Knowledge-Based Framework for Selection of Genomic Data Compression Algorithms
Abstract
1. Introduction
2. Materials and Methods
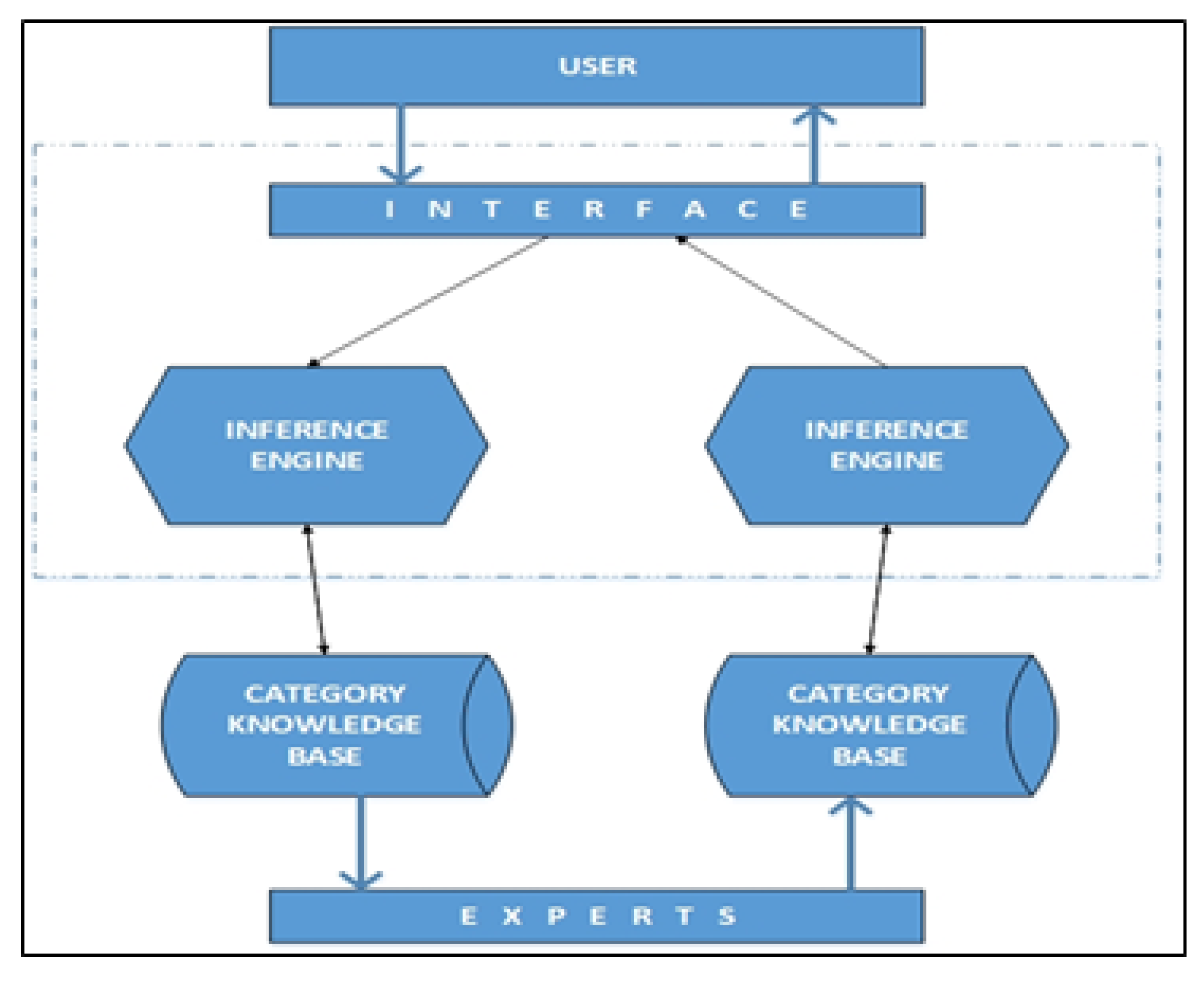
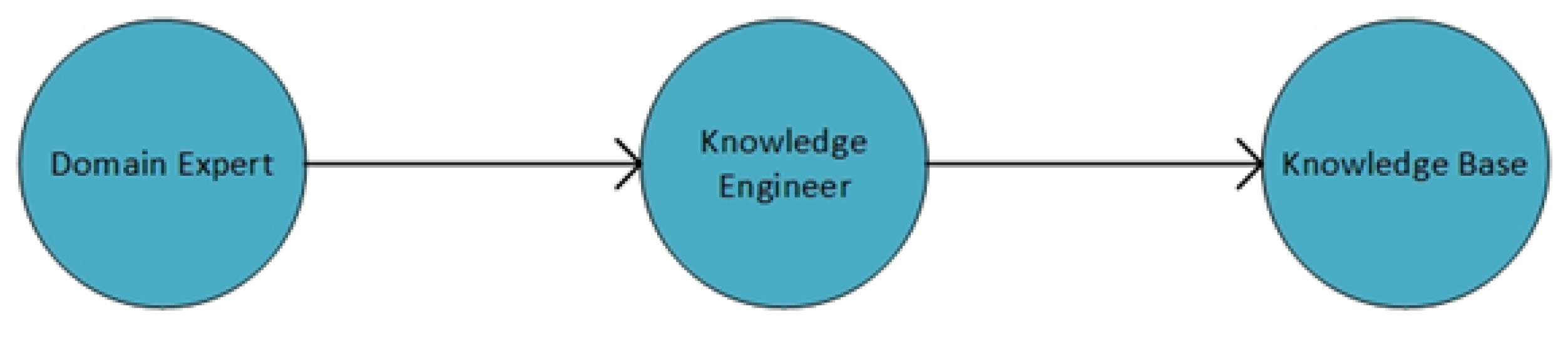
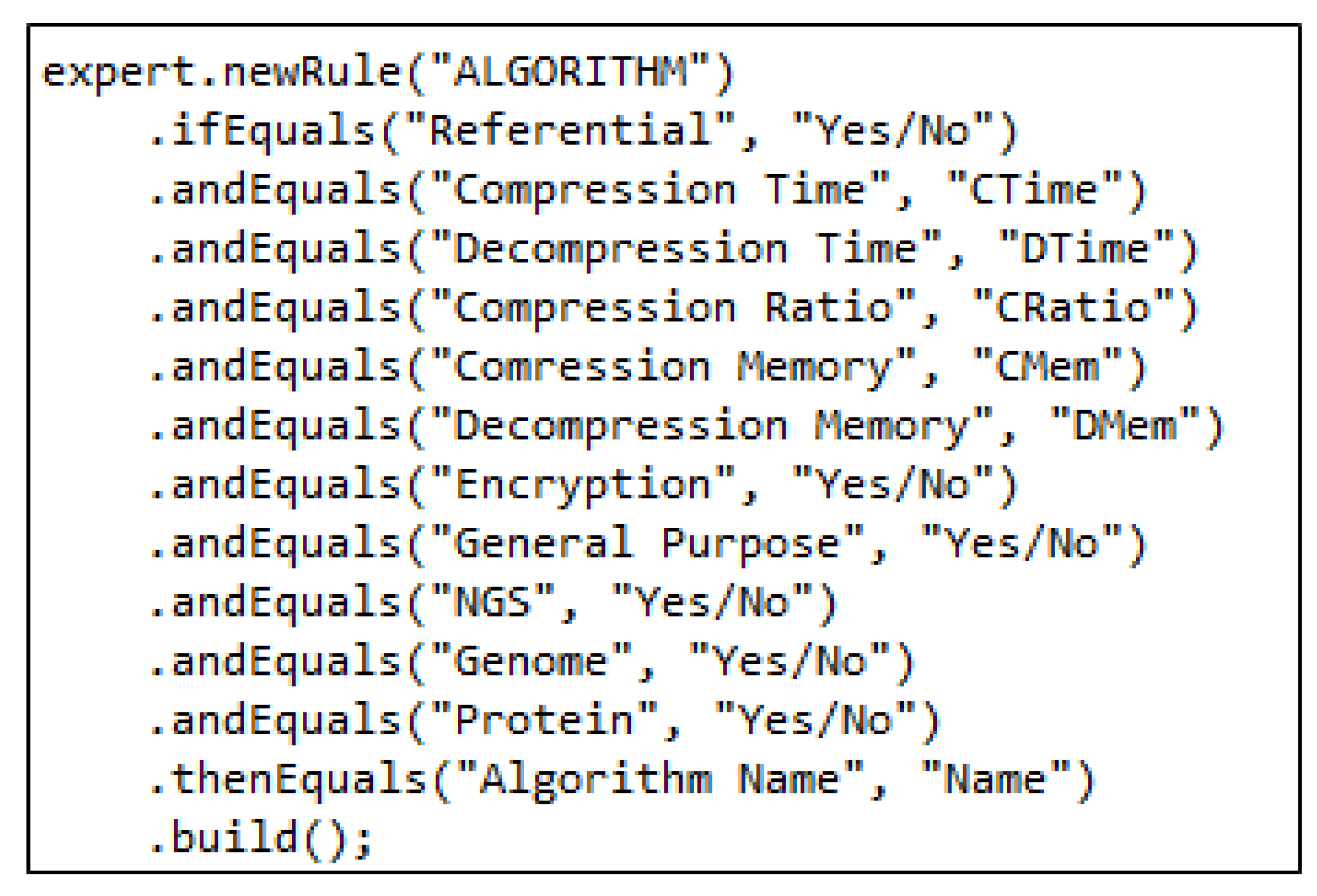
2.1. Knowledge Base
| Algorithm 1: Pseudocode of rule base. |
 |
| Algorithm 2: Pseudocode of rule matching based on properties of each rule. |
 |
2.2. Inference Mechanism
3. Case Studies
3.1. Input Case 1
3.2. Input Case 2
3.3. Input Case 3
3.4. Input Case 4
3.5. Input Case 5
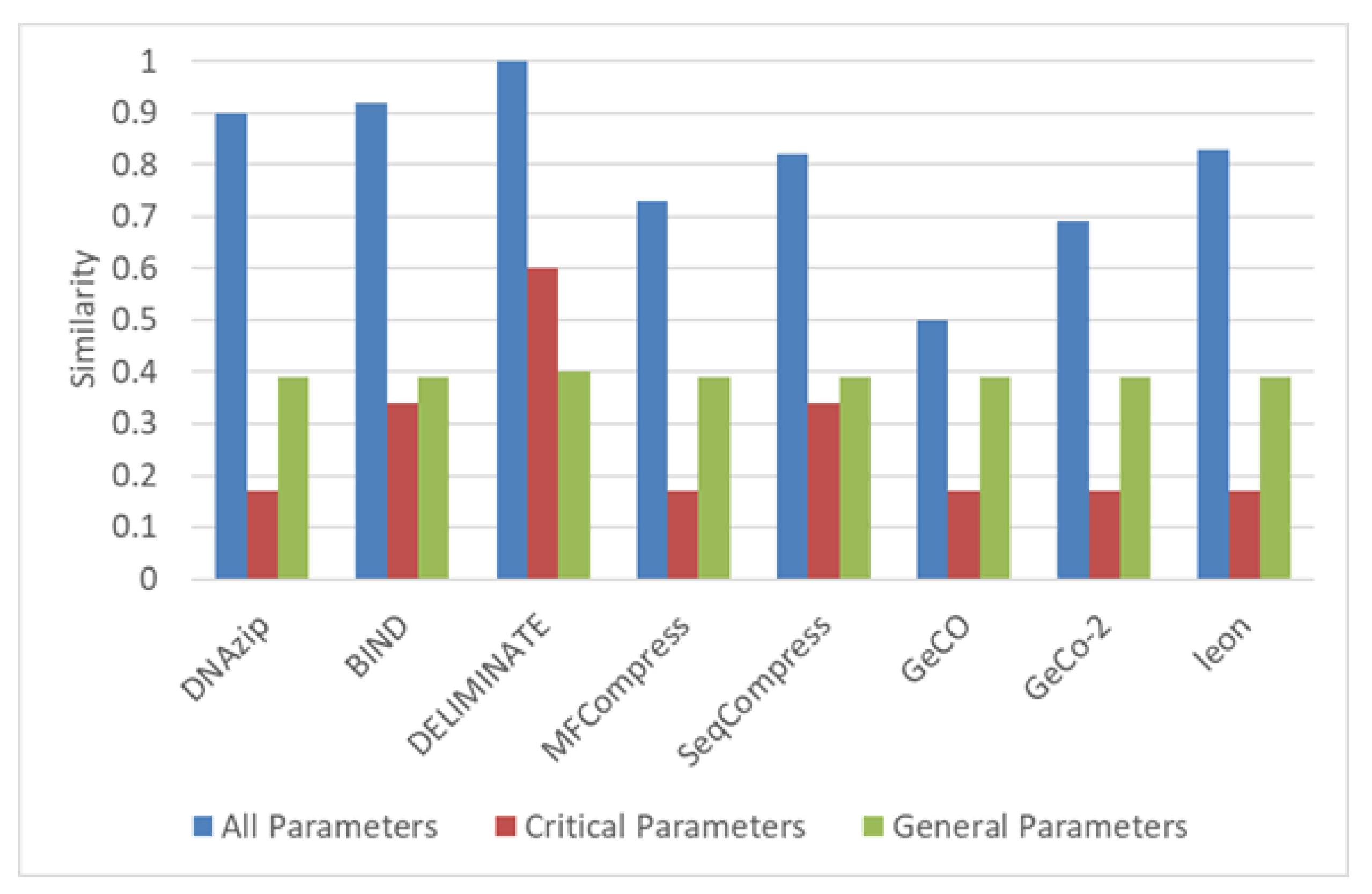
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deorowicz, S.; Grabowski, S. Data compression for sequencing data. Algorithms Mol. Biol. 2013, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Sardaraz, M.; Tahir, M. SCA-NGS: Secure compression algorithm for next generation sequencing data using genetic operators and block sorting. Sci. Prog. 2021, 104, 00368504211023276. [Google Scholar] [CrossRef] [PubMed]
- Gzip Home Page. Available online: https://www.gzip.org (accessed on 16 December 2019).
- Bzip2 Home Page. Available online: http://www.bzip.org/ (accessed on 16 December 2019).
- 7-Zip Home Page. Available online: https://www.7-zip.org/ (accessed on 16 December 2019).
- Chandak, S.; Tatwawadi, K.; Ochoa, I.; Hernaez, M.; Weissman, T. SPRING: A next-generation compressor for FASTQ data. Bioinformatics 2019, 35, 2674–2676. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Haque, M.M.; Bose, T.; Reddy, C.V.S.K.; Mande, S.S. FQC: A novel approach for efficient compression, archival, and dissemination of fastq datasets. J. Bioinform. Comput. Biol. 2015, 13, 1541003. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, I.; Hernaez, M.; Weissman, T. iDoComp: A compression scheme for assembled genomes. Bioinformatics 2015, 31, 626–633. [Google Scholar] [CrossRef]
- Roguski, Ł.; Deorowicz, S. DSRC 2—Industry-oriented compression of FASTQ files. Bioinformatics 2014, 30, 2213–2215. [Google Scholar] [CrossRef]
- Sardaraz, M.; Tahir, M. FCompress: An Algorithm for FASTQ Sequence Data Compression. Curr. Bioinform. 2019, 14, 123–129. [Google Scholar] [CrossRef]
- Sardaraz, M.; Tahir, M.; Ikram, A.A.; Bajwa, H. SeqCompress: An algorithm for biological sequence compression. Genomics 2014, 104, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Sardaraz, M.; Tahir, M.; Ikram, A.A. Advances in high throughput DNA sequence data compression. J. Bioinform. Comput. Biol. 2016, 14, 1630002. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, Y.; Ji, Z.; He, S.; Yang, X. High-throughput DNA sequence data compression. Briefings Bioinform. 2013, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- İpek, M.; Selvi, İ.H.; Findik, F.; Torkul, O.; Cedimoğlu, I. An expert system based material selection approach to manufacturing. Mater. Des. 2013, 47, 331–340. [Google Scholar] [CrossRef]
- Geysen, D.; De Somer, O.; Johansson, C.; Brage, J.; Vanhoudt, D. Operational thermal load forecasting in district heating networks using machine learning and expert advice. Energy Build. 2018, 162, 144–153. [Google Scholar] [CrossRef]
- Raghavendra, U.; Bhandary, S.V.; Gudigar, A.; Acharya, U.R. Novel expert system for glaucoma identification using non-parametric spatial envelope energy spectrum with fundus images. Biocybern. Biomed. Eng. 2018, 38, 170–180. [Google Scholar] [CrossRef]
- Khan, A.R.; Rehman, Z.U.; Amin, H.U. Knowledge-Based systems modeling for software process model selection. Int. J. Adv. Comput. Sci. Appl. 2011, 2, 20–25. [Google Scholar]
- Grobelny, P. The expert system approach in development of loosely coupled software with use of domain specific language. In Proceedings of the 2008 International Multiconference on Computer Science and Information Technology, Wisla, Poland, 20–22 October 2008; IEEE: Piscataway, NJ, USA, 2008; pp. 119–123. [Google Scholar]
- Lujić, R.; Samardžić, I.; Galzina, V. Application of expert systems for selection of installation pipes. Teh. Vjesn. 2015, 22, 241–245. [Google Scholar] [CrossRef]
- Bakeer, H.M.S. Photo Copier Maintenance Knowledge Based System V. 01 Using SL5 Object Language. Int. J. Eng. Inf. Syst. 2017, 1, 116–124. [Google Scholar]
- Rao, R.V.; Davim, J.P. A decision-making framework model for material selection using a combined multiple attribute decision-making method. Int. J. Adv. Manuf. Technol. 2008, 35, 751–760. [Google Scholar] [CrossRef]
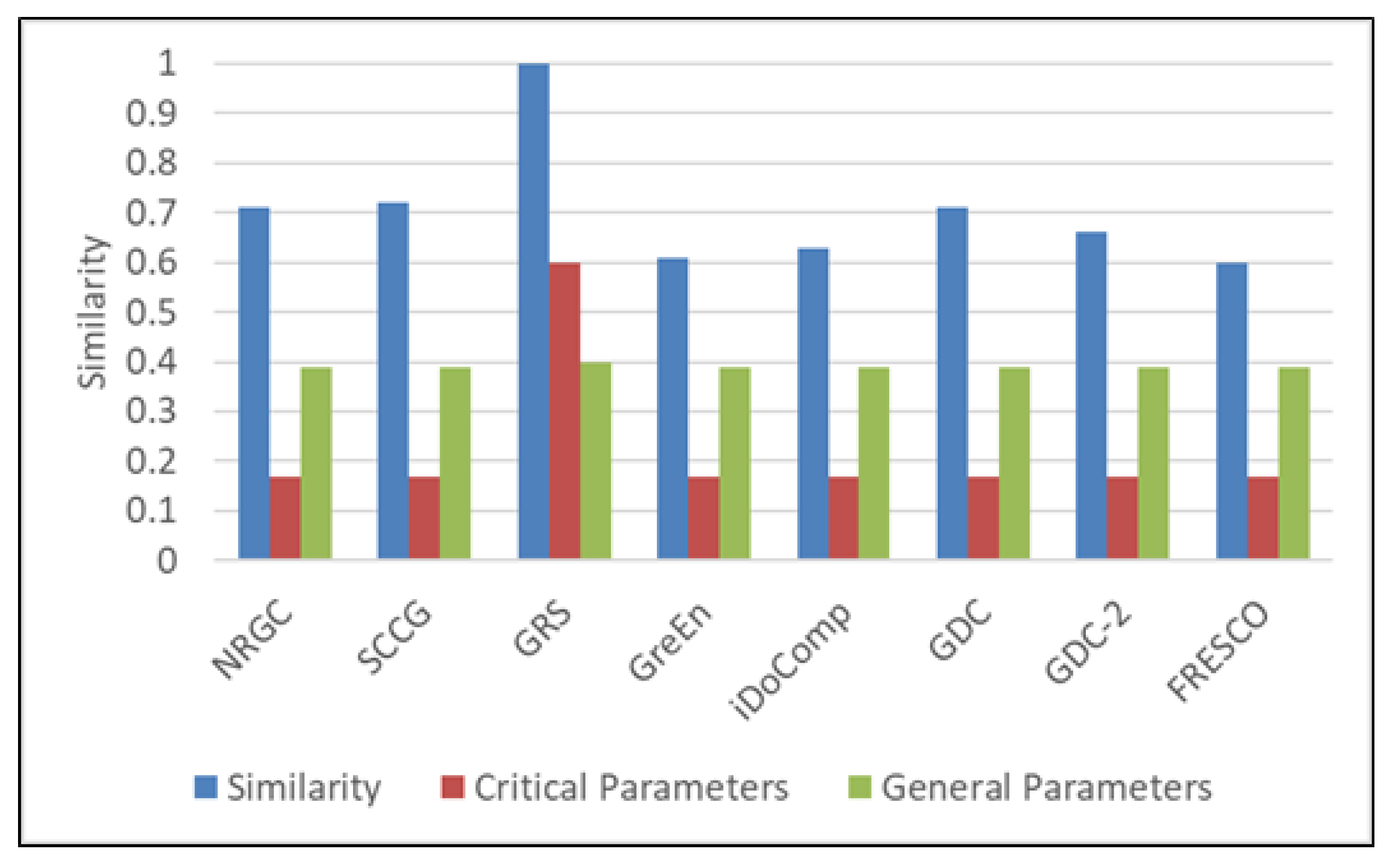
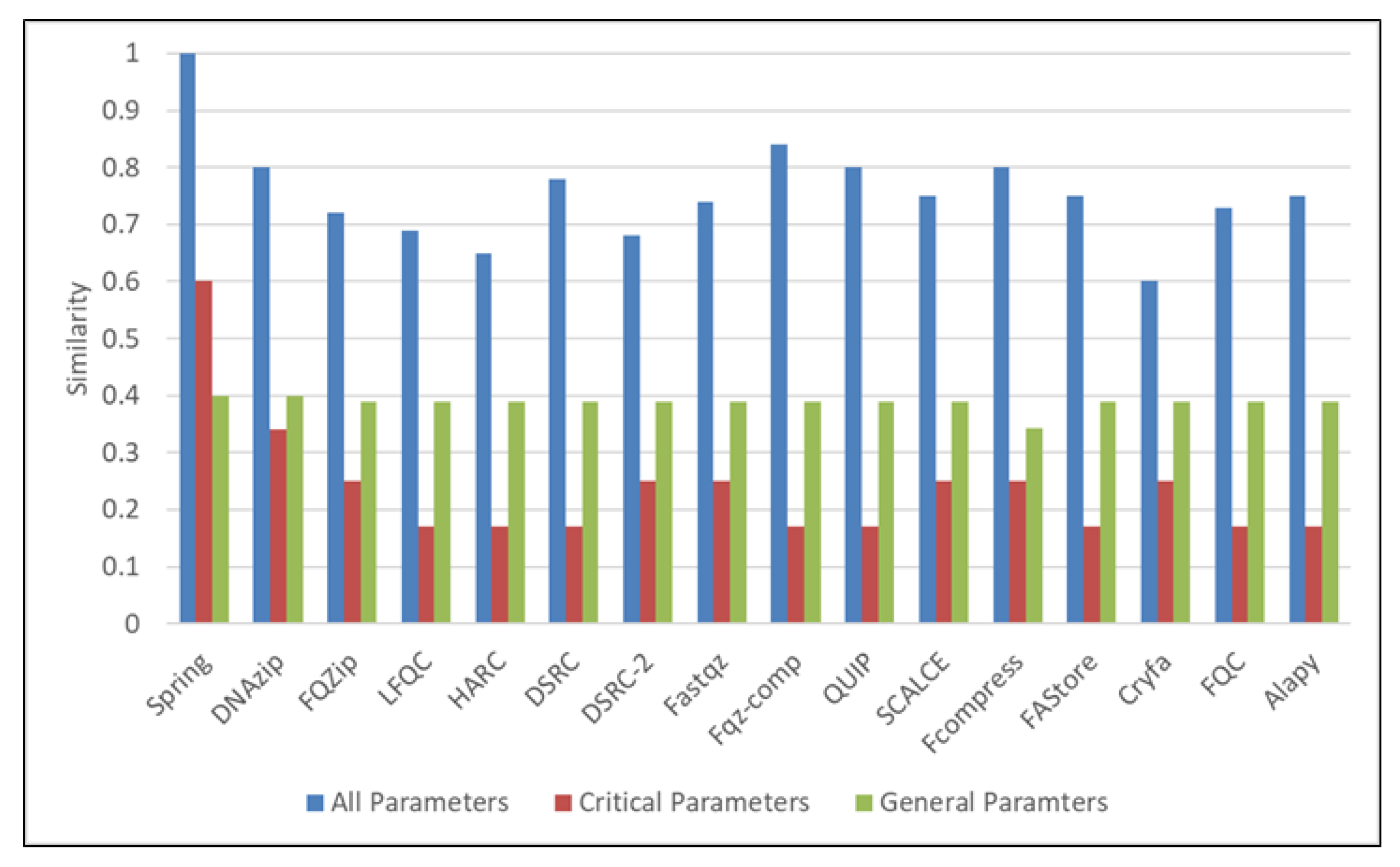
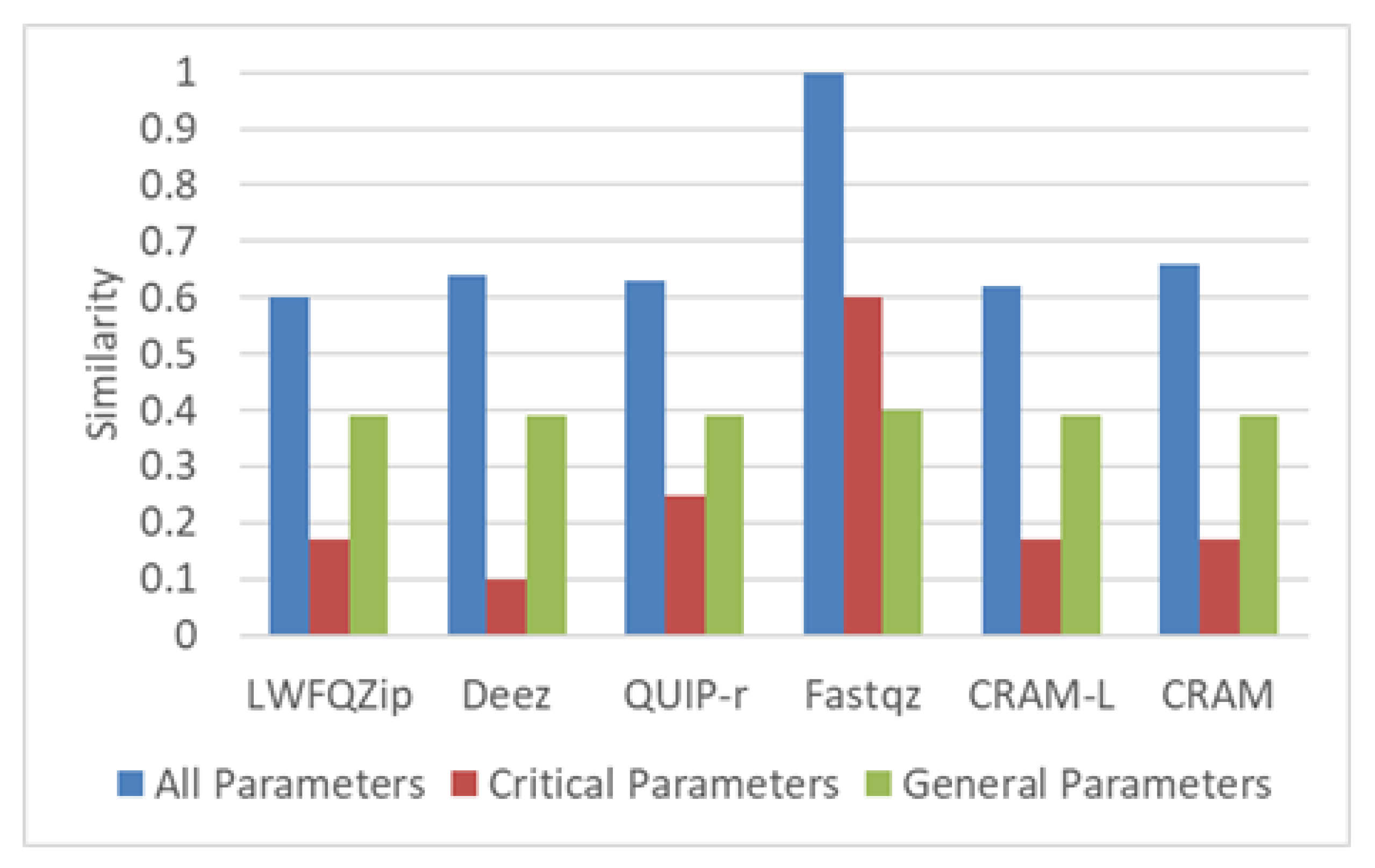
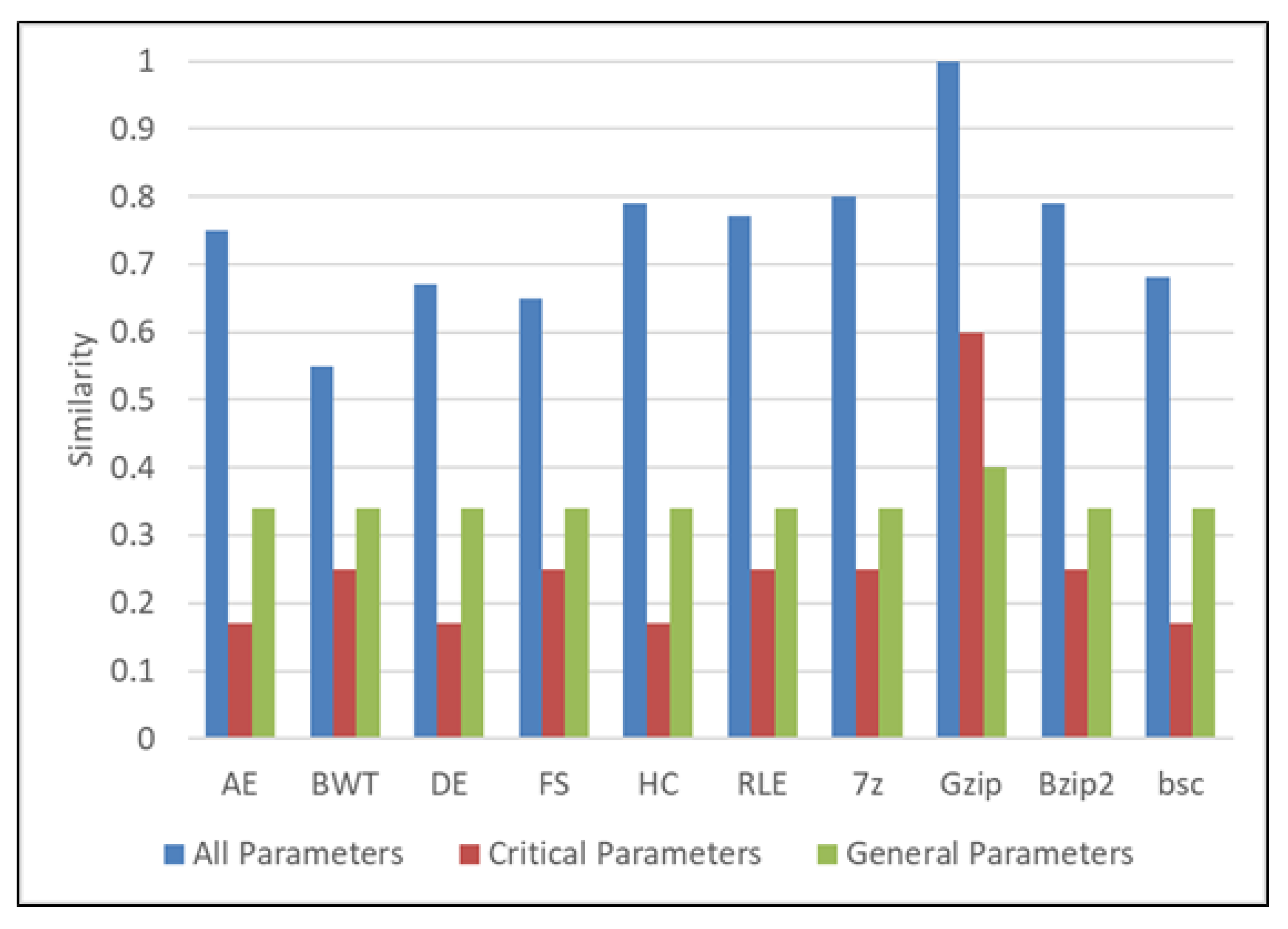
| Parameters | Values |
|---|---|
| Compression Time | Low/ low moderate/moderate/high moderate/ high |
| Compression Memory | Low/ low moderate/moderate/high moderate/ high |
| Compression Ratio | Low/ low moderate/moderate/high moderate/ high |
| Decompression Time | Low/ low moderate/moderate/high moderate/ high |
| Decompression Memory | Low/ low moderate/moderate/high moderate/ high |
| Protein | Yes/ No |
| NGS | Yes/ No |
| Genome | Yes/ No |
| General | Yes/No |
| Encryption | Yes/ No |
| Referential | Yes/No |
| Rule Number | Rules |
|---|---|
| 1 | IF type = NGS AND mode = referential AND Ctime = low moderate AND Cmem = low AND Cratio = high AND Dtime = low moderate AND Dmem = low AND OS = linux AND programming language = C++ AND encryption = no THEN algorithm is Alapy |
| 2 | IF type = genome AND mode = referential = AND Ctime = low moderate AND Cmem = low AND Cratio = moderate AND Dtime = low moderate AND Dmem = low AND encryption = no THEN algorithm is Deliminate |
| n | IF Type = genome AND mode = referential AND Ctime = high AND Cmem = moderate AND Cratio = moderate AND Dtime = high moderate AND Dmem = low moderate AND encryption = no THEN algorithm is GDC |
| Datasets | Species | Read Length | Number of Reads | File Size (MBs) | Reference Genome |
|---|---|---|---|---|---|
| SRR801793 | L. pneumophila | 2 × 100 | 10,812,922 | 2818.11 | NC_018140 |
| ERR022075 | E. Coli | 2 × 101 | 45,440,200 | 11,253.16 | NC_000913 |
| SRR125858 | Homo sapiens | 2 × 76 | 124,815,011 | 52,172.64 | Chr21_GRCh37 |
| Datasets | Species | Genome Length | File size (MBs) | Reference Genome |
|---|---|---|---|---|
| KOREF2009024 | Homo sapiens | 3,069,535,988 | 2986.7 | KOREF 20090131 |
| TAIR10 | A. thaliana | 119,667,743 | 115.56 | TAIR9 |
| NC_017526 | L. pneumophila | 2,682,626 | 2.59 | NC_017525 |
| NC_017652 | E. coli | 5,038,385 | 4.87 | NC_017651 |
| sacCer3 | S. cerevisiae | 12,157,105 | 11.82 | sacCer2 |
| Ce10 | C. elegans | 100,286,070 | 97.55 | Ce6 |
| Input Case | Type | Mode | CT | CM | CRa | DT | DM | Enc |
|---|---|---|---|---|---|---|---|---|
| 1 | Genome | Reference Free | LM | L | M | LM | L | No |
| 2 | Genome | Referential | HM | L | H | H | L | No |
| 3 | NGS | Reference Free | M | HM | LM | L | H | No |
| 4 | NGS | Referential | H | H | M | H | H | No |
| 5 | General | Reference Free | M | M | HM | L | M | No |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alourani, A.; Tahir, M.; Sardaraz, M.; Khan, M.S. Knowledge-Based Framework for Selection of Genomic Data Compression Algorithms. Appl. Sci. 2022, 12, 11360. https://doi.org/10.3390/app122211360
Alourani A, Tahir M, Sardaraz M, Khan MS. Knowledge-Based Framework for Selection of Genomic Data Compression Algorithms. Applied Sciences. 2022; 12(22):11360. https://doi.org/10.3390/app122211360
Chicago/Turabian StyleAlourani, Abdullah, Muhammad Tahir, Muhammad Sardaraz, and Muhammad Saud Khan. 2022. "Knowledge-Based Framework for Selection of Genomic Data Compression Algorithms" Applied Sciences 12, no. 22: 11360. https://doi.org/10.3390/app122211360
APA StyleAlourani, A., Tahir, M., Sardaraz, M., & Khan, M. S. (2022). Knowledge-Based Framework for Selection of Genomic Data Compression Algorithms. Applied Sciences, 12(22), 11360. https://doi.org/10.3390/app122211360







