng. Biotechnol. 2020, 8, 467. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Ayangbenro, A.S.; Glick, B.R.; Babalola, O.O. Plant health: Feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 2019, 103, 1155–1166. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, H.; Sun, X.; Liu, Y.; Yan, W.; Xun, W.; Shen, Q.; Zhang, R. Bacillus velezensis wall teichoic acids are required for biofilm formation and root colonization. Appl. Environ. Microbiol. 2019, 85, e02116–e02118. [Google Scholar] [CrossRef] [PubMed]
- Alori, E.T.; Babalola, O.O. Microbial inoculants for improve crop quality and human health. Front. Microbiol. 2018, 9, 2213. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.; Qin, Y.; Wu, H.; Zuo, W.; He, H.; Tan, J.; Wang, Y.; He, D. Isolation and characterization of phosphorus solubilizing bacteria with multiple phosphorus sources utilizing capability and their potential for lead immobilization in soil. Front. Microbiol. 2020, 11, 752. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, R.; Waqas, M.; Khan, A.L.; Al-Hosni, K.; Kang, S.-M.; Seo, C.-W.; Lee, I.-J. Indoleacetic acid production and plant growth promoting potential of bacterial endophytes isolated from rice (Oryza sativa L.) seeds. Acta Biol. Hung. 2017, 68, 175–186. [Google Scholar] [CrossRef]
- Woo, O.-G.; Kim, H.; Kim, J.-S.; Keum, H.L.; Lee, K.-C.; Sul, W.J.; Lee, J.-H. Bacillus subtilis strain GOT9 confers enhanced tolerance to drought and salt stresses in Arabidopsis thaliana and Brassica campestris. Plant Physiol. Biochem 2020, 148, 359–367. [Google Scholar] [CrossRef]
- Hantke, I.; Schäfer, H.; Janczikowski, A.; Turgay, K. YocM a small heat shock protein can protect Bacillus subtilis cells during salt stress. Mol. Microbiol 2019, 111, 423–440. [Google Scholar] [CrossRef]
- Ribeiro, V.P.; Gomes, E.A.; de Sousa, S.M.; de Paula Lana, U.G.; Coelho, A.M.; Marriel, I.E.; de Oliveira-Paiva, C.A. Co-inoculation with tropical strains of Azospirillum and Bacillus is more efficient than single inoculation for improving plant growth and nutrient uptake in maize. Arch. Microbiol. 2022, 204, 1–13. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Ayangbenro, A.S.; Babalola, O.O. Genomic analysis of endophytic Bacillus cereus T4S and its plant growth-promoting traits. Plants 2021, 10, 1776. [Google Scholar] [CrossRef]
- Santoyo, G.; Guzmán-Guzmán, P.; Parra-Cota, F.I.; Santos-Villalobos, S.d.l.; Orozco-Mosqueda, M.; Glick, B.R. Plant growth stimulation by microbial consortia. Agronomy 2021, 11, 219. [Google Scholar] [CrossRef]
- Jayakumar, A.; Nair, I.C.; Radhakrishnan, E. Environmental adaptations of an extremely plant beneficial Bacillus subtilis Dcl1 identified through the genomic and metabolomic analysis. Microb. Ecol. 2021, 81, 687–702. [Google Scholar] [CrossRef]
- Mukherjee, A.; Gaurav, A.K.; Patel, A.K.; Singh, S.; Chouhan, G.K.; Lepcha, A.; Pereira, A.P.d.A.; Verma, J.P. Unlocking the potential plant growth-promoting properties of chickpea (Cicer arietinum L.) seed endophytes bio-inoculants for improving soil health and crop production. Land Degrad. Dev. 2021, 32, 4362–4374. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Ayilara, M.S.; Ayangbenro, A.S.; Babalola, O.O. Genome mining of three plant growth-promoting Bacillus species from maize rhizosphere. Appl. Biochem. Biotechnol. 2021, 193, 3949–3969. [Google Scholar] [CrossRef] [PubMed]
- Belaouni, H.A.; Compant, S.; Antonielli, L.; Nikolic, B.; Zitouni, A.; Sessitsch, A. In-depth genome analysis of Bacillus sp. BH32, a salt stress-tolerant endophyte obtained from a halophyte in a semiarid region. Appl. Microbiol. Biotechnol. 2022, 106, 3113–3137. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.-L.; Wang, W.-Y.; Zuo, S.-H.; Wu, X.-Q. Genome sequencing of Rahnella victoriana JZ-GX1 provides new insights into molecular and genetic mechanisms of plant growth promotion. Front. Microbiol. 2022, 13, 828990. [Google Scholar] [CrossRef]
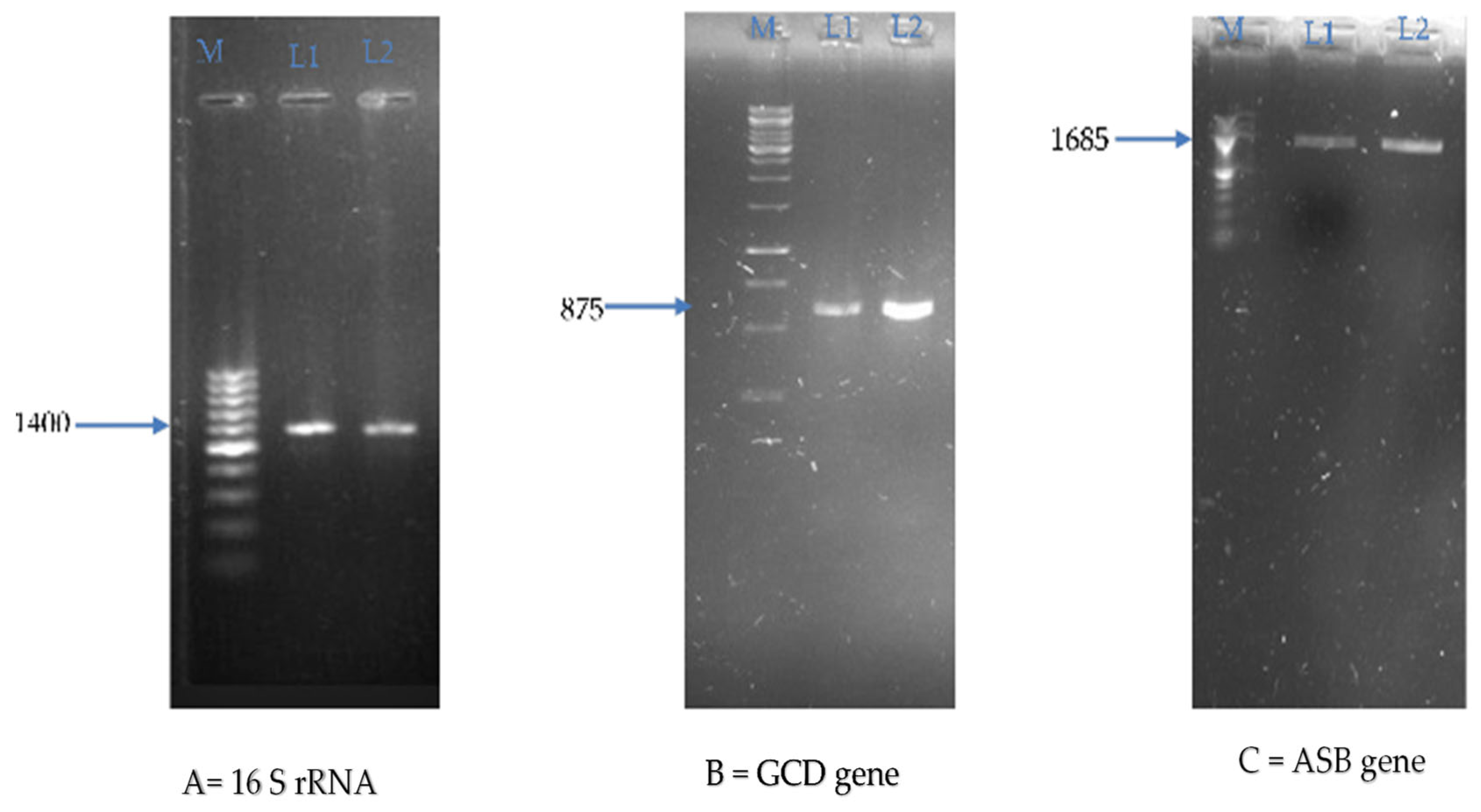

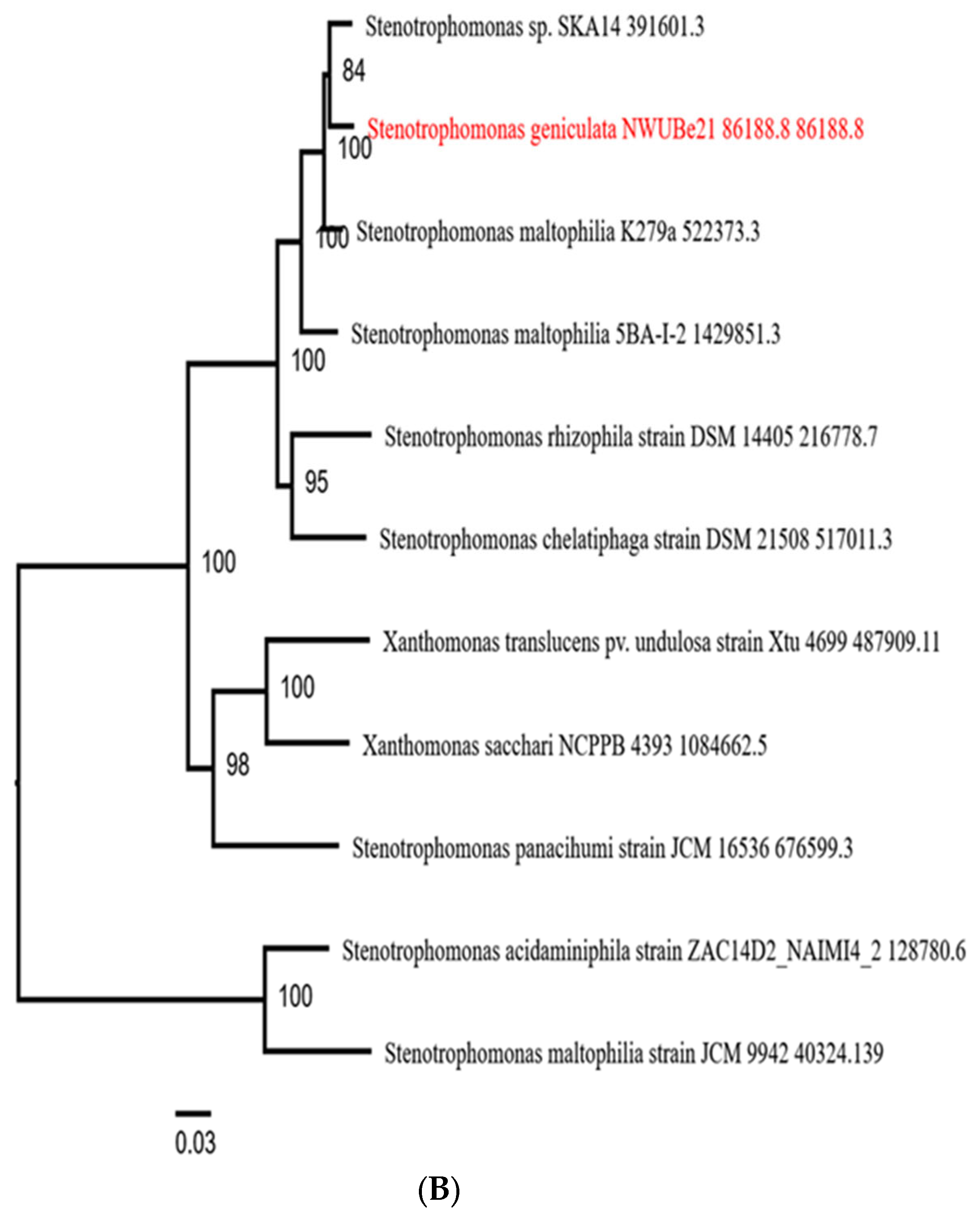
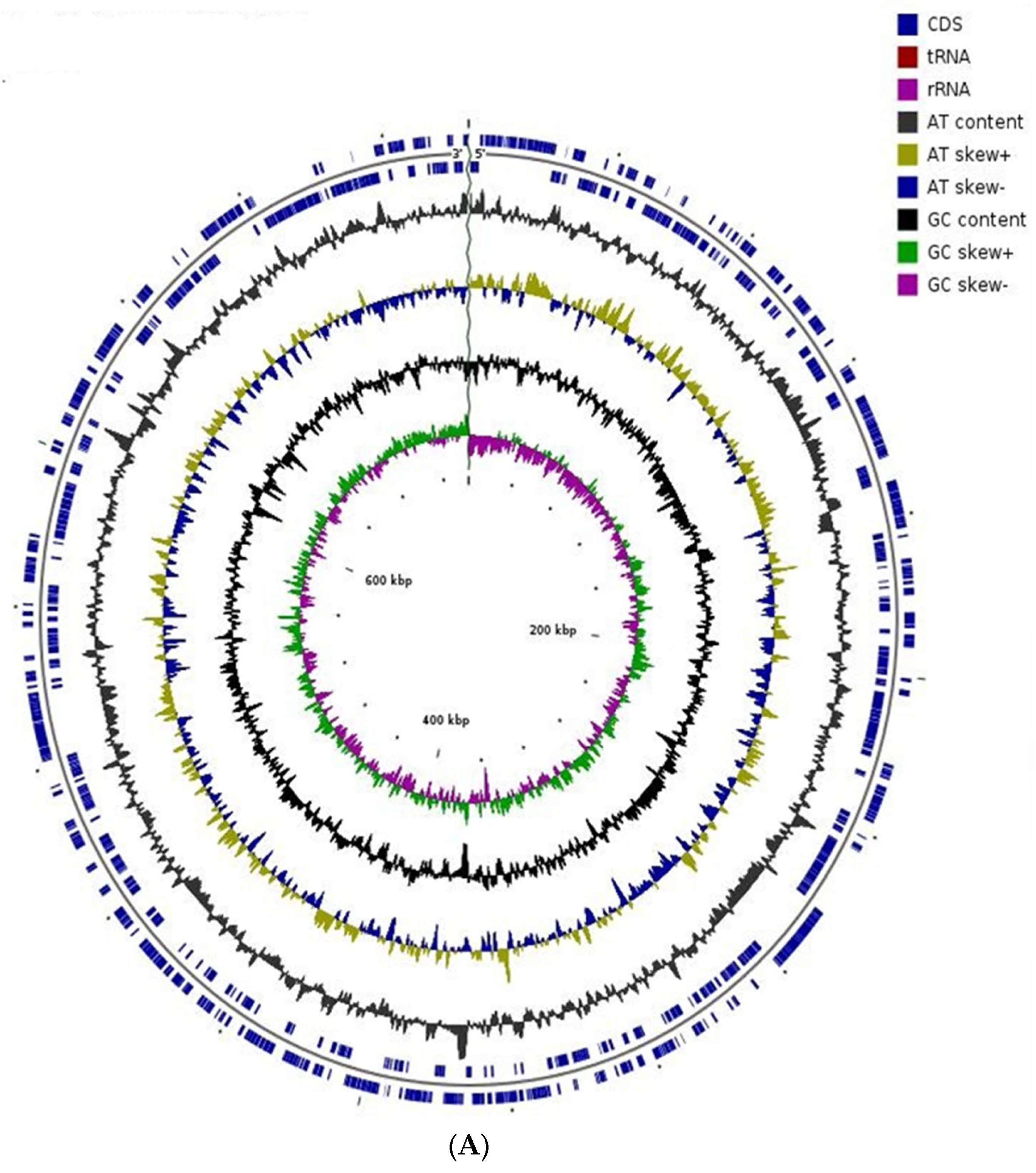
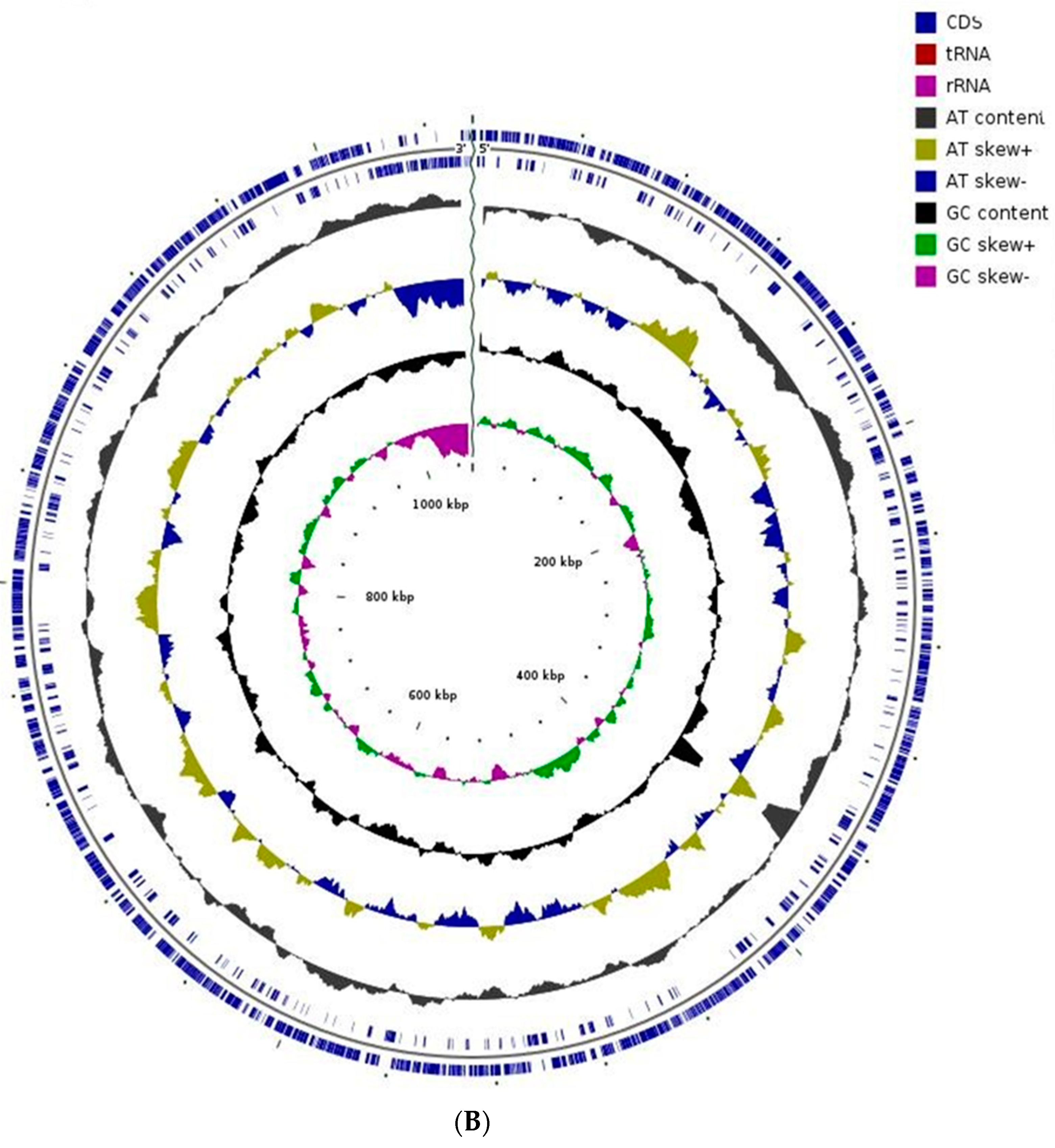
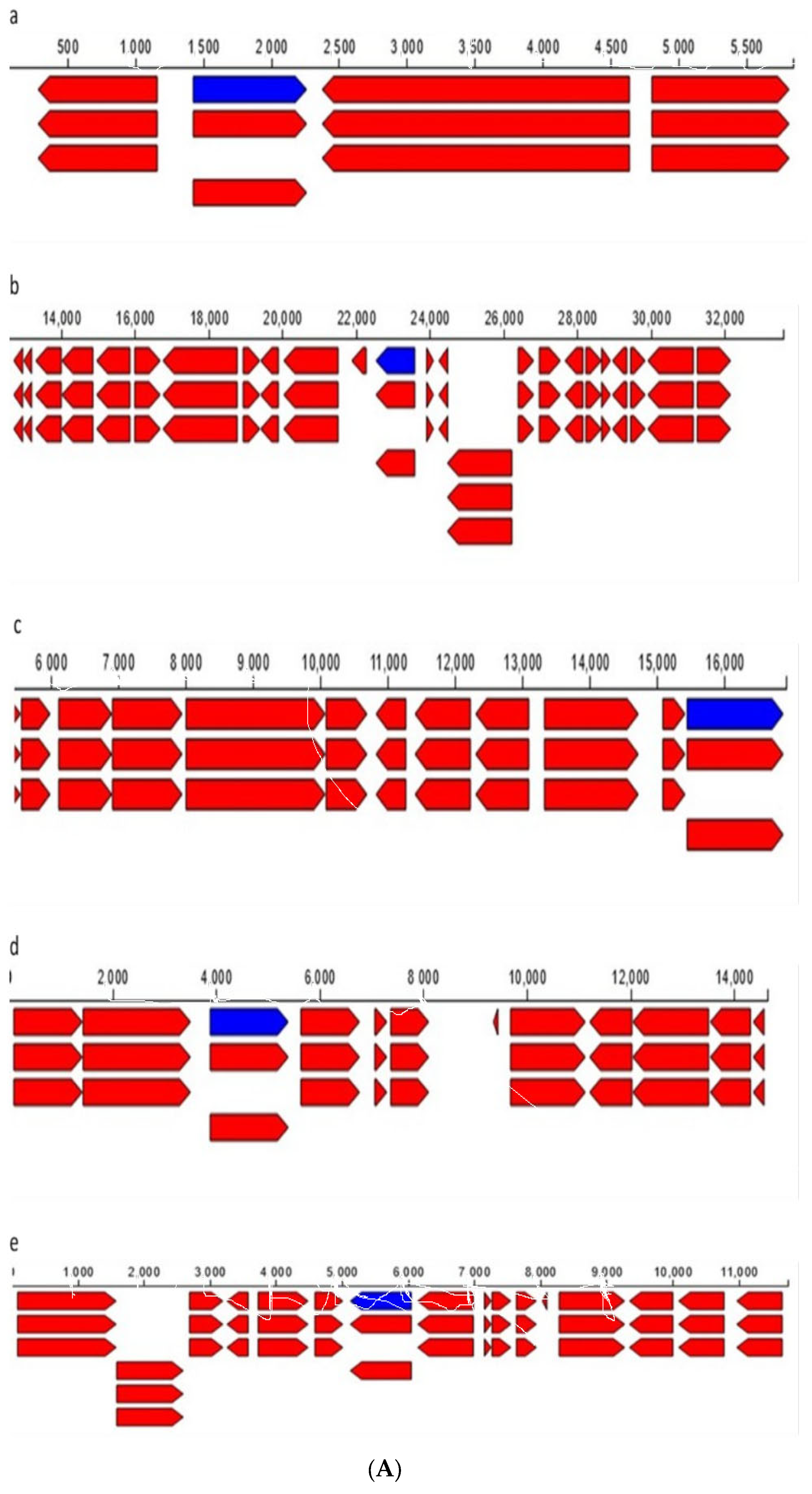
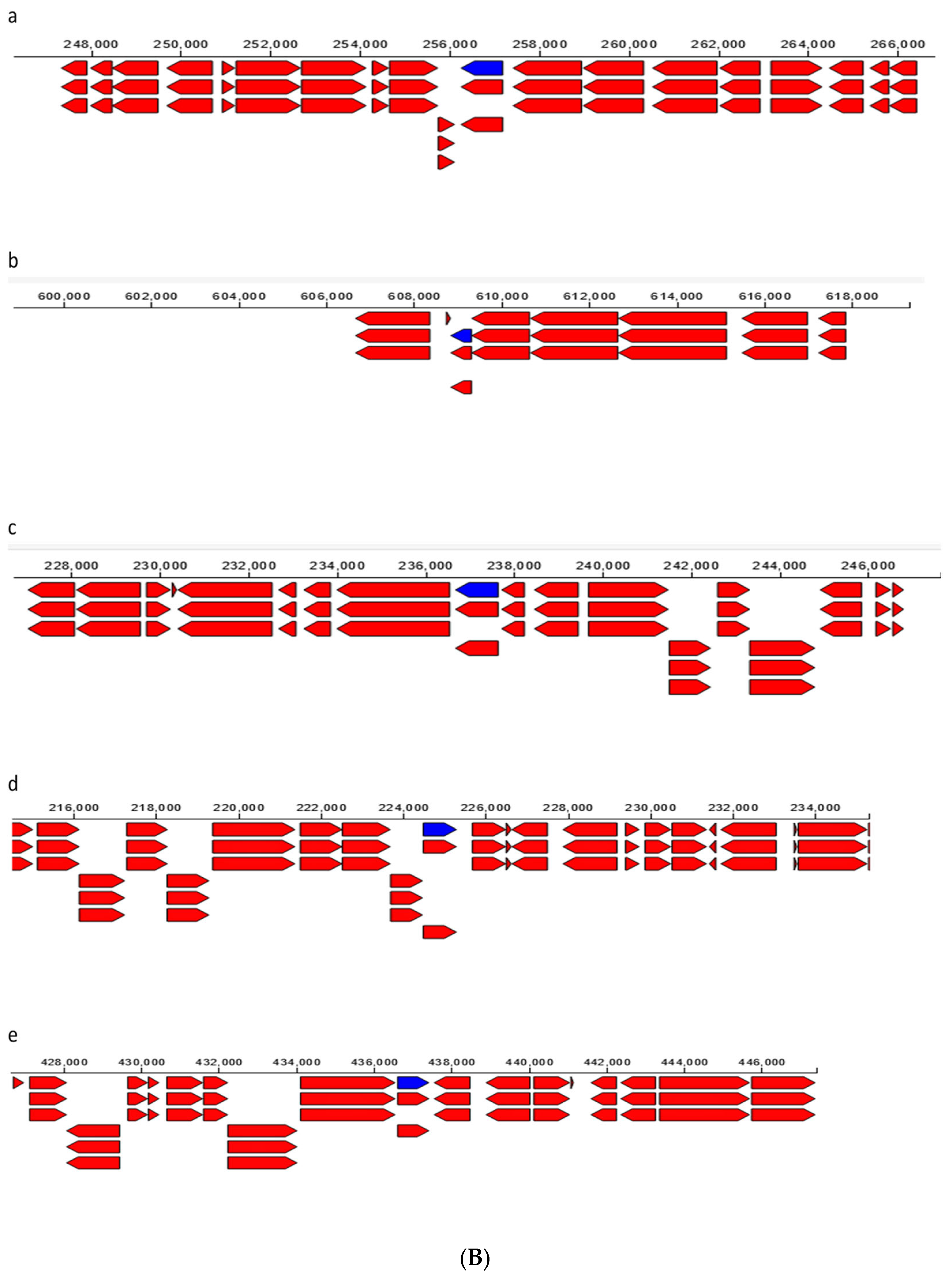
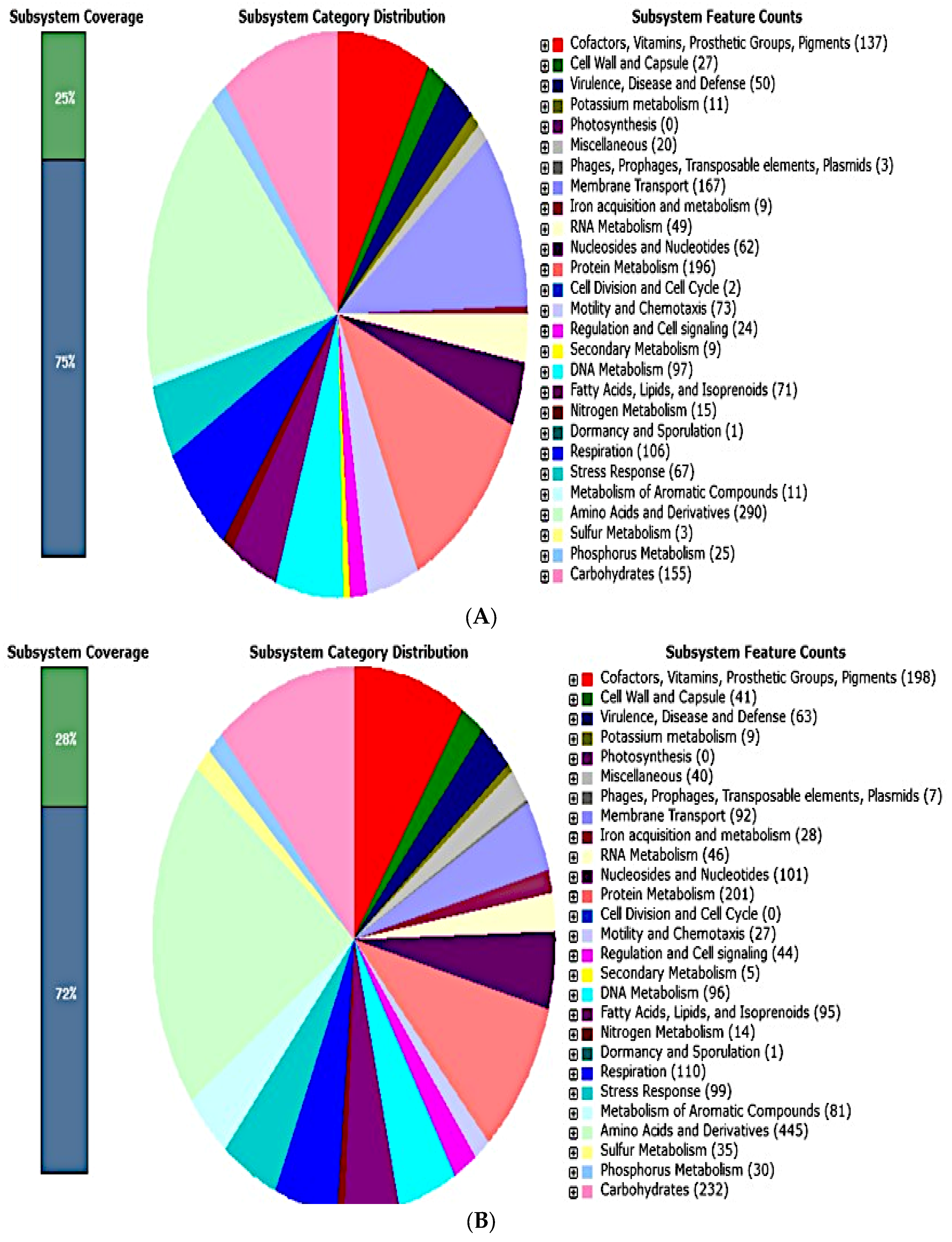
| Endophytic Bacteria | P. carnis NWUBe30 | S. geniculata NWUBe21 | ||
|---|---|---|---|---|
| Plant growth-promoting assay | Qualitative screening output | Quantitative screening output | Qualitative screening output | Quantitative screening output |
| Exopolysaccharide production | ++ | ++ | ||
| Ammonia production | +++ | ++ | ||
| Indole acetic acid (IAA) production | +++ | 17.46 ± 0.08 | +++ | 15.67 ± 0.05 |
| Phosphate solubilization | ++ | 35.85 ± 0.12 | ++ | 32.15 ± 0.05 |
| Siderophore production | ++ | 89.25 ± 0.23 | ++ | 89.60 ± 0.42 |
| Biofilm formation | + | 10.46 ± 0.11 | + | 11.29 ± 0.03 |
| Acds production | + | + | ||
| Hydrogen cyanide production Biochemical traits | ++ | + | ||
| Gram reaction | _ | - | ||
| Shape | R | R | ||
| Citrate utilization | - | - | ||
| Starch | + | + | ||
| Nitrate reduction | - | - | ||
| Fructose | - | - | ||
| Glucose | - | + | ||
| Catalase | + | + | ||
| Oxidase | + | - | ||
| Maltose | + | + | ||
| Genome | S. geniculata Strain NWUBe21 | P. carnis Strain NWUBe30 |
|---|---|---|
| Size | 512,0194 | 5,901,107 |
| GC value | 64.79 | 60.2 |
| N50 | 113,089 | 401,656 |
| L50 | 105 | 6 |
| Number of contigs | 410 | 23 |
| Number of subsystems | 427 | 377 |
| Number of coding sequences | 4986 | 5300 |
| Number of tRNA | 63 | 52 |
| Number of rRNA | 5 | 3 |
| Noncoding repeat | 169 | 189 |
| Genes with SEED annotation | 3934 | 2117 |
| Clusters | Regions | Size (bp) | Most Similar Known Cluster | Percentage Similarity |
|---|---|---|---|---|
| Pseudomonas carnis NWUBe30 | ||||
| RIPP-like | 1.1 | 10,845 | - | - |
| Redox-cofactor | 1.2 | 22,147 | Lankacidin C NRP + Polyketide | 13% |
| Arylpolyene | 2.1 | 43,575 | APE Vf Other | 40% |
| NRPS-like | 2.2 | 25,880 | Fragin NRP | 25% |
| Siderophore | 3.1 | 11,925 | - | - |
| NRPS | 3.2 | 52,896 | Pyoverdin NRP | 10% |
| NAGGN | 3.3 | 14,863 | - | - |
| Hserlactone | 3.4 | 20,575 | - | - |
| NRPS-like betalactone | 4.1 | 43,476 | Pyoverdin NRP | 1% |
| NRPS | 5.1 | 65,892 | Pyoverdin NRP | 11% |
| Terpene | 7.1 | 22,225 | - | - |
| NRPS | 10.1 | 64,235 | Viscosin NRP | 43% |
| RIPP-like | 12.1 | 10,878 | - | - |
| NRPS | 13.1 | 46,269 | Tolaasin I/Tolaasin F NRP: Lipopeptide | 50% |
| Betalactone | 15.1 | 23,174 | Fengycin NRP | 13% |
| Stenotrophomonas geniculata NWUBe21 | ||||
| Lassopeptide | 1.1 | 15,117 | - | - |
| NRPS | 36.1 | 22,883 | Bacillibactin NRP: NRP Siderophore | 80% |
| Arylpolyene | 146.1 | 10,332 | APE Vf Other | 20% |
| RIPP-like | 166.1 | 5818 | - | - |
| RIPP-like | 171.1 | 7325 | - | - |
| Arylpolyene | 358.1 | 3655 | APE Vf Other | 10% |
| Contigs Tag | Product Role | Gene | Pathway |
|---|---|---|---|
| S geniculata NWUBe21 Node_255 | Biofilm PGA outer membrane secretin | PgaA | Biofilm Adhesin biosynthesis |
| Node_21 | Indole-3-Pyruvate decarboxylase | IPDC | Pyruvate metabolism |
| Node_17 | Intermediate for the synthesis of Tryptophan | Indole-3-glycerol phosphate synthase | Tryptophan biosynthesis |
| Node_1 | Glucose-6-phosphate 1-dehydrogenase | GCD | Phosphate pentose |
| Node_120 | Ferri-Bacillibactin esterase (iron transport and uptake) | BesA | Iron metabolism |
| Node_67 | Ammonia/Ammonium transporter | amtB | Ammonia metabolism |
| Node_39 | Phosphoribosylanthranilate isomerase | trpC | Tryptophan biosynthesis |
| Node_89 | Exopolyphosphatase monomer | Ppx | Phosphorus metabolism |
| Node_20 | Alkaline phosphatase | phoA2 | Organic phosphorus metabolism |
| Node_5 | Nitrate reductase | narG | Nitrate metabolism |
| P. carnis NWUBe30 Node_2 | Biofilm PGA synthesis auxiliary protein | PgaD | Biofilm Adhesin biosynthesis |
| Node_5 | Pellicle/biofilm biosynthesis protein | PslA | Biofilm Adhesin biosynthesis |
| Node_5 | Iron siderophore receptor protein | FecA | Iron metabolism |
| Node_2 | Nitrogen regulation protein | NtrC | Nitrogen metabolism |
| Node_10 | Exopolysaccharide protein | ExoZ | Polysaccharide degradation |
| Node_8 | Auxin efflux carrier family protein | Auxin biosynthesis | |
| Node_8 | Tryptophan synthase | trpA | IAA production |
| Node_6 | Nitrogen-fixing | NifU | Nitrogen fixation |
| Node_6 | Ammonia monooxygenase | Ammonia metabolism | |
| Node_1 | Indole-3-glycerol phosphate synthase | Tryptophan biosynthesis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omomowo, O.I.; Babalola, O.O. Genomic Insights into Two Endophytic Strains: Stenotrophomonas geniculata NWUBe21 and Pseudomonas carnis NWUBe30 from Cowpea with Plant Growth-Stimulating Attributes. Appl. Sci. 2022, 12, 12953. https://doi.org/10.3390/app122412953
Omomowo OI, Babalola OO. Genomic Insights into Two Endophytic Strains: Stenotrophomonas geniculata NWUBe21 and Pseudomonas carnis NWUBe30 from Cowpea with Plant Growth-Stimulating Attributes. Applied Sciences. 2022; 12(24):12953. https://doi.org/10.3390/app122412953
Chicago/Turabian StyleOmomowo, Olawale Israel, and Olubukola Oluranti Babalola. 2022. "Genomic Insights into Two Endophytic Strains: Stenotrophomonas geniculata NWUBe21 and Pseudomonas carnis NWUBe30 from Cowpea with Plant Growth-Stimulating Attributes" Applied Sciences 12, no. 24: 12953. https://doi.org/10.3390/app122412953
APA StyleOmomowo, O. I., & Babalola, O. O. (2022). Genomic Insights into Two Endophytic Strains: Stenotrophomonas geniculata NWUBe21 and Pseudomonas carnis NWUBe30 from Cowpea with Plant Growth-Stimulating Attributes. Applied Sciences, 12(24), 12953. https://doi.org/10.3390/app122412953







