1. Introduction
In digital pathology, whole slide images (WSI) are considered the golden standard for primary diagnosis [
1,
2]. The use of computer-assisted image analysis tools is becoming mainstream for automatic quantitative or semi-quantitative analysis for pathologists [
3,
4,
5,
6,
7].
Cancer is one of the critical public health issues around the world. According to the Global Burden of Disease (GBD) study, there have been more than 25 million cancer incidences and 10 million cancer deaths worldwide in 2020. These statistics indicate that cancer incidence expanded by 33% between 2007 and 2017 worldwide [
8] and, for example, is projected to increase more than 92% in 2020 and 2040 in sub-Saharan Africa [
9].
Breast cancer is one of the leading causes of cancer-related deaths worldwide and affects a large number of women today [
10,
11]. Invasive ductal carcinoma (IDC) is the most frequent subtype of all breast cancers [
12]. As stated in [
13], 934,870 new cancer cases and 287,270 cancer deaths in women are projected to occur in the United States in 2022, with an estimated 287,850 new cases of breast cancer (the leading cause, accounting for 31% of new cancer cases) and 43,250 deaths from breast cancer (the second leading estimated cause of cancer death, with breast cancer accounting for 15% in women). Incidence rates for breast cancer in women have slowly increased by about 0.5% per year since the mid-2000s [
14].
Most women diagnosed with breast cancer are over the age of 50, but younger women can also develop breast cancer. About 1 in 8 women will be diagnosed with breast cancer during their lifetime. Survival rates for breast cancer are believed to have increased and the number of related deaths continues to decline, largely due to factors such as earlier detection, a new personalized treatment approach, and a better understanding of the disease [
15,
16].
Histopathology is a technique frequently used to diagnose tumors. It can identify the characteristics of how cancer looks under the microscope. Accurate detection and classification of some types of cancer is a critical task in medical imaging due to the complexity of those tissues [
17,
18], concretely in breast cancer [
19], which is the one that will be analyzed in this paper.
The diagnosis of breast cancer histopathology images with hematoxylin and eosin stained is non-trivial, labor-intensive, and often leads to a disagreement between pathologists [
20,
21]. Computer-assisted diagnosis systems contribute to helping pathologists improve diagnostic consistency and efficiency, especially in breast cancer using histopathological images [
22,
23,
24].
The use of artificial intelligence tools, especially deep learning methods, to classify histopathological images of patients with breast cancer has been extensively and fruitfully analyzed in the literature [
25,
26,
27,
28,
29]. In particular, convolutional neural networks (CNNs) are the most commonly used for histology image analysis tasks [
30,
31,
32,
33].
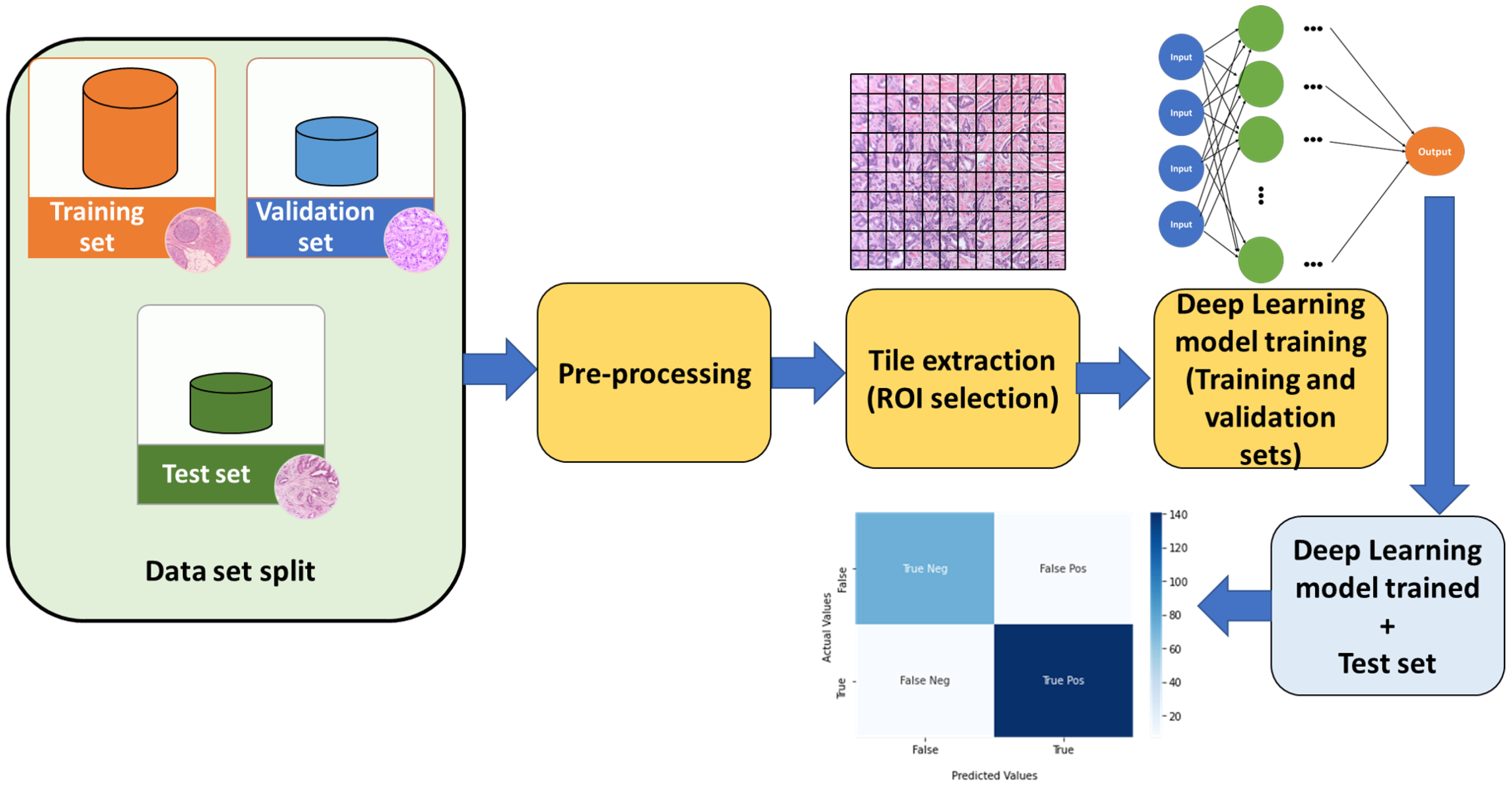
Once the database of the WSI images is available, a phase that is usually performed in different papers presented in the bibliography, is to carry out a pre-processing of said images. The main objective of this pre-processing step is to improve the behavior of the system (several alternatives for pre-processing can be used, among which it is worth mentioning to remove noise, blurring, enhance contrast, and, ultimately, improve the quality of the images to obtain better performance for the final deep learning algorithms). In
Figure 1 a typical classification model pipeline is presented in which common techniques, such as data slipt, image pre-processing, tiling the whole slide images (WSI), and statistical evaluation with a test set, are highlighted.
This paper proposes a comparative study of how different histopathological image preprocessing methods, and various deep learning models, affect the performance of the classification system in breast cancer. The methods used are divided into two groups: based on convolution filters and based on histogram equalization. These preprocessing are tested on a data set containing images of breast cancer.
Our aim with this article is to accurately determine the influence of the different deep learning models and filters (denoted as factors or ways of creating the deep learning system) for analyzing the influence on the behavior (accuracy) of the developed system (in terms of the AUC). To carry out this statistical analysis, the Analysis Of Variance tool (ANOVA), is used [
34,
35,
36,
37].
The rest of this paper is organized as follows. In
Section 2, we present a short overview of some papers employing preprocessing methods in different contexts.
Section 3 describes the dataset used and the preprocessing methods applied. In addition, the deep learning models used and the metrics used for their evaluation are presented.
Section 4 explains how the experiments with the different models and filters were carried out. Finally,
Section 5 provides a discussion of the results and concludes the paper.
2. Related Work
In problems with histopathologic images, it is increasingly common to use some form of preprocessing. In [
38], the authors provide a detailed overview of graph-based deep learning for computational histopathology. They note that stain normalization is a common practice when using standard entity graph pathology workflows. Similarly, in [
39] stain normalization is used as preprocessing for a classification model that predicts not only histological sub-types of endometrial carcinoma, but also molecular sub-types and 18 common genetic mutations based on histopathological images.
A tumor segmentation problem in lung cancer images is addressed in [
40]. In the preprocessing stage, they employ a staining normalization to homogenize the images. Then, they apply another type of Gaussian filter as a color augmentation method to make the models used more robust.
A problem of binary classification of breast cancer images, obtaining an accuracy higher than 98%, is presented in [
41]. The procedure the authors follow for its resolution consists of three stages: preprocessing, segmentation and classification. In the preprocessing stage, the authors use a fuzzy equalization of the histogram of the images to overcome unwanted over-enhancement and noise amplification by preserving local information of the original image.
The segmentation of tumors in Cervical Cancer images was analyzed in [
42]. To improve the quality of the images and thus increase the performance of the models, the development of an automatic framework to preprocess the images was proposed. This framework would include different preprocessing methods such as adaptive histogram equalization, Gaussian filters, or sharpen filters among others.
In [
43], the authors propose a study in which they compare the results of applying different filters to retinal fundus images. The filters used are the following: Contrast Limited Adaptative Histogram Equalization (CLAHE), Adaptative Histogram Equalization (AHE), Median filter, Gaussian filter, Weiner filter, and Adaptive median filters. They also present the advantages and disadvantages of each method, giving a brief explanation of them. To quantify the efficiency of each filter they use the MSE and PSNR measures. Finally, they conclude that the best-performing filter is the adaptive median filter due to its high PSNR and low MSE.
Regarding the different models of deep learning, the alternatives that are frequently found in the literature and that have been successfully applied in histopathology images are VGG16, VGG19, ResNet50, MobilNet, DenseNet121, among others [
44,
45,
46]. For example, in [
47], the authors propose a cascaded deep learning framework for accurate nuclei segmentation in digitized microscopy images of histology slides, based on the VGG16 model and a soft-die loss function. In [
48] a multi-class classification, using the 6B-Net model selected vector and ResNet50 selected vector was fused and applied to breast cancer. In [
44], a review of the popular deep learning algorithms, including convolutional neural networks (CNNs), generative adversarial networks (GANs), and graph neural networks (GNNs) is presented, highlighting the applications in pathology.
3. Materials and Methods
This section is divided into four parts. In the first one, we will give a brief description of the dataset used in the experimentation. In the second, we will detail the preprocessing that has been applied, giving a short description of them. Then we will talk about the deep learning models chosen to carry out the experimentation, ending the section with the metrics that will be used to evaluate the performance of the models.
3.1. Dataset: Breast Histopathology Images
In this paper, the dataset of breast cancer images presented in [
49] will be used. It consists of 277,524 color images of size 50 × 50 px. The images are annotated with a binary label indicating the presence of Invasive Ductal Carcinoma. In this case, a random undersampling balancing of the images has been performed as there are more than twice as many healthy images as cancerous ones (198,738 healthy vs. 78,786 cancerous ones). After the balancing, the images of the train, validation, and test sets were also randomly selected, as shown in
Table 1.
3.2. Preprocessing
The impact of the pre-processing techniques on deep learning frameworks is of great relevance [
50]. There are a large number of proposals and methodologies in pre- and post-image processing in digital pathology [
51]. In this section, we will present the most frequent ones that appear in the bibliography.
3.2.1. Convolution-Based
A convolution is a filter in which the neighborhood of each pixel is organized by weighted average, according to a matrix that we call the convolution kernel [
52]. These kernels are variable in size and shape producing different effects on the image.
Mathematically, the convolution operation can be represented as follows [
53].
Let
be a grayscale image. Let
be the convolution kernel matrix, where
,
with
n,
m non negatives integers. The result of applying the convolution
M to the image
I is the image
where for each
where
,
,
,
.
In this paper we have used some non-parametric filters from the Python Pillow library [
54]. Although they are different in size and the effect they have on the image to which they are applied, they are all based on convolution and the only thing that varies is the kernel.
Table 2 shows the matrices of the filters used, which are as follows:
Blur. Applies a convolution with a kernel of size 5 × 5 which will result in a slightly blurred image. Because of the shape of the kernel, the central pixels will be ignored, thus losing some information.
Contour. Convolute with a 3 × 3 filter. This filter detects the contours of shapes in images.
Detail. It applies a 3 × 3 size convolution, giving greater importance to the central pixel compared to its neighborhood, thus achieving an enhanced image.
Edge Enhance. Applies a convolution with a 3 × 3 kernel. It aims to improve the quality and definition of the edges of an image.
Edge Enhance More. As in the previous filter, a 3 × 3 convolution is applied to improve the definition of the edges. The only difference is that the central value is changed, making the edges better defined than with the previous filter.
Sharpen. This filter convolves a 3 × 3 kernel to generate a cleaner image. It is used to sharpen the edges to improve their quality. It even increases the contrast between the light and dark areas of the image to improve its characteristics.
3.2.2. Histogram-Based
A histogram is a graphical representation that shows the frequency with which a certain intensity appears in an image, i.e., it represents the number of pixels of each intensity in an image. On the X-axis is usually placed the intensity of the pixels while on the Y-axis the frequency with which the pixels appear. The intensity level usually ranges from 0 to 255. For a grayscale image, there is only one histogram, while an RGB color image will have three histograms, one for each color.
The methods in this section are based on applying a transformation on the image, so that the histograms acquire a uniform distribution. This type of transformation is used to improve the contrast of the images [
55].
Histogram Equalization (HE). Histogram equalization is one of the most common ways to enhance the contrast of an image. This is because it is easy to implement and does not require too many computational resources. It is performed by mapping the gray levels of the image based on the probability distribution of the input gray levels. The detailed procedure can be found at [
56]. However, one of the main problems it presents is that it tends to change the brightness of the image significantly and therefore causes the output image to be saturated with very bright or dark intensity values.
Constrast Limited Histogram Equalization (CLAHE). Conventional histogram equalization considers the overall contrast of the image. In many cases, it is not a good idea because when the histogram presents a distribution that is concentrated in a range, it does not work properly [
57]. To solve this problem, adaptive histogram equalization is used, which is carried out as follows. First, the image is divided into small blocks of a predefined size. Then, each of these blocks is histogram equalized as in the previous section. So in an area, the histogram will be limited to a small region unless there is noise in which case it will be amplified. To avoid this behavior of the algorithm, contrast limiting is applied. If any bin in the histogram is above a certain contrast threshold, those pixels are clipped and evenly distributed to other bins before equalization is applied. Then, to remove edge artifacts, bilinear interpolation is applied. The default parameters offered by the OpenCV library for this method have been used, namely: clipLimit = 2.0, tileGridSize = (8,8).
Fuzzy Histogram Equalization (FHE). To perform contrast enhancement of the images by means of fuzzy histogram equalization, the gray level intensities of the image are mapped to a fuzzy space using membership functions to enhance the contrast [
58]. Then the fuzzy plane is mapped to the gray intensities of the image. This process is intended to obtain an image with better contrast than the original image by giving more importance to the gray levels that are closer to the mean than to those that are farther away. Fuzzy image processing consists of three stages: fuzzification, membership plane operations, and defuzzification. The first stage consists of assigning certain membership values to the images based on some attributes of the images such as brightness, homogeneity, etc. Then, using a fuzzy approach, the membership values assigned in the first stage are modified. Finally, in the defuzzification stage, the membership values that have been treated in the previous stage are decoded and transformed back into the gray level plane. In general, dark pixels are assigned low membership values and bright pixels are assigned high membership values [
59]. The pipeline used is from [
60].
An example of the effect of the different filters, both in healthy images (category 0) and in images with cancer (category 1), for the different filters analyzed, is presented in
Figure 2.
3.3. Models Used
Five deep learning models have been selected to use architectures that are diverse in size and performance. We have not tried to make an exhaustive analysis of which models to select, but rather, we have taken models that normally show good performance in image classification tasks and whose architectures are varied in the number of parameters and layers. Thus, the models used were
VGG16 and
VGG19 [
61],
MobileNet [
62],
ResNet50 [
63] and
DenseNet121 [
64]. These five models have been widely used in the bibliography [
65,
66,
67,
68,
69,
70,
71].
VGG16 and
VGG19. Karen Simonyan and Andrew Zisserman proposed the idea of the VGG network in 2013 and presented the current model based on this idea at the 2014 ImageNet Challenge [
61]. VGG is a special convolutional network designed for classification. They named it VGG after the department of the Visual Geometry Group at Oxford College to which they belonged, and because it is a convolutional neural network that is either 16 layers deep (VGG-16) or 19 layers deep (VGG-19). The VGG16 and VGG19 architecture in Deep Learning is a rather simple architecture that uses only blocks consisting of an increasing number of convolutional layers with filters of size 3 × 3. Moreover, to reduce the size of the activation maps obtained, max-pooling blocks are inserted between the convolutional layers, reducing the size of these activation maps by half. Finally, a classification block is used, consisting of two dense layers of 4096 neurons each and a final layer, the output layer, of 1000 neurons.
MobileNet. This model was proposed in 2017 and it is based on a simplified architecture with depth-separable convolutions. The model was originally developed for mobile imaging applications. The architecture of MobileNet initially includes a convolutional layer with 32 filters, followed by 19 layers. MobileNet uses 3 × 3 filters, dropout (an operation in which neurons generate partial learning of the network to avoid overtraining), and normalization during training [
72,
73]
ResNet. This deep learning model is a deep convolutional neural network with 50 layers. The architecture of ResNet-50 starts with a convolution with a kernel size of 7 × 7 and 64 different kernels, all with a step size of 2, so we get one layer. Then there are different convolutional layers with different kernels [
74,
75].
DenseNet-121. The denseNet-121 model of densely connected convolutional networks, proposed in late 2017, consists of networks with blocks where each block has direct access to the gradient of the cost function, the function that provides the error, and the initial input of the block. Of course, there are also convolutional layers and pooling layers [
76].
Table 3 summarizes different deep learning models that were used in this paper alongside pre-trained weights (using information from Keras Applications). Depth refers to the topological depth of the network (including activation layers, batch normalization layers, etc).
3.4. Experimental Settings
A total of three different runs were performed, using a transfer learning technique, for each filter and each model by randomly selecting the training, validation, and test sets.
Different scenarios can be considered for transfer learning. The first scenario would be to have a large dataset (in our case images), but different from the pre-trained model’s dataset and to be able to train the entire model, being useful to initialize the model from a pre-trained model, using its architecture and weights. The second scenario would be when the set of images that you want to train has a certain similarity with the images that were used to train the model, or when you have a small set of images to train the new system. In this case, it is usually chosen to train some layers and leave others frozen. Finally, the third scenario would be to have a small set of images, similar to the pre-trained model’s dataset. In that case, they usually freeze the convolutional base.
The scenario used in this article has been the first scenario: the model is initialized from a pre-trained model (using its architecture and weights), and the entire model is trained/adjusted (no layer was frozen).
Therefore, the procedure that was followed for each filter and deep learning model is the following. First, the model pre-trained with the Imagenet dataset is loaded, removing the last default layers, and replacing them with layers that offer an adequate solution to the problem to be solved (flatten layer). On the other hand, after some experiments in cases where the models suffered from overfitting, measures would be incorporated to avoid its behavior, so we embed regularization kernels and a dropout layer that 20% of the neurons were removed. Finally, a fully connected layer whose activation function is a sigmoid. This results in a single output. Additionally, in this last layer,
and
kernels have been applied to regulate training. These kernels are penalty factors added to the loss function so that abrupt changes in the values of the weights are penalized during training to avoid over-fitting the models. Therefore, the usual transfer learning procedure was used. Relying on prior learning, it avoids starting from scratch and allows us to build accurate models in a time-saving manner [
77,
78,
79].
The input to the different deep learning models is the breast cancer histopathology images. These images received the appropriate scaling for the expected input to each neural network. An Adam optimizer with a learning rate of was used. During training, the binary cross entropy was used as the loss function and a total of 50 epochs were iterated for each model. However, in general, not so many iterations were performed because early stopping with a latency of 15 was used in terms of validation accuracy, with the weights of the best result recovered at the end of training. A batch size of 128 was used.
3.5. Evaluation Metrics
To measure the performance of the models and compare the effect of preprocessing on them, we will use the metrics accuracy, precision, recall, and AUC.
Accuracy. It is a general measure that quantifies the proportion of hits in the classification. Given the number of true positives (
TP), false positives (
FP), true negatives (
TN), and false negatives (
FN), the accuracy is defined as:
Precission. Quantifies actual positives versus all images that have been classified as positive.
Recall. It measures the number of well-classified positives versus actual positives.
AUC. In addition, we will use the area under the curve. Both the ROC curve and the
AUC are a measure of the overall performance of a binary classification model. It measures the ability of the model to correctly classify positives at different thresholds. The higher the
AUC the better the model is at predicting. A perfect model would have an
AUC of 1 and a random classifier would have a value of 0.5 [
80].
4. Experimental Results
Table 4 shows all the information provided by the runs performed. In addition to the Accuracy and AUC metrics considered in the analysis of ANOVA, the complementary measures for Precision and Recall are also provided.
To perform the statistical study, a selection is made from a set of alternatives representative of each of the factors considered for performing the deep learning training procedure. The factors considered in the analysis are listed in
Table 5. By analyzing the different levels of each of these factors, it is possible to determine their influence on the performance of the analyzed system (accuracy) when different alternatives for preprocessing the images and also different deep learning models are presented.
Table 6 gives the two-way variance analysis for the whole set of processing examples of the deep learning system analyzed in this contribution. The ANOVA table containing the sum of squares, degrees of freedom, mean square, test statistics, etc., represents the initial analysis in a compact form. This kind of tabular representation is customarily used to set out the results of the ANOVA calculations. The table ANOVA decomposes the variability of the AUC into contributions due to different factors. Because the sums of squares are of type III (the default), the contribution of each factor is measured after the effects of all other factors have been removed. The P-values test the statistical significance of each factor. Since a P-value is less than 0.05, this factor has a statistically significant effect on the AUC at a 95% confidence level.
Of all the information presented in the table ANOVA, the researcher’s main interest is likely to be focused on the value in the “F-ratio” and “
p-value” columns If the numbers in this column are smaller than the critical value set by the experimenter, the effect is considered significant. This
p-value is often set at 0.05. Any value lower than this leads to significant effects, while any value higher than this leads to non-significant effects [
35]. When effects are found to be significant using the procedure described above, it means that there is more difference between the means than would be expected from chance alone. With respect to the experiment above, this would mean that the preprocessing filter of the WSI images has no significant effect on the accuracy of the system. As can be seen from
Table 6, the model factor is statistically significant.
Thus, a detailed analysis will be performed now for each of the factors examined, using the Multiple Range Test for the two factors (model and filter) in which a multiple comparison procedure is carried out to determine which means are significantly different from which others. The method currently being used to discriminate among the means is Fisher’s least significant difference (LSD) procedure [
36]. A summary of this procedure can be seen in
Table 7.
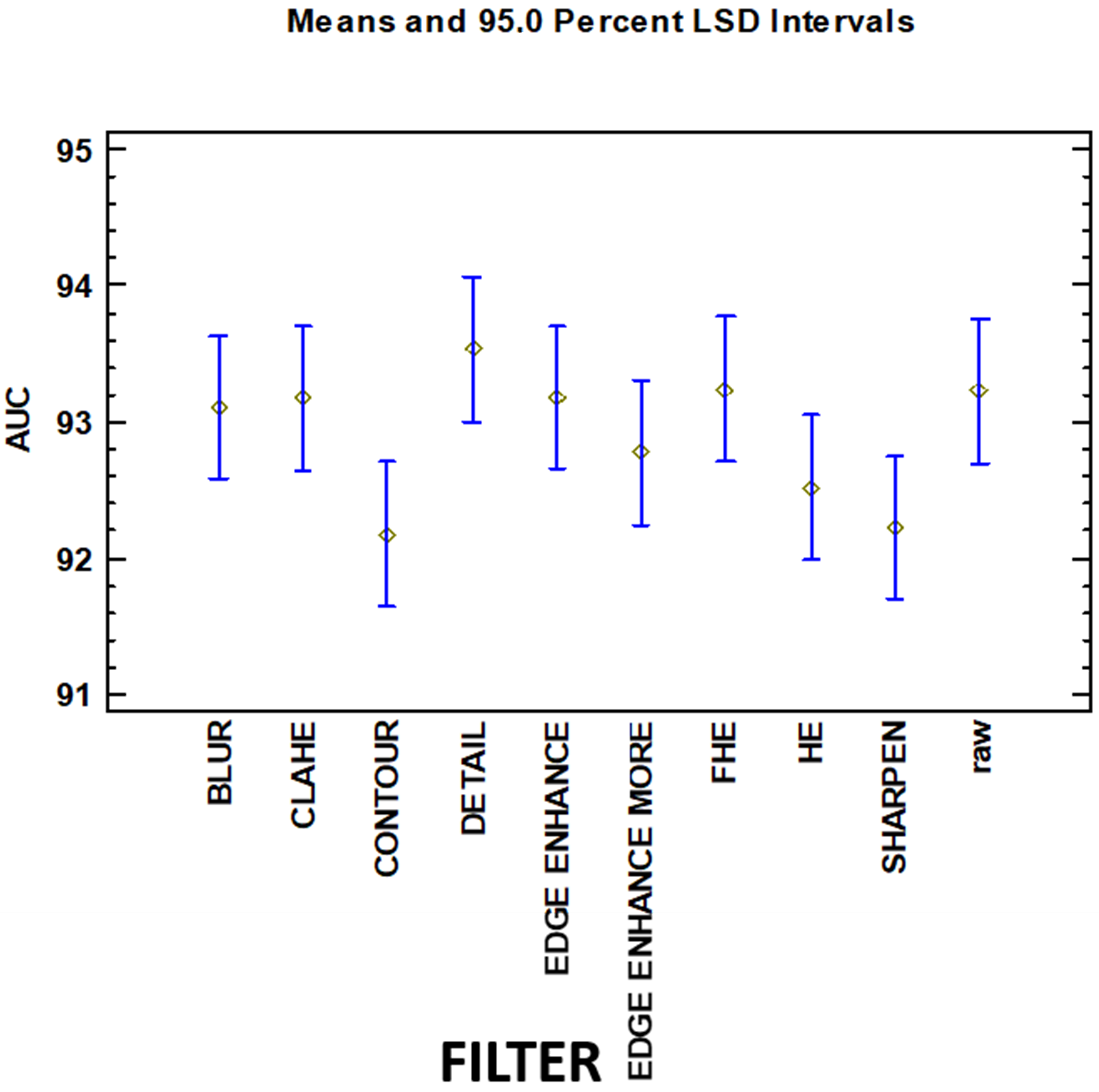
From this test can be observed that there exist three homogenous groups (identified using columns of X’s, with intersection) for the variable filter. All of the groups have intersections (therefore, different levels of these groups have the same behavior over the AUC variable). These groups are:
Contour, Sharpen, HE, Edge Enhance More, Blur, CLAHE, Edge Enhance, Raw
Sharpen, HE, Edge Enhance More, BLUR, CLAHE, EDGE ENHANCE, raw, FHE
HE, Edge Enhance More, BLUR, CLAHE, Edge Enhance, Raw, FHE, Detail
The levels of these 3 groups are not significant from a statistical point of view (analyzing the behavior of the dependent output variable) having an intersection between all of them. This information can be graphically represented in
Figure 3.
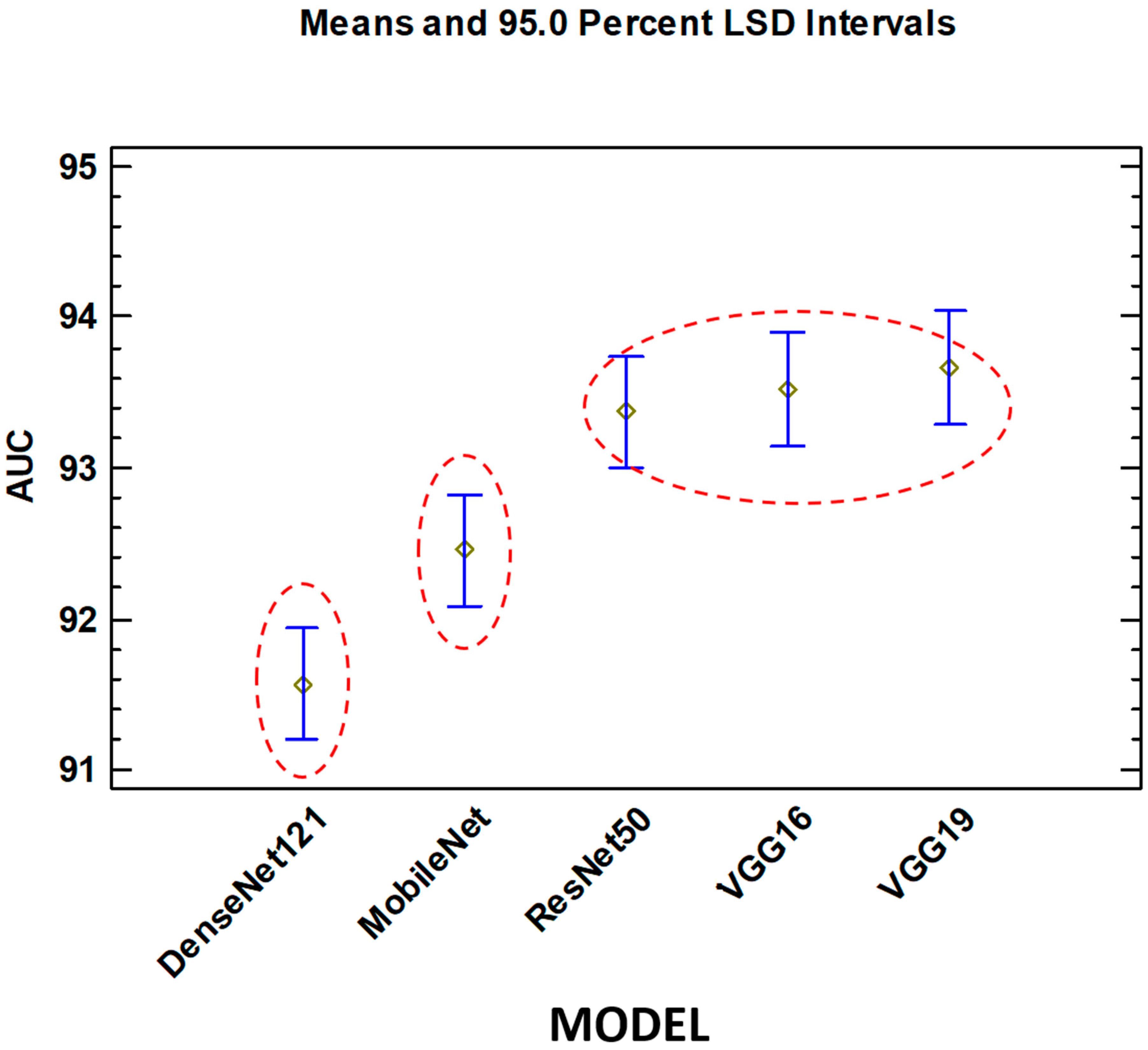
On the other hand, with respect to the Model factor, there are three different groups without intersection.
DenseNet121
MobileNet
ResNet50, VGG16, VGG19
The best group (best AUC) is the third. VGG19 obtains the best results. This factor is statistically significant.
Figure 4 shows that, unlike the filter factor, the groups are well differentiated.
5. Discussion
As mentioned in
Section 2, there are a large number of publications in the literature on the use of deep learning systems in medical image classification, especially in histopathology. Image pre-processing is an important step, and as can be seen from the bibliography, there are a large number of alternatives for its implementation (in this paper we have used the most frequently presented ones in the literature). On the other hand, there are a large number of deep learning models in the literature, and to perform statistical analysis with great rigor and accuracy, all possible combinations of filters and models must be simulated (in several repetitions ), the behavior analyzed, and the error rates measured. The computational cost and time required for each simulation are high. For this reason, it was decided to select these five models that have been widely used in the literature. However, to the best of our knowledge, there is no exhaustive statistical analysis in the literature that attempts to analyze what relevance or impact it has on the behavior of the system to use the different alternatives of deep learning models and preprocessing of histological images for the problem of breast cancer.
As mentioned in
Section 4, the analysis of the
p-value in
Table 6 yields a value of 0.146 for the filter technique and a value of 0.0000 for the model of deep learning. This means that the choice of the different deep learning model alternatives has a statistically significant influence on the behavior of the system (measured by the AUC index). On the other hand, for the filter effect, it can be stated that it is not statistically significant, and therefore, the different methods have similar behavior to obtain the AUC value. This means that the designer of a deep learning system, for the problem of histopathological cancer images studied, must focus more attention on the deep learning model to be used, than on the processing systems used. The three homogeneous groups of filter types intersect each other, therefore, from a statistical point of view they are equivalent in terms of their behavior on the AUC output variable. It is also important to note that different convolution-based and histogram-based filter alternatives have been used. These alternatives, of these two major types of pre-processing, are mixed in the three homogeneous groups of
Table 7. No statistically significant difference was found when using convolution-based or histogram-based types, both methods have similar behavior for the output variable.
For the deep learning model type factor, there are also three groups, but, in this case, there are statistically significant differences between them. For this particular problem, and when analyzing and discussing the results, it can be seen that the group that achieves the best results (consisting of VGG16, VGG19, and ResNet50) are deep learning models that have a large number of parameters and therefore their size (measured in MB) and complexity is high. This does not carry over to the depth parameter (depth refers to the topological depth of the network, including activation layers, batch normalization layers, etc).
As might be initially expected, VGG16 and VGG19 behave similarly from a statistical point of view, and both produce the best results. While it is true that both have a larger number of parameters out of the five deep learning models analyzed, their depth topology is still the lowest. As strengths of this paper, we must highlight the novelty and robustness of performing a comprehensive statistical analysis of the impact that the different pre-processing algorithms and deep learning models have on the classification of histopathological images in breast cancer. As a possible weakness of the study, we point out that it would have been interesting to analyze other models of deep learning systems, preprocessing, and even other pathologies. This weakness is due to the high computational cost and time required to run multiple simulations for each of the combinations of filters and deep learning models.
6. Conclusions and Future Works
There is an extensive bibliography on the use of artificially intelligent systems for the classification and decision support of histopathological images. In particular, various deep learning models and image preprocessing algorithms have been proposed and used to obtain accurate systems. However, it is necessary to perform a comprehensive statistical analysis of the influence of the preprocessing stage and deep learning models on the system behavior.
In this paper, it was proposed to use 50 different combinations (5 deep learning models and 10 image preprocessing alternatives) to statistically analyze the behavior of the system accurately through the ANOVA analysis. Accuracy, precision, recall, and AUC metrics were used to quantify the performance of the models. Using the ANOVA test, it was shown that the different alternatives of the filter have similar behavior in the accuracy indexes of the system. Furthermore, with the ANOVA test, it became clear that the choice of the architecture of the deep learning models used is statistically relevant. In future work, we propose to perform a similar statistical analysis and examine a wider variety of architectures to determine which models perform best.