Breast Cancer Detection in Thermography Using Convolutional Neural Networks (CNNs) with Deep Attention Mechanisms
Abstract
Featured Application
Abstract
1. Introduction
2. Deep Learning and Convolutional Neural Network (CNN)
3. Deep Attention Mechanisms
- Soft Attention: A series of elements are used to calculate categorical distribution. The resulting possibilities reflect the importance of each element and are used as weights to generate context-aware encoding that represents the weighted total of all elements [10]. It identifies how much attention should be spent on each element, considering the interdependency between the deep neural network’s mechanism and the target, by assigning a weight of 0 to 1 to each input element. The attention layers calculate weights using softmax functions, making the overall attentional model deterministic and differentiable. Soft attention has the ability to act both spatially and temporally. The primary function of the spatial context is to extract the features or weight of the most essential features. It adjusts the weights of all samples in sliding time windows for the temporal context, as samples at various periods contribute differently. Soft mechanisms have a high processing cost while being deterministic and differentiable [11].
- Hard Attention: From the input sequence, a subset of elements is chosen. The hard AMs force the model to focus solely on the important elements, ignoring all others, where the weight assigned to an input part is either 0 or 1. As a result, the objective is non-differentiable since the input elements are either observed or not. The procedure entails making a series of decisions on which parts to highlight. For example, in the temporal context, the model attends to a portion of the input to acquire information, deciding where to focus in the following step based on the known information. Based on this information, a neural network may decide. Hard-attention mechanisms are represented by stochastic processes since there is no ground truth to suggest the optimal selection policy. Reinforcement learning approaches are required to train models with hard attention since the model is not differentiable [11]. In fact, various natural language processing tasks rely solely on very sparse tokens from long text input. Hard attention is well-suited for these tasks because it overcomes the weakness of soft attention in long sequences [10]. When compared to soft mechanisms, inference time and computational costs are reduced whenever the complete input is not stored or processed [11].
- Self-Attention: The interdependence between the input elements of the mechanism is measured because it allows the input to interact with the other “self” and decide what it should pay more attention to. The parallel computation capacity for a long input is one of the self-attention layer’s primary benefits over soft and hard mechanisms. This mechanism layer uses simple and readily parallelizable matrix calculations to check the attention of all the identical input elements [11].
4. Related Work
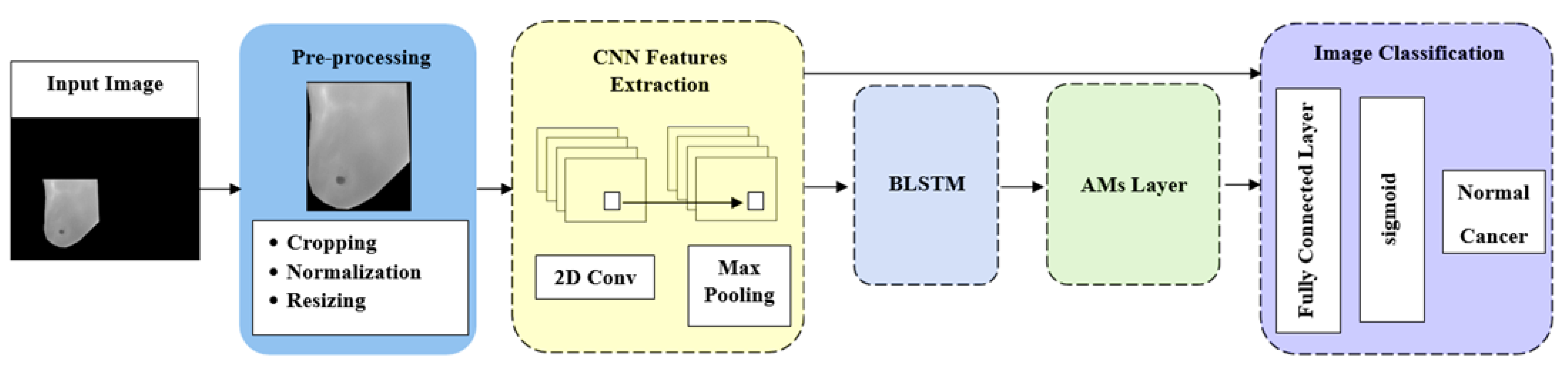
5. Materials and Methods
5.1. Preprocessing
5.2. Feature Extraction
- Image input layer: The image input layer is the connection layer to the network, as it contains the channel size and the input image size. Based on the database containing the grayscale images, the channel size was 1, and the size of the post-processing images was 56 × 56, so the input layer was 56 × 56 × 1.
- Convolution layer: A set of filters in the CNN processed the input image to generate activation maps. By stacking the activation maps along the depth dimension, the layer output was produced. Three parameters determined the convolution layer structure; the first parameter determined the filter size, and the second parameter determined the feature mappings. Then, via a Stride, we slid the filter over the following receptive area of the same input image and repeated the procedure. The same procedure was repeated until we completed the entire image. The input for the following layer will be the output.
- Max pooling layer: After convolution, the spatial volume of the input image was reduced using a pooling layer. It was employed between two convolution layers. Max pooling is a reduced feature map that retains the majority of the dominant features from the previous feature map after redundant data have been eliminated.
- Dropout layer: Overfitting may be avoided by using the dropout technique, which also offers a way of roughly combining many distinct neural network architectures efficiently and exponentially. Dropout refers to units in a neural network that are dropping, both hidden and visible. Dropping a unit out entails removing it temporarily from the network.
5.3. BLSTM Layer
5.4. AMs Layer
- SL layer: Processing sequential data with an attention technique that takes each timestamp’s context into account. In this paper, it is implemented by package keras-self attention with multiplicative type, by using the following equation [33]:
- SF layer: Discredits irrelevant areas by multiplying the corresponding features map with a low weight. Accordingly, a high attention area keeps the original value while low-attention areas get closer to 0 (become dark in the visualization). We use the hidden state from the previous time step to compute a weight for covering each sub-section of an image. We compute a score to measure how much attention as the following equation [34]:We pass to a softmax for normalization to compute the weight .With softmax, adds up to 1, and we use it to compute a weighted average for
- HD layer: This forces the model to focus solely on the important elements, ignoring all others, where the weight assigned to an input part is either 0 or 1. As a result, the objective is non-differentiable since the input elements are either observed or not. To compute HD, using as a sample rate to pick one as the input to the next layer, instead of a weighted average as in SF [34].
5.5. Fully Connected Layer
5.6. Sigmoid Layer
5.7. Classification Layer
- Rotation
- Brightness alteration
5.8. Evaluation Metrics
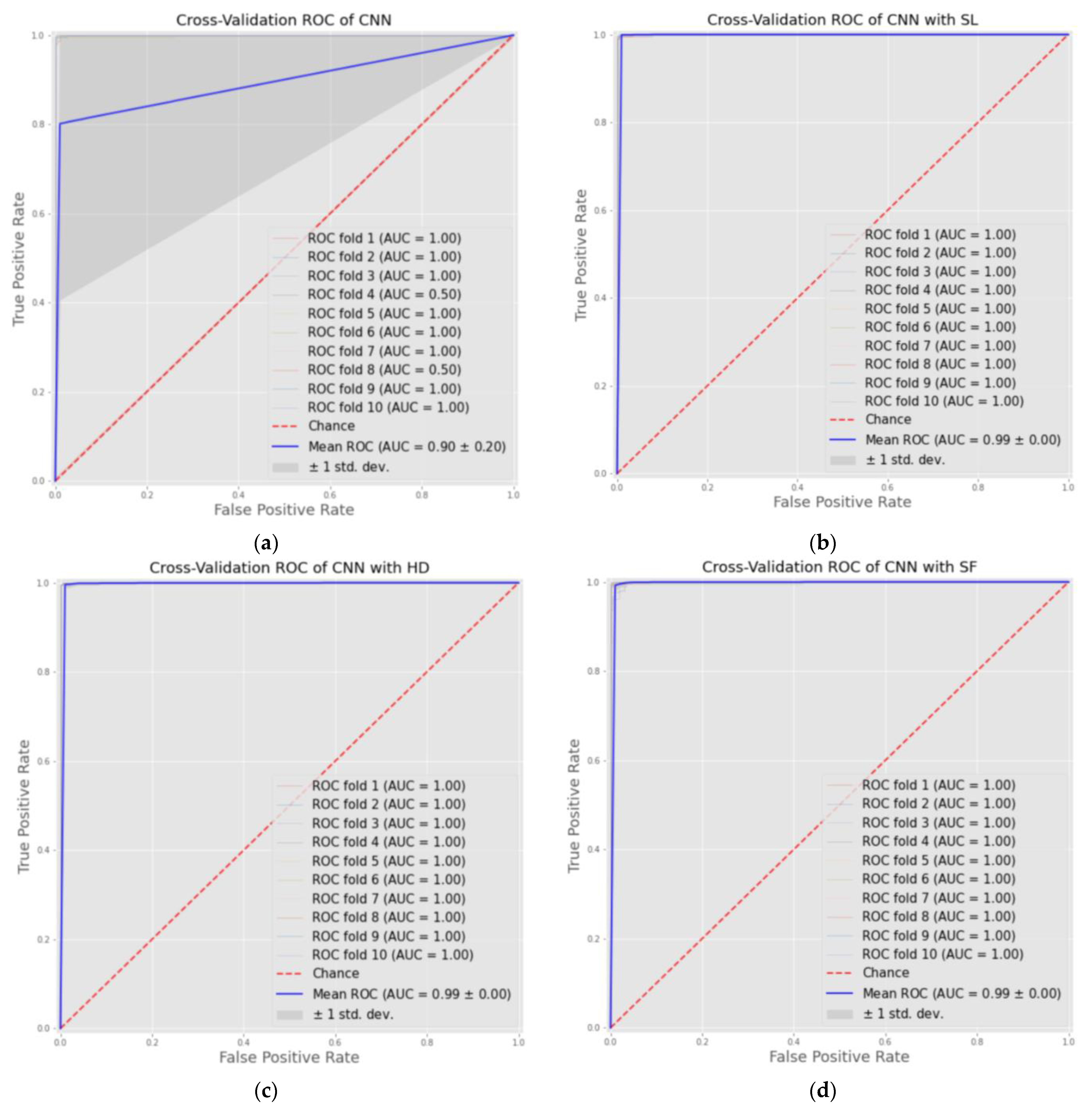
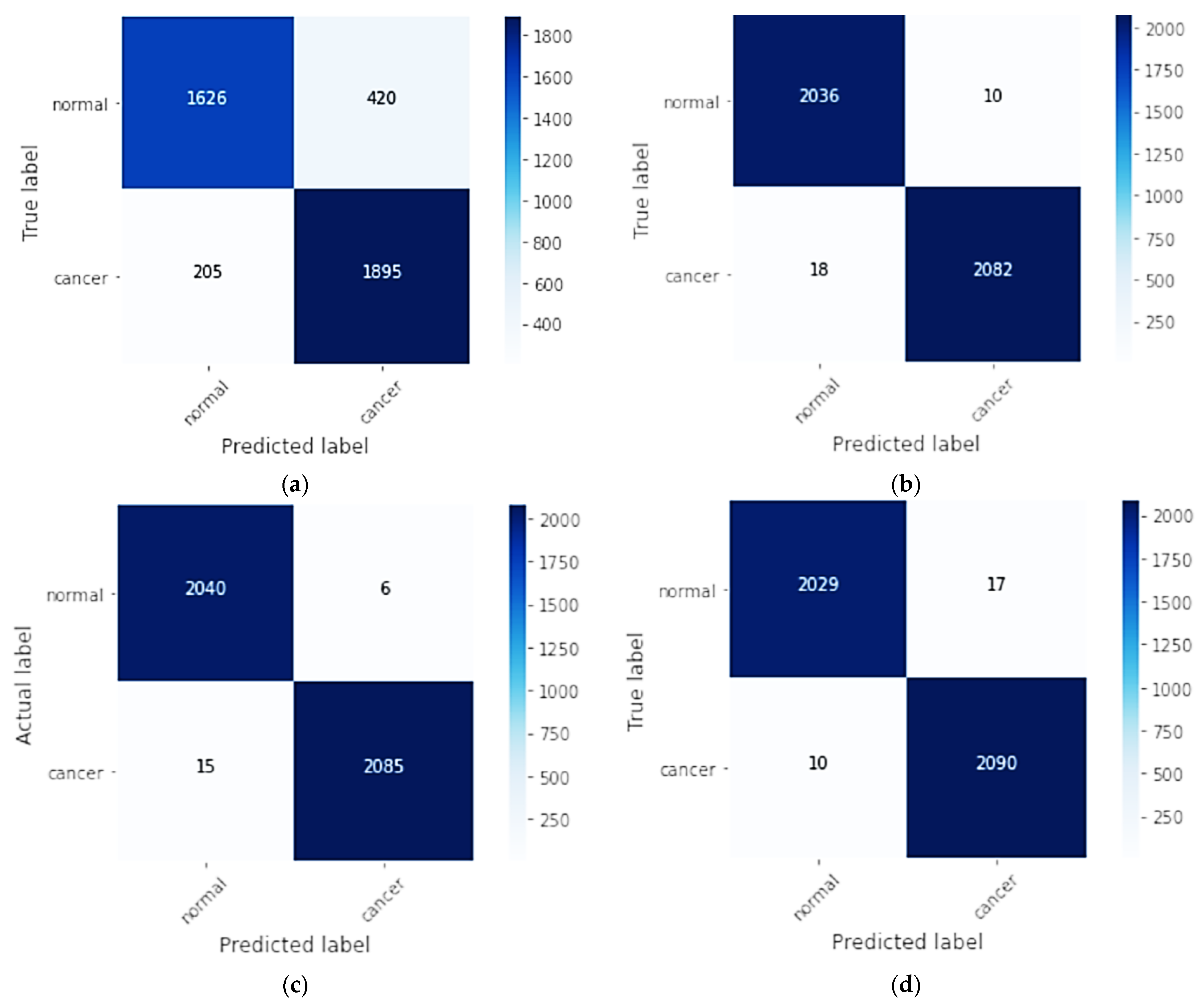
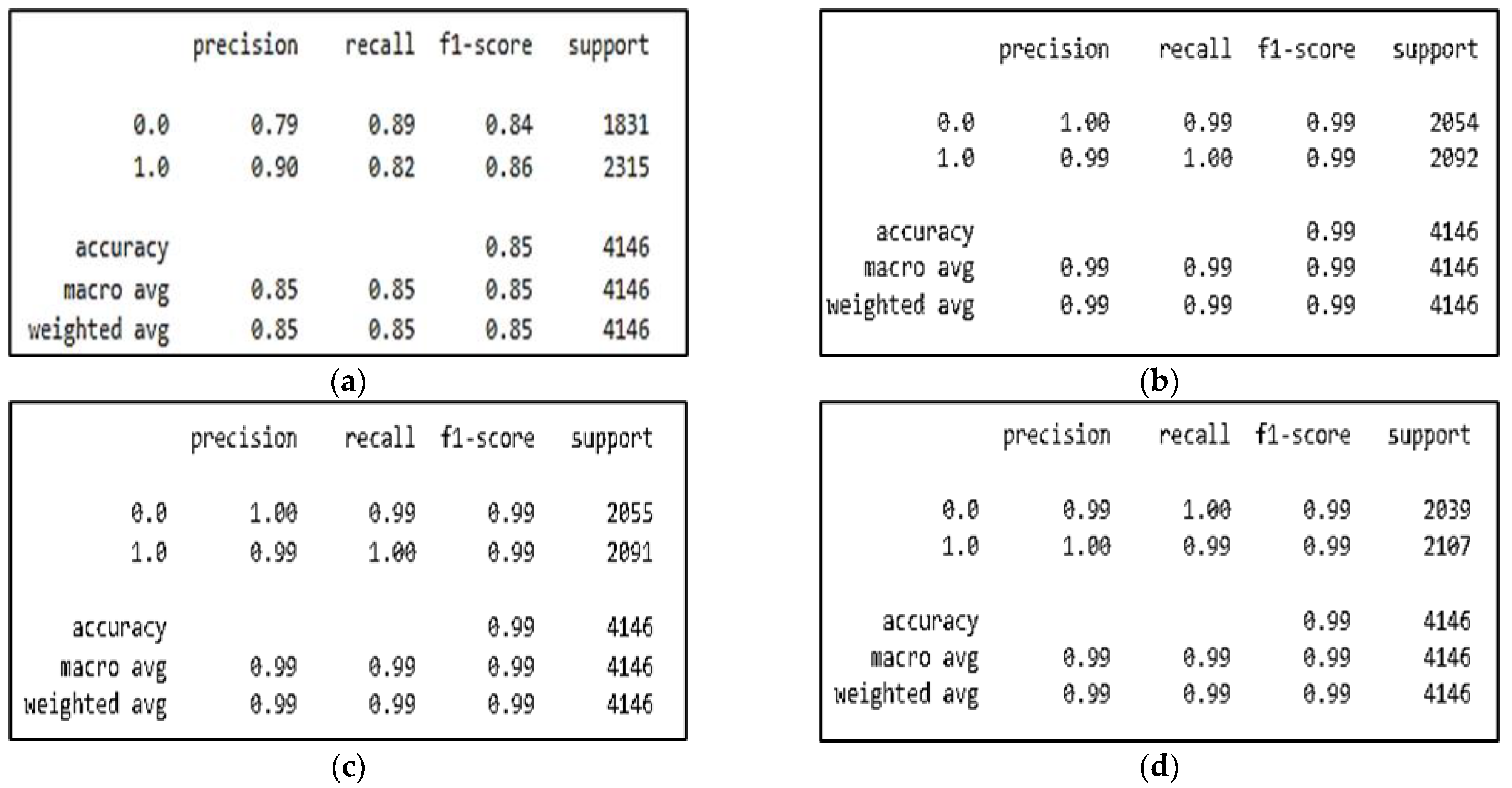
6. Results
6.1. Experimental Settings
6.2. Classification Results
6.3. Comparison with Other Models
6.3.1. Comparison with Methods Using DL on Different Types of Images
6.3.2. Comparison with Methods Using DL on Thermal Images
6.3.3. Comparison with Methods Using DL and AMs on Different Types of Images
7. Conclusions and Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Rajinikanth, V.; Kadry, S.; Taniar, D.; Damaševičius, R.; Rauf, H.T. Breast-Cancer Detection Using Thermal Images with Marine-Predators-Algorithm Selected Features. In Proceedings of the 2021 Seventh International Conference on Bio Signals, Images, and Instrumentation (ICBSII), Chennai, India, 25–27 March 2021; pp. 1–6. [Google Scholar]
- Singh, D.; Singh, A.K. Role of Image Thermography in Early Breast Cancer Detection- Past, Present and Future. Comput. Methods Programs Biomed. 2020, 183, 105074. [Google Scholar] [CrossRef] [PubMed]
- Parisky, Y.R.; Skinner, K.A.; Cothren, R.; DeWittey, R.L.; Birbeck, J.S.; Conti, P.S.; Rich, J.K.; Dougherty, W.R. Computerized Thermal Breast Imaging Revisited: An Adjunctive Tool to Mammography. In Proceedings of the 20th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Hong Kong, China, 1 November 1998; Volume 2, pp. 919–921. [Google Scholar]
- Wu, P.; Qu, H.; Yi, J.; Huang, Q.; Chen, C.; Metaxas, D. Deep Attentive Feature Learning for Histopathology Image Classification. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1865–1868. [Google Scholar]
- Melekoodappattu, J.G.; Subbian, P.S. Automated Breast Cancer Detection Using Hybrid Extreme Learning Machine Classifier. J. Ambient. Intell. Hum. Comput. 2020. [Google Scholar] [CrossRef]
- Palminteri, S.; Pessiglione, M. Chapter Five—Reinforcement Learning and Tourette Syndrome. In International Review of Neurobiology; Martino, D., Cavanna, A.E., Eds.; Advances in the Neurochemistry and Neuropharmacology of Tourette Syndrome; Academic Press: Cambridge, MA, USA, 2013; Volume 112, pp. 131–153. [Google Scholar]
- Almajalid, R.; Shan, J.; Du, Y.; Zhang, M. Development of a Deep-Learning-Based Method for Breast Ultrasound Image Segmentation. In Proceedings of the 2018 17th IEEE International Conference on Machine Learning and Applications (ICMLA), Orlando, FL, USA, 17–20 December 2018; pp. 1103–1108. [Google Scholar]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional Neural Networks: An Overview and Application in Radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; Wang, P.; Tansey, K.; Han, D.; Zhang, J.; Zhang, S.; Li, H. A Deep Learning Framework under Attention Mechanism for Wheat Yield Estimation Using Remotely Sensed Indices in the Guanzhong Plain, PR China. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102375. [Google Scholar] [CrossRef]
- Shen, T.; Zhou, T.; Long, G.; Jiang, J.; Wang, S.; Zhang, C. Reinforced Self-Attention Network: A Hybrid of Hard and Soft Attention for Sequence Modeling. In Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence, Stockholm, Sweden, 13–19 July 2018; International Joint Conferences on Artificial Intelligence Organization: Stockholm, Sweden, 2018; pp. 4345–4352. [Google Scholar]
- de Correia, A.S.; Colombini, E.L. Attention, Please! A Survey of Neural Attention Models in Deep Learning. arXiv 2021, arXiv:2103.16775. [Google Scholar] [CrossRef]
- Nahid, A.-A.; Mehrabi, M.A.; Kong, Y. Histopathological Breast Cancer Image Classification by Deep Neural Network Techniques Guided by Local Clustering. Biomed Res. Int. 2018, 2018, 2362108. [Google Scholar] [CrossRef] [PubMed]
- Rashed, E.; El Seoud, M.S.A. Deep Learning Approach for Breast Cancer Diagnosis. In Proceedings of the 2019 8th International Conference on Software and Information Engineering, Cairo, Egypt, 9 April 2019; ACM: Cairo, Egypt, 2019; pp. 243–247. [Google Scholar]
- Arslan, A.K.; Yaşar, Ş.; Çolak, C. Breast Cancer Classification Using a Constructed Convolutional Neural Network on the Basis of the Histopathological Images by an Interactive Web-Based Interface. In Proceedings of the 2019 3rd International Symposium on Multidisciplinary Studies and Innovative Technologies (ISMSIT), Ankara, Turkey, 11–13 October 2019; pp. 1–5. [Google Scholar]
- Platania, R.; Shams, S.; Yang, S.; Zhang, J.; Lee, K.; Park, S.-J. Automated Breast Cancer Diagnosis Using Deep Learning and Region of Interest Detection (BC-DROID). In Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Boston, MA, USA, 20 August 2017; ACM: Boston, MA, USA, 2017; pp. 536–543. [Google Scholar]
- Patil, R.S.; Biradar, N. Automated Mammogram Breast Cancer Detection Using the Optimized Combination of Convolutional and Recurrent Neural Network. Evol. Intel. 2021, 14, 1459–1474. [Google Scholar] [CrossRef]
- Santana, M.A.; Pereira, J.M.S.; Silva, F.L.; Lima, N.M.; Sousa, F.N.; Arruda, G.M.S.; de Lima, R.C.F.; Silva, W.W.A.; Santos, W.P. Breast Cancer Diagnosis Based on Mammary Thermography and Extreme Learning Machines. Res. Biomed. Eng. 2018, 34, 45–53. [Google Scholar] [CrossRef]
- Gogoi, U.R.; Bhowmik, M.K.; Ghosh, A.K.; Bhattacharjee, D.; Majumdar, G. Discriminative Feature Selection for Breast Abnormality Detection and Accurate Classification of Thermograms. In Proceedings of the 2017 International Conference on Innovations in Electronics, Signal Processing and Communication (IESC), Shillong, India, 6–7 April 2017; pp. 39–44. [Google Scholar]
- Ekici, S.; Jawzal, H. Breast Cancer Diagnosis Using Thermography and Convolutional Neural Networks. Med. Hypotheses 2020, 137, 109542. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Prakash, A.; Roy, S.K.; Sharan, P.; Mathur, N. Breast Cancer Detection Using Thermal Images and Deep Learning. In Proceedings of the 2020 7th International Conference on Computing for Sustainable Global Development (INDIACom), Bvicam, New Delhi, 12–14 March 2020; pp. 211–216. [Google Scholar]
- Mambou, S.J.; Maresova, P.; Krejcar, O.; Selamat, A.; Kuca, K. Breast Cancer Detection Using Infrared Thermal Imaging and a Deep Learning Model. Sensors 2018, 18, 2799. [Google Scholar] [CrossRef] [PubMed]
- Mookiah, M.R.K.; Acharya, U.R.; Ng, E.Y.K. Data Mining Technique for Breast Cancer Detection in Thermograms Using Hybrid Feature Extraction Strategy. Quant. InfraRed Thermogr. J. 2012, 9, 151–165. [Google Scholar] [CrossRef]
- Tello-Mijares, S.; Woo, F.; Flores, F. Breast Cancer Identification via Thermography Image Segmentation with a Gradient Vector Flow and a Convolutional Neural Network. J. Healthc. Eng. 2019, 2019, e9807619. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cauce, R.; Pérez-Martín, J.; Luque, M. Multi-Input Convolutional Neural Network for Breast Cancer Detection Using Thermal Images and Clinical Data. Comput. Methods Programs Biomed. 2021, 204, 106045. [Google Scholar] [CrossRef] [PubMed]
- de Freitas Oliveira Baffa, M.; Grassano Lattari, L. Convolutional Neural Networks for Static and Dynamic Breast Infrared Imaging Classification. In Proceedings of the 2018 31st SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI), Paraná, Brazil, 29 October–1 November 2018; pp. 174–181. [Google Scholar]
- Yao, H.; Zhang, X.; Zhou, X.; Liu, S. Parallel Structure Deep Neural Network Using CNN and RNN with an Attention Mechanism for Breast Cancer Histology Image Classification. Cancers 2019, 11, 1901. [Google Scholar] [CrossRef] [PubMed]
- Toğaçar, M.; Özkurt, K.B.; Ergen, B.; Cömert, Z. BreastNet: A Novel Convolutional Neural Network Model through Histopathological Images for the Diagnosis of Breast Cancer. Phys. A Stat. Mech. Appl. 2020, 545, 123592. [Google Scholar] [CrossRef]
- Fan, M.; Chakraborti, T.; Chang, E.I.-C.; Xu, Y.; Rittscher, J. Fine-Grained Multi-Instance Classification in Microscopy Through Deep Attention. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 169–173. [Google Scholar]
- Xu, B.; Liu, J.; Hou, X.; Liu, B.; Garibaldi, J.; Ellis, I.O.; Green, A.; Shen, L.; Qiu, G. Attention by Selection: A Deep Selective Attention Approach to Breast Cancer Classification. IEEE Trans. Med. Imaging 2020, 39, 1930–1941. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, R.; Jethanandani, M.; Sarkar, R. DAN: Breast Cancer Classification from High-Resolution Histology Images Using Deep Attention Network. In Proceedings of the Innovations in Computational Intelligence and Computer Vision; Sharma, M.K., Dhaka, V.S., Perumal, T., Dey, N., Tavares, J.M.R.S., Eds.; Springer: Singapore, 2021; pp. 319–326. [Google Scholar]
- Deng, J.; Ma, Y.; Li, D.; Zhao, J.; Liu, Y.; Zhang, H. Classification of Breast Density Categories Based on SE-Attention Neural Networks. Comput. Methods Programs Biomed. 2020, 193, 105489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.; Qian, B.; Liu, X.; Li, X.; Wang, X.; Yin, C.; Lv, X.; Song, L.; Wang, L. Classifying Breast Cancer Histopathological Images Using a Robust Artificial Neural Network Architecture. In Proceedings of the Bioinformatics and Biomedical Engineering; Rojas, I., Valenzuela, O., Rojas, F., Ortuño, F., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 204–215. [Google Scholar]
- CyberZHG. Keras-Self-Attention: Attention Mechanism for Processing Sequential Data That Considers the Context for Each Timestamp; GitHub, Inc.: San Francisco, CA, USA, 2021. [Google Scholar]
- Soft & Hard Attention. Available online: https://jhui.github.io/2017/03/15/Soft-and-hard-attention/ (accessed on 12 December 2022).
- Silva, L.F.; Saade, D.C.M.; Sequeiros, G.O.; Silva, A.C.; Paiva, A.C.; Bravo, R.S.; Conci, A. A New Database for Breast Research with Infrared Image. J. Med. Imaging Health Inform. 2014, 4, 92–100. [Google Scholar] [CrossRef]
- Murugan, R.; Goel, T. E-DiCoNet: Extreme Learning Machine Based Classifier for Diagnosis of COVID-19 Using Deep Convolutional Network. J. Ambient. Intell. Hum. Comput. 2021, 12, 8887–8898. [Google Scholar] [CrossRef]
- McHugh, M.L. Interrater Reliability: The Kappa Statistic. Biochem. Med. 2012, 22, 276–282. [Google Scholar] [CrossRef]
| Ref | Approaches | Imaging Modalities | Datasets | Results |
|---|---|---|---|---|
| [12] | CNN-LSTM | Histopathology | BreakHis | Acc = 91% Prec = 96% |
| [13] | U-Net CNN | Mammography | CBIS-DDSM mass images CBIS-DDSM microcalcification images | Acc = 94.31% |
| [14] | CNN | Histopathology | BreakHis | Acc = 91.4% |
| [15] | CNN | Mammography | DDSM | Acc = 93.5% AUC = 0.92315 |
| [16] | CNN-RNN | Mammography | Mammogram Image dataset | Acc = 90.59% Sn = 92.42% Sp = 89.88% |
| [17] | Bayes Network Naïve Bayes SVM Decision Tree J48 MLP RF RT ELM | Thermal | University Hospital of the Federal University of Pernambuco | Acc = 76.01% |
| [18] | SVM KNN DT ANN | Thermal | DBT-TU-JU DMR-IR | Acc = 84.29% Acc = 87.50% |
| [19] | CNNs-Bayes optimization algorithm | Thermal | DMI | Acc: 98.95% |
| [20] | DCNNs | Thermal | DMR-IR | Acc = 95.8% Sn = 99.40% Sp = 76.3% |
| [21] | DNN | Thermal | DMR-IR | Conf (Sick) = 78% Conf (Healthy) = 94% |
| [22] | DT Fuzzy Sugeno Naïve Bayes K Nearest Neighbor Gaussian Mixture Model Probabilistic Neural Network | Thermal | Singapore General Hospital NEC-Avio Thermo TVS2000 MkIIST System | Acc = 93.30% Sn = 86.70% Sp = 100% |
| [23] | CNNs-GVF | Thermal | DMR-IR | Acc = 100% Sn = 100% Sp = 100% |
| [24] | multi-input CNN | Thermal | DMR-IR | Acc = 97% Sn = 83% Sp = 100% AUC = 0.99 |
| [25] | CNNs | Thermal | DMR-IR | Acc(color) = 98% Acc(grayscale) = 95% Acc(color) = 95% Acc(grayscale) = 92% |
| [26] | CNNs+RNNs+AM | Histopathology | BACH2018 Bioimaging2015 Extended Bioimaging2015 | Acc = 92% Acc = 96% Acc = 97.5% |
| [27] | CNNs+AM | Histopathology | BreakHis | Acc = 98.51% Sn = 98.70% F1-score = 98.28% |
| [4] | CNNs+AM | Histopathology | BreakHis | Acc = 99.3% |
| [28] | CNNs+AM | Histopathology | Peking Union Hospital Benchmark | Acc = 96% |
| [29] | Deep attention network | Histopathology | BreakHis | Acc = 98% |
| [30] | CNNs+BLSTM+AM | Histopathology | ICIAR 2018 | Acc(2-class) = 96.25% Acc(4-class) = 85.50% |
| [31] | CNNs+ SE-Attention | Mammography | New Benchmarking dataset | Acc = 92.17% |
| [32] | ResNet-CBAM | Histopathology | BreakHis | Acc = 92.6% Sn = 94.7% Sp = 88.9% F1-score = 94.1% AUC = 0.918 |
| Dataset | Size | Normal Breasts | Abnormal Breasts |
|---|---|---|---|
| DMR-IR-Original | 1542 | 762 | 780 |
| DMR-IR-Augmented | 2604 | 1284 | 1320 |
| Total | 4146 | 2046 | 2100 |
| Models | Accuracy (%) | Specificity (%) | Sensitivity (Recall) (%) | Precision (%) | F1-Score (%) | AUC | Cohens Kappa |
|---|---|---|---|---|---|---|---|
| CNN | 84.92% | 89.61% | 89.61% | 90.23% | 83.91% | 0.851 | 0.69 |
| CNN-SL | 99.32% | 99.52% | 99.52% | 99.14% | 99.32% | 0.999 | 0.98 |
| CNN-HD | 99.49% | 99.71% | 99.71% | 99.28% | 99.49% | 0.999 | 0.98 |
| CNN-SF | 99.34% | 99.21% | 99.21% | 99.52 % | 99.36% | 0.999 | 0.98 |
| Ref | Approaches | Imaging Modalities | Accuracy | Specificity | Sensitivity (Recall) | Precision | F1-Score | AUC |
|---|---|---|---|---|---|---|---|---|
| A- Approaches using DL on different types of images | ||||||||
| [12] | CNN- LSTM | Histopathology | 91% | - | - | 96% | - | |
| [13] | U-Net CNN | Mammography | 94.31% | - | - | - | - | |
| [14] | CNN | Histopathology | 91.4% | - | - | - | - | |
| [15] | CNN | Mammography | 93.5% | - | - | - | - | 0.9931 |
| [16] | CNN-RNN | Mammography | 90.59% | 89.88% | 92.42% | - | - | |
| B- Approaches using DL on thermal images | ||||||||
| [19] | CNNs-Bayes optimization algorithm | Thermal | 98.95% | - | - | - | - | |
| [20] | DCNNs | Thermal | 95.8% | 76.3% | 99.40% | - | - | |
| [24] | multi-input CNN | Thermal | 97% | 100% | 83% | - | - | 0.99 |
| [25] | CNNs | Thermal | Static (color) = 98% Static (grayscale) = 95% Dynamic (color) = 95% Dynamic (grayscale) = 92% | - | - | - | - | |
| C- Approaches using DL with AM | ||||||||
| [26] | CNNs + RNNs + AM | Histopathology | -Acc = 92% -Acc = 96% -Acc = 97.5% | - | - | - | - | |
| [27] | CNNs + AM | Histopathology | 98.51% | - | 98.70% | - | 98.28% | |
| [4] | CNNs + AM | Histopathology | 99.3% | - | - | - | - | |
| [28] | CNNs + AM | Histopathology | 96% | - | - | - | - | |
| [29] | Deepattention network | Histopathology | 98% | - | - | - | - | |
| [30] | CNNs + BLSTM + AM | Histopathology | 96.25% | - | - | - | - | |
| [31] | CNNs + SE-Attention | Mammography | 92.17% | - | - | - | - | |
| [32] | ResNet -CBAM | Histopathology | 92.6% | 88.9% | 94.7% | 94.1% | 0.918 | |
| D- Proposed approach with different DL methods and AM on thermal images | ||||||||
| Our Proposed Approaches | CNN | Thermal | 84.92% | 89.61% | 89.61% | 90.23% | 83.91% | 0.851 |
| CNN-SL | Thermal | 99.32% | 99.52% | 99.52% | 99.14% | 99.32% | 0.999 | |
| CNN-HD | Thermal | 99.49% | 99.71% | 99.71% | 99.28% | 99.49% | 0.999 | |
| CNN-SF | Thermal | 99.34% | 99.21% | 99.21% | 99.52 % | 99.36% | 0.999 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshehri, A.; AlSaeed, D. Breast Cancer Detection in Thermography Using Convolutional Neural Networks (CNNs) with Deep Attention Mechanisms. Appl. Sci. 2022, 12, 12922. https://doi.org/10.3390/app122412922
Alshehri A, AlSaeed D. Breast Cancer Detection in Thermography Using Convolutional Neural Networks (CNNs) with Deep Attention Mechanisms. Applied Sciences. 2022; 12(24):12922. https://doi.org/10.3390/app122412922
Chicago/Turabian StyleAlshehri, Alia, and Duaa AlSaeed. 2022. "Breast Cancer Detection in Thermography Using Convolutional Neural Networks (CNNs) with Deep Attention Mechanisms" Applied Sciences 12, no. 24: 12922. https://doi.org/10.3390/app122412922
APA StyleAlshehri, A., & AlSaeed, D. (2022). Breast Cancer Detection in Thermography Using Convolutional Neural Networks (CNNs) with Deep Attention Mechanisms. Applied Sciences, 12(24), 12922. https://doi.org/10.3390/app122412922





