Abstract
The development of new sequencing technologies has led to a significant increase in biological data. The exponential increase in data has exceeded increases in computing power. The storage and analysis of the huge amount of data poses challenges for researchers. Data compression is used to reduce the size of data, which ultimately reduces the cost of data transmission over the Internet. The field comprises experts from two domains, i.e., computer scientists and biological scientists. Computer scientists develop programs to solve biological problems, whereas biologists use these programs. Computer programs need parameters that are usually provided as input by the users. Users need to know different parameters while operating these programs. Users need to configure parameters manually, which leads to being more time-consuming and increased chances of errors. The program selected by the user may not be an efficient solution according to the desired parameter. This paper focuses on automatic program selection for biological data compression. Forward chaining is employed to develop an expert system to solve this problem. The system takes different parameters related to compression programs from the user and selects compression programs according to the desired parameters. The proposed solution is evaluated by testing it with benchmark datasets using programs available in the literature.
1. Introduction
Genomic data are increasing rapidly, which creates research challenges in the field of bioinformatics. There has been tremendous development in computer hardware and processing power in the last few decades. At the same time, the production cost of biological data has also been reduced significantly. The pace of data production has exceeded the processing capabilities of computer hardware. Some projects recently generated data in terabytes. This huge amount of generated data needs to be stored and analyzed meaningfully [1].
To handle the exponentially growing amount of data, we need some techniques that can dramatically control the size of the data without losing details. Compression is a technique that helps us in dealing with such huge data. Compression programs encode, modify, or decrease the number of bits to represent the data, which ultimately saves storage space, reduces the time needed to perform operations on these data, and also reduces the cost of transferring data over the Internet [2].
General-purpose compressors such as Gzip [3], Bzip2 [4], and 7z [5] are not suitable for biological data compression. These techniques cannot make use of the characteristics of biological data. To solve this problem, many compression programs have been developed with a better compression ratio than general-purpose algorithms. There exists strong literature on algorithms for both genome and NGS data [6,7,8,9,10,11]. The two commonly used methods for compression are lossless and lossy compression. In the former method, the data can be retransformed to their original form. In the latter, the data cannot be retransformed to the original form once they are compressed. It is important to understand the targeted parameters and data of a particular compression program. The compression programs for compressing video files are different than the compressing programs for the compression of text data. The programs developed for text data also target different types of text data, e.g., general text compressors and compressors for biological data.
Comprehensive work has been conducted by many researchers in the field of bioinformatics to handle, analyze, and compress genomic data. Different researchers have developed compression tools for different types of biological data. The programs work on different parameters such as compression ratio, compression/decompression time, memory requirements, etc. Users need to be aware of the relationship between different parameters while selecting a program for compression. Some compression algorithms are not tuned for the compression of DNA sequences and do not give the desired compression results. Different implementations of Huffman coding use DNA sequence features to yield a better compression ratio. These implementations focus on the concept of selecting frequent repetitions to force a skewed Huffman tree and to construct multiple Huffman trees at the time of encoding.
Compression programs are categorized based on the type of data. Two main categories include NGS data and genome data. The tools are further categorized into referential or reference-free compressors. Referential compressors need a reference sequence for compression and decompression, whereas reference-free compressors do not need any other sequence except the target sequence. Readers are referred to review articles for further details of programs in different categories [1,12,13].
Expert systems refer to the application of Artificial Intelligence (AI) having two main components, i.e., a knowledge base and an inference engine. With the massive development of computer technology, the use of machines has increased significantly. With the rapid development in technology, machines are used as experts instead of human experts, which are time-consuming and expensive [14]. The knowledge is integrated into the knowledge base to develop expert systems that can give expert advice in the same way as the human expert of a specific domain. In expert systems, the error rate is also minimum as compared to human experts who deal with a lot of other data of different domains or different purposes.
The literature on expert systems exists in a variety of domains. For efficient thermal load forecasting, an expert system is proposed in [15]. The system combines the different unique mechanisms that are data-driven for thermal load forecasting with the incorporation of machine learning to obtain the best method for thermal load forecasting. To detect glaucoma, an expert system is presented in [16]. The technique is helpful in accurately detecting glaucoma using optic disk localization and non-parametric gist descriptors. Some researchers have worked on expert systems that guide the users and automate the process of selecting suitable software process models according to requirements. The proposed model uses AI to support the software development process by handling this issue using some expert systems [17]. Expert systems can advise engineers in the field of programming without thorough understanding of programming languages. Using the deductive databases and domain knowledge, the users can use and integrate the earlier development of software constituents to implement different functions [18].
Another expert system is proposed based on some input parameters for the selection of installation pipes, where the output is the type of installation pipe. The decision tree is the model of the solution based on the possible combinations of attributes and rules to predict the solution using intelligent databases [19]. An expert system has been proposed to identify and rectify the problem in the photocopying machine through an easy questionnaire. The questions can be answered with either yes or no. The expert system hence attempts to correct the errors by analyzing the answers given by the users. The system is fruitful and a quick problem solver in case manpower is unavailable [20].
Expert systems have also been used for material selection. The availability of different materials, designers, and engineers use different evaluation and selection criteria. Logically, material selection depends on the selection of multiple attributes. Materials are selected by considering attribute quantification and positive points [21].
The users of compression programs face difficulties while selecting different compression programs from a pool of available programs according to their needs. The process is time-consuming and costly in terms of storage, and the chances of unsuitable solutions are high. Users also need knowledge of different parameters related to a program. This process needs to be optimized and automated to make it easier for users, even if they have less knowledge of computers. In this article, we present an expert system that can overcome this problem. We propose an expert system that asks the user to select different parameters, and the system selects suitable programs according to the users’ requirements. The program then asks the user to enter the desired parameters for the compression algorithm selected. The data are compressed according to the given input.
2. Materials and Methods
The selection of the desired and appropriate algorithm for the compression of biological data is a complex task because different algorithms are tuned for different purposes having different properties. The compression algorithms related to biological data have many features, so it is difficult for a user to select the suitable compression algorithm. Biologists are not aware of all these details of compressing algorithms as compared to computer scientists. Often, they need computer experts or detailed user manuals to teach them about the details of the groups of compression algorithms.
This article presents an expert system to select the appropriate algorithm from different algorithms based on the requirements of the user. The classification of algorithms, appropriate features for the algorithms, and the choice of the algorithm are given in a rule-based approach. The architecture of the proposed framework is shown in Figure 1. The expert system consists of a knowledge base and an inference mechanism. Each component is discussed in the following sub-sections.
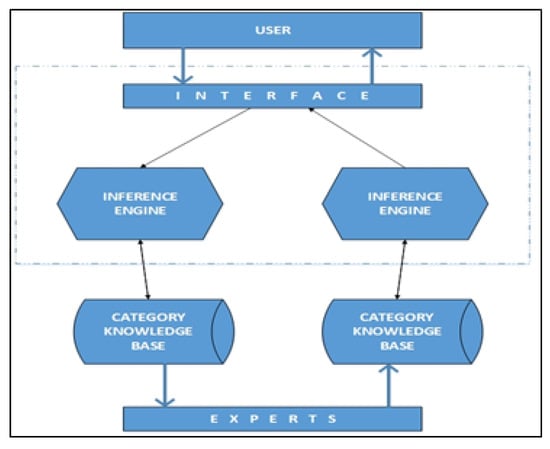
Figure 1.
Architecture of the proposed framework.
2.1. Knowledge Base
The knowledge base is created with the knowledge or information of an expert of a specific domain or existing knowledge. The information is then converted into the knowledge base according to the structure of the expert system, as shown in Figure 2. In this study, we created the knowledge base by conducting experiments and acquiring knowledge from the literature. We have created the rule-based structure separated into two sets, i.e., features that are essential for each category of algorithms and the algorithms that have different parameters. The features required for the algorithms in each category have been determined. The parameters used to select the algorithms have specific values for certain categories. Table 1 shows the values of each parameter. The number of features required for each category is the same, but parameter selection against each category may be different as per the user requirements.
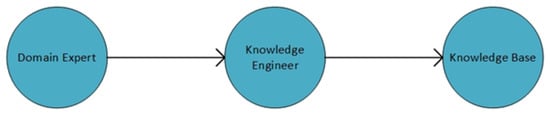
Figure 2.
Structure of the knowledge base.

Table 1.
Values used for the selection of each parameter for all algorithms.
There are two main categories of compressors for biological data, i.e., genome and NGS. Each category has further subcategories, i.e., reference-free and referential. In addition to specialized compression techniques, general-purpose compressors can also be used for genomic data compression. Different parameters such as compression time, compression ratio, memory consumption, etc., are measured for each algorithm. Users may opt for some set or combination of parameters for a particular category. We have used the literature to extract the parameters and specifications of each algorithm. The desired information obtained is converted and expressed according to the rule-based expert system approach in terms of the rule base. Parameters and categories described in Table 1 are transformed into the form of IF-THEN rules [21]. Values and categories are displayed in a rule base and algorithms that are associated with requirements and categories, while the values of their properties are shown in a separate rule base. It is projected that property values and their occurrence are gained from the rule base. These values are found, and then the algorithm and its properties are selected for equivalent property values from the rule base, as shown in Figure 3. Algorithm 1 shows the procedure of how the system accepts the input from the user in the form of parameters and then how the system matches the rules. The rule-matching procedure is shown in Algorithm 2.
| Algorithm 1: Pseudocode of rule base. |
 |
| Algorithm 2: Pseudocode of rule matching based on properties of each rule. |
 |
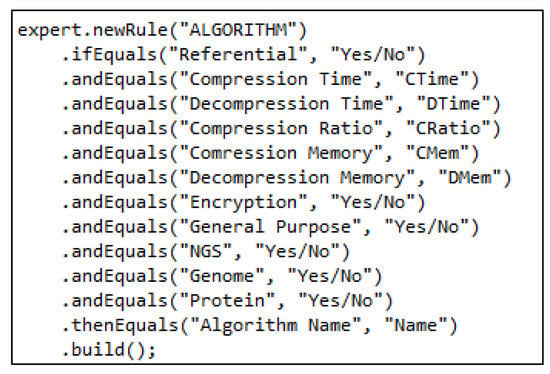
Figure 3.
An example rule.
2.2. Inference Mechanism
Obtaining the results based on some information or data given to the system related to the available situations is called inference. The data-driven method exploring the rules where the antecedents are found true and which uses this information to conclude subsequent information is called forward chaining. In the beginning, the system prompts the user to input the values of parameters. The inference engine decides to select a particular branch based on two parameters, i.e., type and mode. Based on this decision, the inference engine decides whether to scan the rule further or move to the next rule. In the end, the system selects algorithms according to the desired parameters. Some example rules are shown in Table 2. For example, if the user enters algorithm type “NGS” and sets the mode as referential, the proposed system looks for the rules where the antecedent is true for these two. If antecedents are true, the system goes deeper into that rule to check all other conditions. If antecedents are not true, the system does not go further into that rule and moves toward the next rule. This process continues until our system finds the best-fit rules. When the system finds the initial antecedents, the system matches all other rules. In case exact matches of the user input and the rules are found, the system displays the name of the algorithms. In addition, the similarity between the user input and the rules for all other cases is also displayed to the user. The system also shows the similarity of critical and general parameters that help the user in selecting the algorithms where exact matches are not found. The similarity is calculated as the Levenshiton distance of the user input and conditions in rules. The similarity is calculated with Equation (1). We have categorized the parameters into critical and general. Literature is used for categorization, i.e., targeted parameters by different researchers, e.g., compression ratio and time are targeted in all algorithms. We consider the first five parameters in Table 1 as critical. These parameters strongly influence the choice of a user for selecting an algorithm. The other parameters in Table 1 are considered general. These parameters do not strongly influence the choice of a user. During the calculation of the weight of parameters, weights of 60% and 40% are given to critical and general parameters, respectively:
where is the distance between the first i characters of a and the first j characters of b.

Table 2.
Example rules.
3. Case Studies
In this section, we discuss the experimental results of different compressors and the proposed framework. In the first phase, we conducted experiments on compression programs to generate datasets to test the proposed system. Next, we use the generated data to validate the proposed framework. Different compression programs available in literature are used for the first phase. Benchmark datasets are used for experiments. The datasets include data on three species, i.e., human, plant, and bacteria, of varying sizes ranging from 3.0 GB to 54 GB. The datasets are from different platforms such as Illumina, 464, and SOLid. We also used genome datasets of different species with varying sizes. We used datasets of different sizes and platforms to help us to better evaluate the experimental results. The details of the NGS and genome datasets are shown in Table 3 and Table 4, respectively, along with their reference genomes.

Table 3.
NGS datasets used for experiments.

Table 4.
Genome datasets used for experiments.
Compression tools of different categories were included in the experiments. Categories include NGS reference-free, NGS referential, genome reference-free, genome referential, and general-purpose compressors for both NGS and genome data. All tools were compiled with the respective compilers and environments. In case where the source code of a tool is not available, precompiled binaries were used. Compressed file sizes only include necessary files that are needed for decompression. Compression memory is the peak memory usage for a specific program. All programs were executed on a computer equipped with Core i5 7th generation CPU, 16 GB RAM, and Ubuntu 16.04 operating system. Using the above experimental setup and datasets, the values of compression and decompression times, compression and decompression memory requirements, and compression ratio by running each program were noted. The results of NGS reference-free, NGS referential, and general-purpose compressors for NGS data are shown in Supplementary Tables S1–S3, respectively. The results of genome reference-free, referential, and general-purpose compression programs on genome data are reported in Supplementary Tables S4–S6, respectively. The results are shown for each parameter against each dataset, as well as the average of all datasets for all algorithms. For other parameters (type, mode, operating system, programming language, and encryption), we find their values from the respective articles, as these parameters do not need to be calculated by running the programs. Supplementary Table S7 shows the values with respect to each algorithm for all categories of algorithms. Based on the data in supplementary Tables S1 through S7, we categorized the results of NGS reference-free, referential, and general-purpose compression programs against the calculated parameters into five levels (low, low moderate, moderate, high moderate, high), as shown in Supplementary Table S7.
Using these experimental results, we use hierarchical clustering to categorize the values for the parameters into the following five categories, i.e., low, low moderate, moderate, high moderate, and high. We use clustering to avoid any human errors in categorizing the categories manually. Each parameter in the experimental results was used to obtain the category value of each parameter for all algorithms. We evaluate the system using different input cases, for which the results are known to check the system behavior on different input cases. We examine the similarity of different compression algorithms against the related input case for the desired algorithm category using forward chaining. Forward chaining is used to not allow the system to go through all rules completely in depth. It is a data-driven method, so it moves according to the data provided and then uses the other related data from the rules to further move on to the goal-oriented rules. For example, if the user selects the NGS reference-based category, then the proposed system only displays the similarities of the algorithms of the NGS reference-based category by considering the related rules, excluding all other algorithms.
3.1. Input Case 1
In this case, the parameters shown in input case 1 from Table 5 are given as input. The system displays the programs that match exactly with the input parameters. In addition, the system also generates the similarity of each algorithm with the given input. The system also produces the weightage of all the algorithms that are non-referential and belongs to the file type genome. The results are shown in Figure 4. It can be observed that the similarity of DELIMINATE algorithm is 1, which means it exactly matches the input. DELIMINATE also has the exact match of critical and general parameters. GeCO has the lowest similarity and lowest matches in critical parameters as well.

Table 5.
Five input cases used to validate the proposed framework. H, M, and L mean high, moderate, and low, respectively.
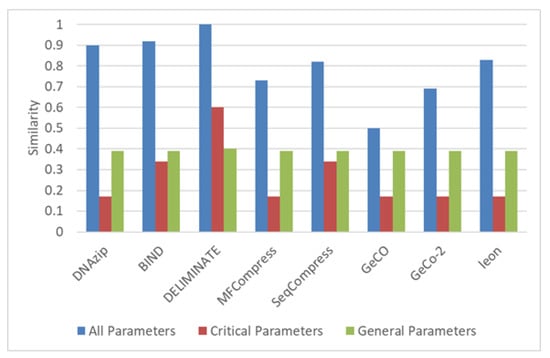
Figure 4.
Similarity and weighting results of genome non-referential algorithms.
3.2. Input Case 2
In this case, the user provides the input parameters shown in input case 2 of Table 5, where the algorithm type is genome with referential mode. The results are shown in Figure 5. It can be observed that the similarity of the GRS algorithm is 1, i.e., it exactly matches the user input. GreEn and FRESCO have the least similarity and least matches in critical parameters.
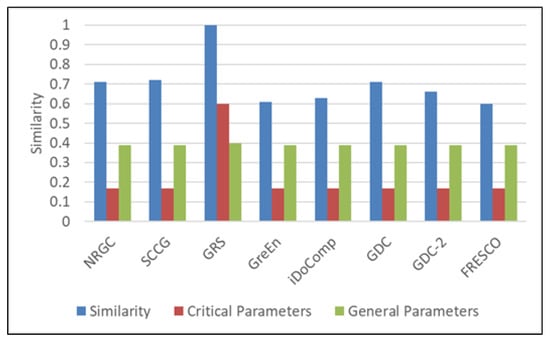
Figure 5.
Similarity and weighting results of genome referential algorithms.
3.3. Input Case 3
In the next case, the input consists of the NGS file type in reference-free mode with other values, as shown in input case 3 in Table 5. The results are shown in Figure 6. The results show that the Spring algorithm has the highest similarity with the highest matches of both critical and general parameters. Cryfa shows the lowest similarity and matches in critical parameters.
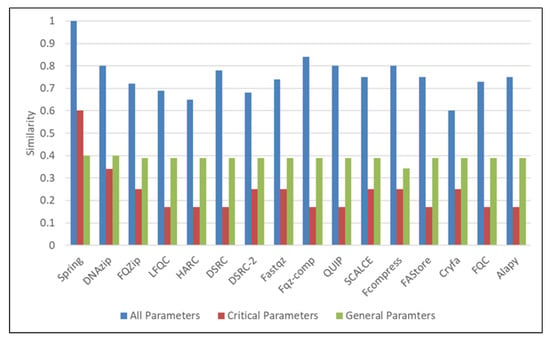
Figure 6.
Similarity and weighting results of NGS non-referential algorithms.
3.4. Input Case 4
In this case, the user provides the input shown in input case 4 of Table 5. In this input, the algorithm type is NGS and the mode is referential. The results are plotted in Figure 7. The Fastqz algorithm has the similarity of 1, i.e., it exactly matches the input requirements. LWFQzip has the lowest similarity measure.
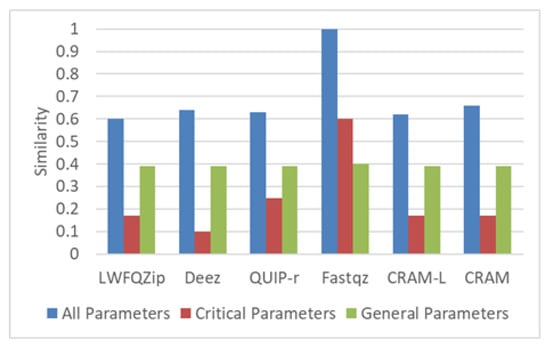
Figure 7.
Similarity and weighting results of NGS referential algorithms.
3.5. Input Case 5
In this case, the user provides the input with the general purpose algorithm type in non-referential mode, with parameters shown in input case 5 of Table 5. Figure 8 shows the results. In this scenario, the similarity of GZip matches the input provided. BWT has the lowest similarity.
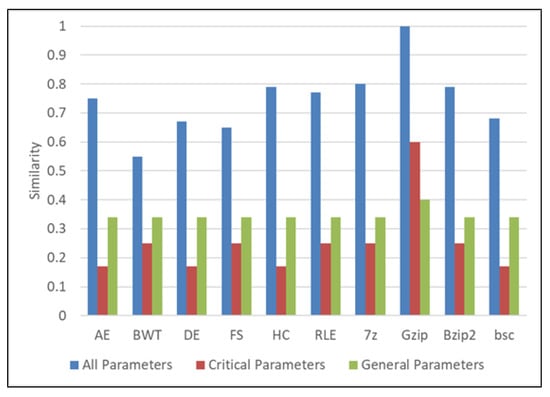
Figure 8.
Similarity and weighting results of general-purpose algorithms. AE refers to Arithmetic Encoding, DE refers to Delta Encoding, HC refers to Huffman Coding, and RLE refers to Run Length Encoding.
4. Conclusions
The size of biological data has grown rapidly in the last two decades, exceeding the increase in computing power. Data compression techniques are used to reduce the size of such huge data. A variety of specialized compression tools have been proposed in the literature. The algorithms work on different parameters. The selection of parameters is critical to take advantage of the most relevant algorithm. In this article, an expert system is presented that selects the best data-compression algorithm for biological data based on different parameters. The framework is based on the knowledge base and inference using forward chaining. We have generated datasets by executing different compression programs on benchmark datasets. The datasets are used to validate the proposed framework. The proposed framework is validated with different case studies. The results show that the proposed framework can be helpful for researchers to select compression algorithms for biological data according to the desired parameters. In addition to these parameters, different features in data can be helpful in selecting algorithms according to the input data. In the future, we plan to use machine learning to make the framework extend to more features and datasets.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/app122211360/s1, Table S1: Experimental results of reference free compression tools on NGS data. We could not run DNAZip program on large datasets due to high memory requirements; Table S2: Experimental results of referential compression tools on NGS data; Table S3: Experimental results of general-purpose compression tools on NGS data; Table S4: Experimental results of reference free compression tools on genomic data; Table S5: Experimental results of referential compression tools on genomic data; Table S6: Experimental results of general-purpose compression tools on genomic data; Table S7: Categorization of different programs based on various parameters. H, M and L refer to high, moderate, and low, respectively. CT, CM, DT, DM, and CR, refer to compression time, compression memory, decompression time, decompression memory, and compression ratio respectively.
Author Contributions
Conceptualization, A.A. and M.S.; methodology, A.A. and M.S.K.; software, M.S. and M.S.K.; validation, M.S.K., M.T., M.S. and A.A.; formal analysis, M.S., M.T. and M.S.K.; investigation, M.S.; resources, M.S. and M.T.; data curation, A.A. and M.T.; writing—original draft preparation, M.S. and M.S.K.; writing—review and editing, M.S., M.T. and A.A.; visualization, M.T.; supervision, M.S.; project administration, A.A. and M.S.; funding acquisition, A.A. All authors have read and agreed to the published version of the manuscript.
Funding
The authors extend their appreciation to the deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (IFP-2020-75).
Data Availability Statement
The datasets used are available publicly.
Acknowledgments
The authors extend their appreciation to the deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number (IFP-2020-75).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Deorowicz, S.; Grabowski, S. Data compression for sequencing data. Algorithms Mol. Biol. 2013, 8, 25. [Google Scholar] [CrossRef] [PubMed]
- Sardaraz, M.; Tahir, M. SCA-NGS: Secure compression algorithm for next generation sequencing data using genetic operators and block sorting. Sci. Prog. 2021, 104, 00368504211023276. [Google Scholar] [CrossRef] [PubMed]
- Gzip Home Page. Available online: https://www.gzip.org (accessed on 16 December 2019).
- Bzip2 Home Page. Available online: http://www.bzip.org/ (accessed on 16 December 2019).
- 7-Zip Home Page. Available online: https://www.7-zip.org/ (accessed on 16 December 2019).
- Chandak, S.; Tatwawadi, K.; Ochoa, I.; Hernaez, M.; Weissman, T. SPRING: A next-generation compressor for FASTQ data. Bioinformatics 2019, 35, 2674–2676. [Google Scholar] [CrossRef] [PubMed]
- Dutta, A.; Haque, M.M.; Bose, T.; Reddy, C.V.S.K.; Mande, S.S. FQC: A novel approach for efficient compression, archival, and dissemination of fastq datasets. J. Bioinform. Comput. Biol. 2015, 13, 1541003. [Google Scholar] [CrossRef] [PubMed]
- Ochoa, I.; Hernaez, M.; Weissman, T. iDoComp: A compression scheme for assembled genomes. Bioinformatics 2015, 31, 626–633. [Google Scholar] [CrossRef]
- Roguski, Ł.; Deorowicz, S. DSRC 2—Industry-oriented compression of FASTQ files. Bioinformatics 2014, 30, 2213–2215. [Google Scholar] [CrossRef]
- Sardaraz, M.; Tahir, M. FCompress: An Algorithm for FASTQ Sequence Data Compression. Curr. Bioinform. 2019, 14, 123–129. [Google Scholar] [CrossRef]
- Sardaraz, M.; Tahir, M.; Ikram, A.A.; Bajwa, H. SeqCompress: An algorithm for biological sequence compression. Genomics 2014, 104, 225–228. [Google Scholar] [CrossRef] [PubMed]
- Sardaraz, M.; Tahir, M.; Ikram, A.A. Advances in high throughput DNA sequence data compression. J. Bioinform. Comput. Biol. 2016, 14, 1630002. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Zhang, Y.; Ji, Z.; He, S.; Yang, X. High-throughput DNA sequence data compression. Briefings Bioinform. 2013, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- İpek, M.; Selvi, İ.H.; Findik, F.; Torkul, O.; Cedimoğlu, I. An expert system based material selection approach to manufacturing. Mater. Des. 2013, 47, 331–340. [Google Scholar] [CrossRef]
- Geysen, D.; De Somer, O.; Johansson, C.; Brage, J.; Vanhoudt, D. Operational thermal load forecasting in district heating networks using machine learning and expert advice. Energy Build. 2018, 162, 144–153. [Google Scholar] [CrossRef]
- Raghavendra, U.; Bhandary, S.V.; Gudigar, A.; Acharya, U.R. Novel expert system for glaucoma identification using non-parametric spatial envelope energy spectrum with fundus images. Biocybern. Biomed. Eng. 2018, 38, 170–180. [Google Scholar] [CrossRef]
- Khan, A.R.; Rehman, Z.U.; Amin, H.U. Knowledge-Based systems modeling for software process model selection. Int. J. Adv. Comput. Sci. Appl. 2011, 2, 20–25. [Google Scholar]
- Grobelny, P. The expert system approach in development of loosely coupled software with use of domain specific language. In Proceedings of the 2008 International Multiconference on Computer Science and Information Technology, Wisla, Poland, 20–22 October 2008; IEEE: Piscataway, NJ, USA, 2008; pp. 119–123. [Google Scholar]
- Lujić, R.; Samardžić, I.; Galzina, V. Application of expert systems for selection of installation pipes. Teh. Vjesn. 2015, 22, 241–245. [Google Scholar] [CrossRef]
- Bakeer, H.M.S. Photo Copier Maintenance Knowledge Based System V. 01 Using SL5 Object Language. Int. J. Eng. Inf. Syst. 2017, 1, 116–124. [Google Scholar]
- Rao, R.V.; Davim, J.P. A decision-making framework model for material selection using a combined multiple attribute decision-making method. Int. J. Adv. Manuf. Technol. 2008, 35, 751–760. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).