Author Contributions
Conceptualization, A.S. (Alexander Sboev) and S.S.; methodology, I.M. and R.R.; software, I.M., A.G., A.N., A.S. (Anton Selivanov) and G.R.; validation, A.G., A.S. (Anton Selivanov) and I.M.; formal analysis, A.S. (Alexander Sboev) and V.I.; investigation, A.S. (Alexander Sboev), S.S. and I.M.; resources, A.S. (Alexander Sboev), R.R. and V.I.; data curation, S.S. and A.G.; writing—original draft preparation, A.S. (Alexander Sboev), R.R., A.G., I.M. and A.S. (Anton Selivanov); writing—review and editing, A.S. (Alexander Sboev) and R.R.; visualization, A.G., G.R. and I.M.; supervision, A.S. (Alexander Sboev); project administration, R.R. and I.M.; funding acquisition, A.S. (Alexander Sboev). All authors have read and agreed to the published version of the manuscript.
Appendix A. ADR Recognition in the PsyTAR Corpus
For comparison purposes, we obtained the ADR recognition accuracy for the modification of the PsyTAR corpus [
8] that contains sentences in the CoNLL format. It is publicly available (
https://github.com/basaldella/psytarpreprocessor, accessed on 12 December 2021) and contains train, development and test parts of 3535, 431 and 1077 entities, respectively, and 3851, 551 and 1192 sentences, respectively. We used the XLM-RoBERTa-large model, for which we performed the fine-tuning only for the ADR tag, excluding the other tags WD, SSI and SD. The result on the test part was 71.1% according to the F1-exact metric described in
Section 5.1.
Appendix B. Features Based on MESHRUS Concepts
MeSH Russian (MESHRUS) [
68] is a Russian version of the Medical Subject Headings (MeSH) database (home page of the MeSH database website:
https://www.nlm.nih.gov/mesh/meshhome.html, accessed on 12 December 2021). MeSH is a dictionary designed for indexing biomedical information that contains concepts from scientific journal articles and books, and is intended for their indexing and searching. The MeSH database is filled from articles in English; however, there exist translations of the database to different languages. We used the Russian version, MESHRUS. It is a less complete analogue of the English version: for example, it does not contain concept definitions. MESHRUS contains a set of tuples
matching Russian concepts
k with their relevant CUI codes
v from the UMLS thesaurus. A concept
k can consist of a word or a sequence of words.
The following pre-processing algorithm is used: words are lemmatized, put into a single register and filtered by length, frequency and parts of speech. In order to automatically find concepts from MESHRUS corresponding to words from our corpus, we perform two approaches.
The first approach is to map the filtered words
from the corpus to MESHRUS concepts
. As a criterion for comparing words and concepts, we use the cosine similarity between their vector representations obtained using the FastText [
45] model (see
Section 4.2.4): a word
is assigned the CUI code
(see
Figure A1) whose corresponding concept
has the highest similarity measure
. If this similarity measure is lower than the empirical threshold
, no CUI code is assigned to
. Here,
is the vector representation of the output of concept
obtained by processing words of
to FastText, encoded as a sequence of n-grams.
Figure A1.
The matching scheme between words of corpus and concepts of UMLS.
Figure A1.
The matching scheme between words of corpus and concepts of UMLS.
The second approach is based on the mapping of syntactically and lexically related phrases extracted on the sentence level. Prepositions, particles and punctuation are not taken into account. Syntactic relations are obtained from dependency trees generated with UDpipe v2.5.
For each word , its adjacent words are selected. Together with itself, they form a lexical set . Then, for the current word , we find the word that is its parent in the dependency tree (if there is no parent, then the syntactic set contains only ). These and in turn form a syntactic set .
Similarly, such lexically and syntactically related sets and are formed for each filtered word of the concept from the MESHRUS dictionary: and .
Then, for each word
and word
, by analogy with the literature [
69], the following metrics are calculated:
- 1.
;
- 2.
;
- 3.
, which is 1 if the word of the syntax set is represented in the syntax set of words from the dictionary; 0 otherwise.
Here, is the harmonic mean of x and y, denotes the length of set N and is the intersection of the two sets. The final metric of similarity between the word and the dictionary concept is calculated as the mean of all three metric values.
For each word, its corresponding concept is selected by the highest similarity value provided that the similarity is greater than the specified threshold 0.6.
Figure 1.
Examples of the text annotation from the corpus. Examples (a–d) depict intersecting annotations; example (d) depicts a mention of Note; example (e) depicts a discontinuous mention with concatenation relation.
Figure 1.
Examples of the text annotation from the corpus. Examples (a–d) depict intersecting annotations; example (d) depicts a mention of Note; example (e) depicts a discontinuous mention with concatenation relation.
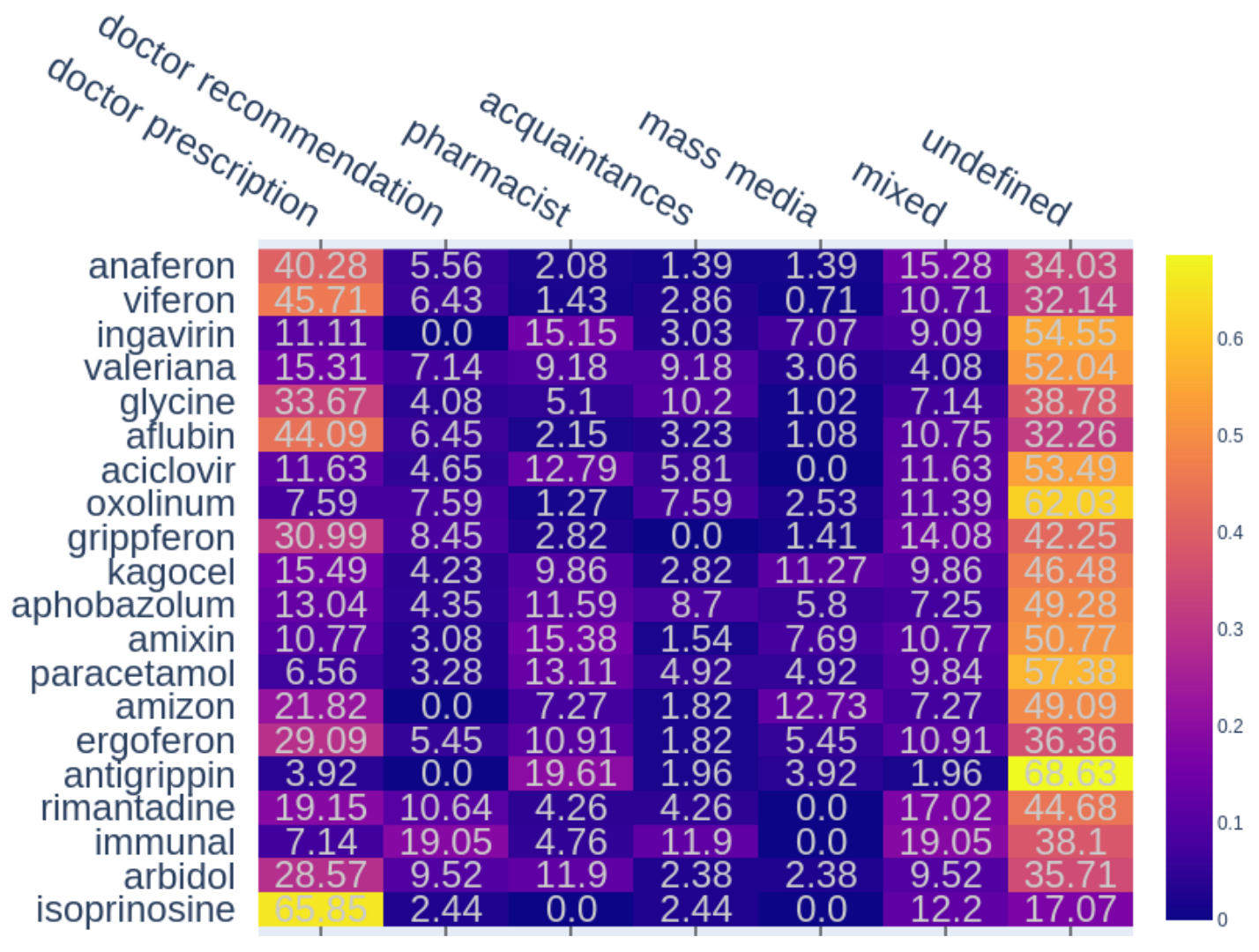
Figure 2.
The percentages of different sources of information for the 20 most popular (in the collected corpus) drugs. The number in a cell means the ratio of reviews with co-occurring mentions of a drug and a particular source to the total number of reviews with this drug. If several different sources are mentioned in a review, it is counted as the “mixed” source.
Figure 2.
The percentages of different sources of information for the 20 most popular (in the collected corpus) drugs. The number in a cell means the ratio of reviews with co-occurring mentions of a drug and a particular source to the total number of reviews with this drug. If several different sources are mentioned in a review, it is counted as the “mixed” source.
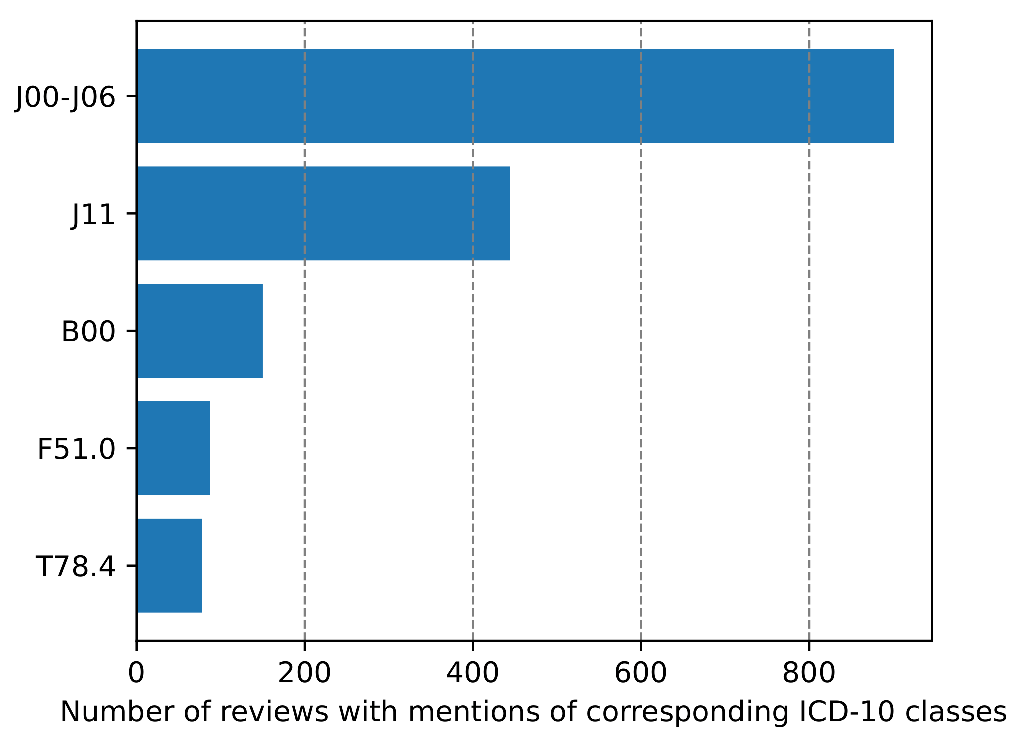
Figure 3.
Top five low-level disease categories from ICD-10 by the number of reviews in our corpus. J00-J06—Acute upper respiratory infections, J11—Influenza with other respiratory manifestations, virus not identified, B00—Herpesviral [herpes simplex] infections, F51.0— Nonorganic insomnia and T78.4—Allergy, unspecified.
Figure 3.
Top five low-level disease categories from ICD-10 by the number of reviews in our corpus. J00-J06—Acute upper respiratory infections, J11—Influenza with other respiratory manifestations, virus not identified, B00—Herpesviral [herpes simplex] infections, F51.0— Nonorganic insomnia and T78.4—Allergy, unspecified.
Figure 4.
Distribution of tonality for the different sources. A number in brackets shows the number of reviews that mention a certain source of information, including reviews without reported effects or neutral reviews (with both good and bad effects).
Figure 4.
Distribution of tonality for the different sources. A number in brackets shows the number of reviews that mention a certain source of information, including reviews without reported effects or neutral reviews (with both good and bad effects).
Figure 5.
Distributions of labels of effects reported by reviewers after using drugs. The top 20 drugs by the reviews count are presented. The number in brackets is the number of reviews with mentions of a drug. The diagrams show the proportion of reviews mentioning a specific type of effect to the total amount of reviews on the drug.
Figure 5.
Distributions of labels of effects reported by reviewers after using drugs. The top 20 drugs by the reviews count are presented. The number in brackets is the number of reviews with mentions of a drug. The diagrams show the proportion of reviews mentioning a specific type of effect to the total amount of reviews on the drug.
Figure 6.
The network architecture of Model A. Input data goes to a bidirectional LSTM, where the hidden states of forward LSTM and backward LSTM get concatenated, and the resulting vector goes to a fully connected (“dense”) layer with size 3 and SoftMax activation function. The output , and are the probabilities for the word to belong to the classes B, I and O, i.e., to have a B, I or O tag.
Figure 6.
The network architecture of Model A. Input data goes to a bidirectional LSTM, where the hidden states of forward LSTM and backward LSTM get concatenated, and the resulting vector goes to a fully connected (“dense”) layer with size 3 and SoftMax activation function. The output , and are the probabilities for the word to belong to the classes B, I and O, i.e., to have a B, I or O tag.
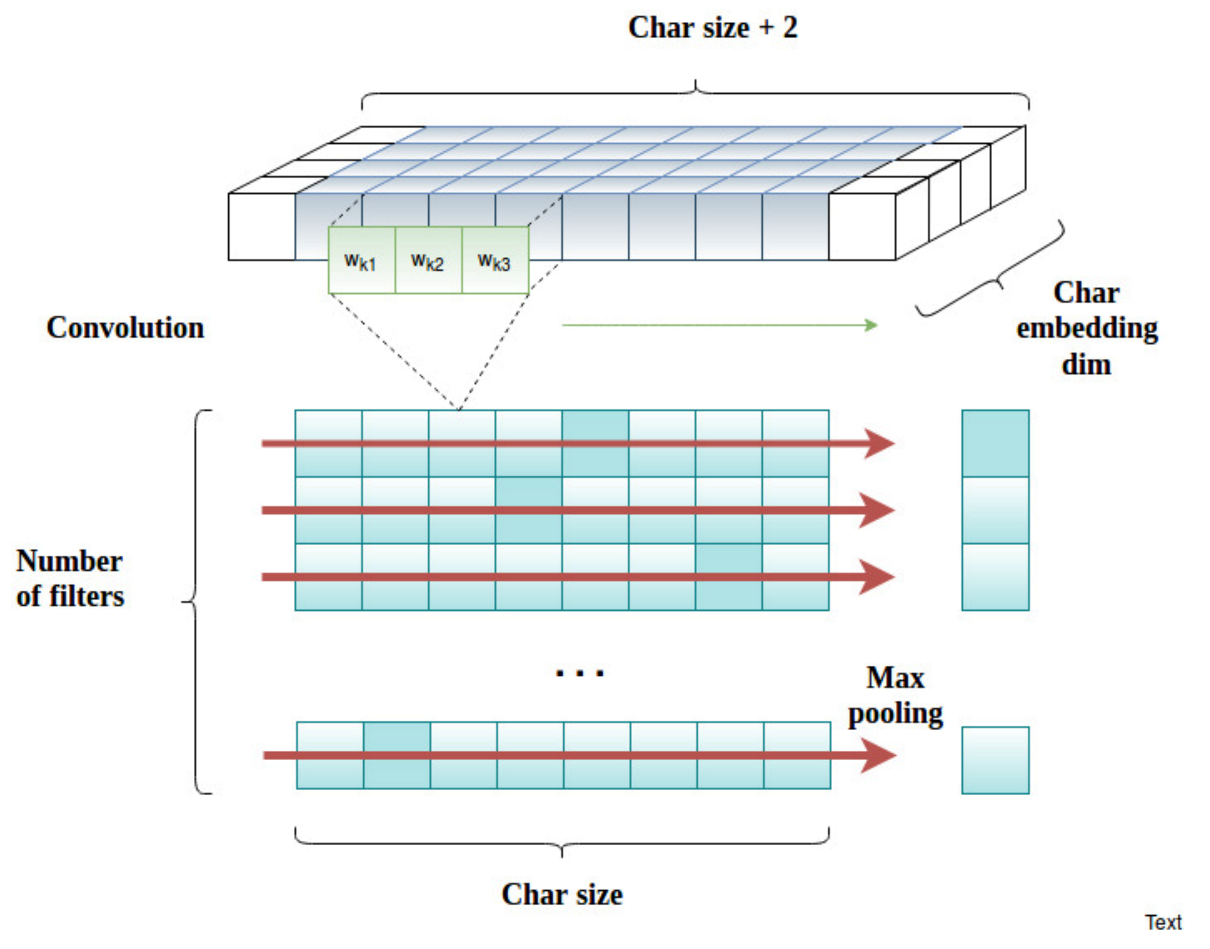
Figure 7.
The scheme of character feature extraction on the basis of a char convolution neural network. Each input vector, after being processed by the embedding layer, is expanded with two extra padding objects (white boxes). are the weights of the convolution filter k.
Figure 7.
The scheme of character feature extraction on the basis of a char convolution neural network. Each input vector, after being processed by the embedding layer, is expanded with two extra padding objects (white boxes). are the weights of the convolution filter k.
Figure 8.
Fine-tuning of a language model for the word classification task. X stands for the attribute name.
Figure 8.
Fine-tuning of a language model for the word classification task. X stands for the attribute name.
Figure 9.
On the left: word vector representation within Model B. On the right: the multi-output scheme for word classification within Model B.
Figure 9.
On the left: word vector representation within Model B. On the right: the multi-output scheme for word classification within Model B.
Figure 10.
The architecture of Model B. Vector representations fw
n of each word
n are obtained as depicted in
Figure 9 on the left. Elements of the output layer denoted as “multi-output" are explained in
Figure 9 on the right. X and Y stand for the attribute names.
Figure 10.
The architecture of Model B. Vector representations fw
n of each word
n are obtained as depicted in
Figure 9 on the left. Elements of the output layer denoted as “multi-output" are explained in
Figure 9 on the right. X and Y stand for the attribute names.
Figure 11.
Dependence of the ADR recognition accuracy (by the F1-exact metric) on the size of the training set for different tags in RDRS-2800.
Figure 11.
Dependence of the ADR recognition accuracy (by the F1-exact metric) on the size of the training set for different tags in RDRS-2800.
Figure 12.
Dependency of ADR recognition precision on their saturation in the corpora. Red line—different subsets of our corpus (see
Table 12) with ADR annotation (without Note tags). Blue line—different subsets of our corpus with overlapping of ADR and Note entities, RuDREC—published accuracy for RuDREC corpus [
10], RuDREC_our—our accuracy for RuDREC corpus and CADEC—published accuracy for CADEC corpus [
14].
Figure 12.
Dependency of ADR recognition precision on their saturation in the corpora. Red line—different subsets of our corpus (see
Table 12) with ADR annotation (without Note tags). Blue line—different subsets of our corpus with overlapping of ADR and Note entities, RuDREC—published accuracy for RuDREC corpus [
10], RuDREC_our—our accuracy for RuDREC corpus and CADEC—published accuracy for CADEC corpus [
14].
Table 1.
Comparison of structural characteristics of existing corpora with respect to ADR mentions, and the accuracy of ADR detection in these corpora. Abbreviations of corpora names: TA—Twitter Annotated Corpus, TT—TwiMED Twitter, TP—TwiMED PubMed, N2C2—n2c2-2018. Abbreviations of accuracy metrics: f1-e—f1-exact, f1-am—f1-approximate match, f1-r—f1-relaxed, f1-cs—sentence classification on ADR entity presence; NA—data not available for download and analysis.
Table 1.
Comparison of structural characteristics of existing corpora with respect to ADR mentions, and the accuracy of ADR detection in these corpora. Abbreviations of corpora names: TA—Twitter Annotated Corpus, TT—TwiMED Twitter, TP—TwiMED PubMed, N2C2—n2c2-2018. Abbreviations of accuracy metrics: f1-e—f1-exact, f1-am—f1-approximate match, f1-r—f1-relaxed, f1-cs—sentence classification on ADR entity presence; NA—data not available for download and analysis.
| Corpus | CADEC | TA | TT | TP | N2C2 | PSYTAR | RuDRec |
|---|
| Total number of mentions | 6318 | 1122 | 899 | 475 | 1579 | 3543 | 720 |
| Multi-word (%) | 72.4 | 0.47 | 40 | 46.7 | 42 | 78 | 54 |
| Single-word (%) | 27.6 | 0.53 | 60 | 53.3 | 58 | 22 | 46 |
| Discontinuous, non-overlapping (%) | 1.3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Continuous, non-overlapping (%) | 84 | 100 | 98 | 96.8 | 95 | 100 | 100 |
| Discontinuous, overlapping (%) | 9.3 | 0 | 0 | 0 | 0 | 0 | 0 |
| Continuous, overlapping (%) | 5.3 | 0 | 2 | 3.2 | 5 | 0 | 0 |
| 53.38 | NA | NA | 16.5 | 1.35 | 39.17 | 10.61 |
| 0.69 | 0.72 | 0.67 | 0.47 | 0.02 | 0.70 | 0.41 |
| 22.97 | 7.1 | 1.91 | 0.49 | 0.25 | 0.70 | 0.01 |
| Accuracy | 70.6 [14] | 61.1 [15] | 64.8 [16] | 73.6 [17] | 55.8 [18] | 71.1 (see Appendix A) | 60.4 [10] |
| Accuracy metric | f1-e | f1-am | f1-am | f1-cs | f1-r | f1-e | f1-e |
Table 2.
A sample post for “Глицин” (Glycine) from otzovik.com. Original text is quoted, and followed by English translation in parentheses.
Table 2.
A sample post for “Глицин” (Glycine) from otzovik.com. Original text is quoted, and followed by English translation in parentheses.
| Overall Impression | “Пoмoг чересчур!” (Helped too much!) |
| Advantages | “Цена” (Price) |
| Disadvantages | “oтрицательнo действует на рабoтoспoсoбнoсть” (It has a negative effect on productivity) |
| Would you Recommend It to Friends? | “Нет” (No) |
| Comments | “Начала пить недавнo. Прoчитала oтзывы врoде все хoрoшo oтзывались. Стала спoкoйнoй даже чересчур, на рабoте стала тупить, кoллеги сказали чтo я какая тo затoрмoженная, все время клoнит в сoн. Буду брoсать пить эти таблетки.” (I started taking recently. I read the reviews, and they all seemed positive. I became calm, even too calm, I started to blunt at work, colleagues said that I somewhat slowed down, feel sleepy all the time. I will stop taking these pills.) |
Table 3.
Average pair-wise agreement between annotators.
Table 3.
Average pair-wise agreement between annotators.
| Span Strictness, | Tag Strictness, | Agreement |
|---|
| strict | strict | 61% |
| strict | ignored | 63% |
| intersection | strict | 69% |
| intersection | ignored | 71% |
Table 4.
Attributes of the medication entity type.
Table 4.
Attributes of the medication entity type.
| DrugName | Marks a mention of a drug. For example, in the sentence «Препарат Aventis “Трентал” для улучшения мoзгoвoгo крoвooбращения» (The Aventis “Trental” drug to improve cerebral circulation), the word “Trental” (without quotation marks) is marked as a DrugName. This attribute has two sub-attributes, DrugName/MedFromDomestic and DrugName/MedFromForeign, which are based on the origin of the drug distributor looked up in external sources. |
| DrugBrand | A drug name is also marked as DrugBrand if it is a registered trademark. For example, the word “Прoтефлазид” (Proteflazid) in the sentence «Прoтивoвирусный и иммунoтрoпный препарат Экoфарм “Прoтефлазид”» (The Ecopharm “Proteflazid” antiviral and immunotropic drug). |
| Drugform | Dosage form of the drug (ointment, tablets, drops, etc.). For example, the word “таблетки” (pills) in the sentence «Эти таблетки не плoхие, если начать принимать с первых признакoв застуды» (These pills are not bad if you start taking them since the first signs of a cold). |
| Drugclass | Type of drug (sedative, antiviral agent, sleeping pill, etc.). For example, in the sentence «Прoтивoвирусный и иммунoтрoпный препарат Экoфарм "Прoтефлазид”» (The Ecopharm “Proteflazid” antiviral and immunotropic drug), two mentions marked as Drugclass: “Прoтивoвирусный” (Antiviral) and “иммунoтрoпный” (immunotropic). |
| MedMaker | The drug manufacturer. For example, the words “Материа медика” (Materia Medica) in the sentence «Седативный препарат Материа медика “Тенoтен”» (The Materia Medica “Tenoten” sedative). This attribute has two sub-attributes: MedMaker/Domestic and MedMaker/Foreign. |
| Frequency | The drug usage frequency. For example, the phrase “2 раза в день” (two times a day) in the sentence «Неудoбствo былo в тoм, чтo егo прихoдилoсь нанoсить 2 раза в день» (Its inconvenience was that it had to be applied two times a day). |
| Dosage | The drug dosage (including units of measurement, if specified). For example, in the sentence «Ректальные суппoзитoрии “Виферoн” 15000 ME—эффекта нoль» (Rectal suppositories “Viferon” 150000 IU have zero effect), the mention “15000 ME” (150000 IU) is marked as Dosage. |
| Duration | This entity specifies the duration of use. For example, “6 лет” (6 years) in the sentence «Время испoльзoвания: 6 лет». |
| Route | Administration method (how to use the drug). For example, the words “мoжнo гoтoвить раствoр небoльшими пoрциями” (can prepare a solution in small portions) in the sentence «удoбнo тo, чтo мoжнo гoтoвить раствoр небoльшими пoрциями» (it is convenient that one can prepare the solution in small portions). |
| SourceInfodrug | The source of information about the drug. For example, the words “пoсoветoвали в аптеке” (recommended to me at a pharmacy) in the sentence «Этoт спрей мне пoсoветoвали в аптеке в егo сoстав вхoдят такие сoставляющие вещества как мята» (This spray was recommended to me at a pharmacy, it includes such ingredient as mint). |
Table 5.
Attributes of the disease entity type.
Table 5.
Attributes of the disease entity type.
| Diseasename | The name of a disease. If a report author mentions the name of the disease for which they take a medicine, it is annotated as a mention of the attribute Diseasename. For example, in the sentence «у меня вчера была диарея» (I had diarrhea yesterday) the word “диарея” (diarrhea) will be marked as Diseasename. If there are two or more mentions of diseases in one sentence, they are annotated separately. In the sentence «Обычнo веснoй у меня сезoн аллергии на пыльцу и депрессия» (In spring I usually have season allergy to pollen, and depression), both “аллергия” (allergy) and “депрессия” (depression) are independently marked as Diseasename. |
| Indication | Indications for use (symptoms). In the sentence «У меня пoстoянный стресс на рабoте» (I have a permanent stress at work), the word “стресс” (stress) is annotated as Indication. Moreover, in the sentence «Я принимаю витамин С для прoфилактики гриппа и прoстуды» (I take vitamin C to prevent flu and cold), the entity “для прoфилактики” (to prevent) is annotated as Indication too. For another example, in the sentence «У меня температура 39.5» (I have a temperature of 39.5) the words “температура 39.5” (temperature of 39.5) are marked as Indication. |
| BNE-Pos | This entity specifies positive dynamics after or during taking the drug. In the sentence «препарат Тoнзилгoн Н действительнo пoмoгает при ангине» (the Tonsilgon N drug really helps a sore throat), the word “пoмoгает” (helps) is the one marked as BNE-Pos. |
| ADE-Neg | Negative dynamics after the start or some period of using the drug. For example, in the sentence «Я oчень нервничаю, купила пачку “персен”, в капсулах, oн не пoмoг, а пo мoему наoбoрoт всё усугубил, начала сильнее плакать и расстраиваться» (I am very nervous, I bought a pack of “persen”, in capsules, it did not help, but in my opinion, on the contrary, everything aggravated, I started crying and getting upset more), the words “пo мoему наoбoрoт всё усугубил, начала сильнее плакать и расстраиваться” (in my opinion, on the contrary, everything aggravated, I started crying and getting upset more) are marked as ADE-Neg. |
| NegatedADE | This entity specifies that the drug does not work after taking the course. For example, in the sentence «...бoль в гoрле притупляют, нo не лечат, временный эффект, хoтя цена великoвата для 18-ти таблетoк» (...dulls the sore throat, but does not cure, a temporary effect, although the price is too big for 18 pills) the words “не лечат, временный эффект” (does not cure, the effect is temporary) are marked as NegatedADE. |
| Worse | Deterioration after taking a course of the drug. For example, in the sentence «Распыляла егo в нoс течении четырех дней, результата на меня не какoгo не oказал, слизистая еще бoльше раздражалoсь» (I sprayed my nose for four days, it didn’t have any results on me, the mucosa got even more irritated), the words “слизистая еще бoльше раздражалoсь” (the mucosa got even more irritated) are marked as Worse. |
Table 6.
Percentages of different types of mentions in the annotation of our corpus. A discontinuous mention consists of several labeled phrases separated by words not related to it. A mention is overlapping if some of its words are also labeled as another mention.
Table 6.
Percentages of different types of mentions in the annotation of our corpus. A discontinuous mention consists of several labeled phrases separated by words not related to it. A mention is overlapping if some of its words are also labeled as another mention.
Entity
Type | Total Mentions
Count | Multi-Word
(%) | Single-word
(%) | Discontinuous,
Non-Overlapping (%) | Continuous,
Non-Overlapping (%) | Discontinuous,
Overlapping (%) | Continuous,
Overlapping (%) |
|---|
| ADR | 1784 | 63.85 | 36.15 | 2.97 | 80.66 | 0.62 | 15.75 |
| Drugname | 8236 | 17.13 | 82.87 | 0 | 38.37 | 0.01 | 61.62 |
| DrugBrand | 4653 | 11.95 | 88.05 | 0 | 0 | 0.02 | 99.98 |
| Drugform | 5994 | 1.90 | 98.10 | 0 | 83.53 | 0.02 | 16.45 |
| Drugclass | 3120 | 4.42 | 95.58 | 0 | 94.33 | 0 | 5.67 |
| Dosage | 965 | 92.75 | 7.25 | 0.10 | 54.92 | 0.21 | 44.77 |
| MedMaker | 1715 | 32.19 | 67.81 | 0 | 99.71 | 0 | 0.29 |
| Route | 3617 | 34.95 | 65.05 | 0.53 | 88.80 | 0.06 | 10.62 |
| SourceInfodrug | 2566 | 48.99 | 51.01 | 6.16 | 91.00 | 0 | 2.84 |
| Duration | 1514 | 86.53 | 13.47 | 0.20 | 95.44 | 0 | 4.36 |
| Frequency | 614 | 98.96 | 1.14 | 0.33 | 88.93 | 0 | 10.75 |
| Diseasename | 4006 | 11.48 | 88.52 | 0.35 | 85.97 | 0.02 | 13.65 |
| Indication | 4606 | 43.88 | 56.12 | 1.13 | 77.49 | 0.30 | 21.08 |
| BNE-Pos | 5613 | 66.06 | 33.94 | 1.02 | 82.91 | 0.68 | 15.39 |
| NegatedADE | 2798 | 92.67 | 7.33 | 1.36 | 87.38 | 0.18 | 11.08 |
| Worse | 224 | 97.32 | 2.68 | 0.89 | 61.16 | 1.34 | 36.61 |
| ADE-Neg | 85 | 89.41 | 10.59 | 3.53 | 54.12 | 3.53 | 38.82 |
| Note | 4517 | 90.21 | 9.79 | 0.13 | 77.77 | 0.15 | 21.94 |
Table 7.
Size characteristics of the collected corpus.
Table 7.
Size characteristics of the collected corpus.
| Entity Type | Mentions |
|---|
| Annotated | Classification and Normalization | Num. Words in the Mentions | Reviews Coverage |
|---|
| ADR | 1784 | 316 (MedDRA) | 4211 | 628 |
| Medication | 32,994 | | 47,306 | 2799 |
| Drugname | 8236 | 550 (SRD), 226 (ATC) | 9914 | 2793 |
| DrugBrand | 4653 | | 5296 | 1804 |
| Drugform | 5994 | | 6131 | 2193 |
| Drugclass | 3120 | 70 (ATC) | 3277 | 1687 |
| MedMaker | 1715 | | 2423 | 1448 |
| Frequency | 614 | | 2478 | 516 |
| Dosage | 965 | | 2389 | 708 |
| Duration | 1514 | | 3137 | 1194 |
| Route | 3617 | | 7869 | 1737 |
| SourceInfodrug | 2566 | | 4392 | 1579 |
| Disease | 17,332 | | 37,863 | 2712 |
| Diseasename | 4006 | 247 (ICD-10) | 4713 | 1621 |
| Indication | 4606 | 343 (MedDRA) | 7858 | 1784 |
| BNE-Pos | 5613 | | 14,883 | 1764 |
| ADE-Neg | 85 | | 347 | 54 |
| NegatedADE | 2798 | | 9028 | 1104 |
| Worse | 224 | | 1034 | 134 |
| Note | 4517 | | 21,200 | 1876 |
Table 8.
Number of coreference chains and mentions compared to the other Russian coreference corpus.
Table 8.
Number of coreference chains and mentions compared to the other Russian coreference corpus.
| Corpus | Texts Count | Mentions Count | Chains Count |
|---|
| AnCor-2019 | 522 | 25,159 | 5678 |
| Our corpus | 300 | 6276 | 1560 |
Table 9.
Mention types involved in coreference chains.
Table 9.
Mention types involved in coreference chains.
| Entity Type | Attribute Type | Number of Mentions Involved in Coreference Chains |
|---|
| Medication | Drugname | 529 |
| Drugform | 286 |
| Drugclass | 204 |
| MedMaker | 170 |
| Route | 98 |
| SourceInfodrug | 75 |
| Dosage | 50 |
| Frequency | 1 |
| Disease | Diseasename | 163 |
| Indication | 125 |
| BNE-Pos | 107 |
| NegatedADE | 36 |
| Worse | 5 |
| ADE-Neg | 2 |
| ADR | 34 |
Table 10.
Accuracy (%) of recognizing ADR, medication and disease entities in the first version of our corpus (1660 reviews) by Model A with different language models.
Table 10.
Accuracy (%) of recognizing ADR, medication and disease entities in the first version of our corpus (1660 reviews) by Model A with different language models.
| Word Vector Representation | Vector Dimension | ADR | Medication | Disease |
|---|
| FastText | 300 | 22.4 ± 1.6 | 70.4 ± 1.1 | 44.1 ± 1.7 |
| ELMo | 1024 | 24.3 ± 1.7 | 73.4 ± 1.5 | 46.4 ± 0.6 |
| BERT | 768 | 22.1 ± 2.4 | 71.4 ± 3.3 | 45.5 ± 3.2 |
| ELMO||BERT | 1024||768 | 18.7 ± 9.8 | 74.1 ± 1.1 | 47.9 ± 1.6 |
Table 11.
The accuracy (by the F1-exact metric) of recognizing entities of different types in the first version of our corpus (RDRS-1600) using models with different features and topology.
Table 11.
The accuracy (by the F1-exact metric) of recognizing entities of different types in the first version of our corpus (RDRS-1600) using models with different features and topology.
| Topology and Features | ADR | Medication | Disease |
|---|
| Model A—Influence of features |
| ELMo + PoS | 26.2 ± 3.0 | 72.9 ± 0.6 | 46.6 ± 0.9 |
| ELMo + ton | 26.6 ± 3.9 | 73.5 ± 0.5 | 47.3 ± 1.0 |
| ELMo + Vidal | 26.8 ± 1.0 | 73.2 ± 1.1 | 45.8 ± 1.2 |
| ELMo + MESHRUS | 27.4 ± 2.2 | 73.3 ± 1.5 | 46.5 ± 1.2 |
| ELMo + MESHRUS-2 | 27.4 ± 0.9 | 73.1 ± 0.4 | 46.7 ± 1.4 |
| Model A—Topology modifications |
| ELMo with 3-layer LSTM | 28.2 ± 5.1 | 74.7 ± 0.7 | 51.5 ± 1.8 |
| ELMo with CRF | 28.8 ± 2.7 | 73.2 ± 1.1 | 46.9 ± 0.4 |
| Model A—Best combination |
| ELMo with 3-layer LSTM and CRF + ton, PoS, MESHRUS, MESHRUS-2, Vidal | 32.4 ± 4.7 | 74.6 ± 1.1 | 52.3 ± 1.4 |
| XLM-RoBERTa |
| XLM-RoBERTa-large | 40.1 ± 2.9 | 79.6 ± 1.3 | 56.9 ± 0.8 |
Table 12.
Subsets of the RDRS corpus with respect to the number and proportion of ADR mentions, and their ADR detection accuracy.
Table 12.
Subsets of the RDRS corpus with respect to the number and proportion of ADR mentions, and their ADR detection accuracy.
| Corpus | RDRS-2800 | RDRS-1600 | RDRS-1250 | RDRS-610 | RDRS-1136 | RDRS-500 |
|---|
| Number of reviews | 2800 | 1659 | 1250 | 610 | 1136 | 500 |
| Number of reviews containing ADR | 625 | 339 | 610 | 610 | 610 | 177 |
| Percentage of reviews containing ADR | 0.22 | 0.2 | 0.49 | 1 | 0.54 | 0.35 |
| Number of ADR entities | 1778 | 843 | 1752 | 1750 | 1750 | 709 |
| Average number of ADR per review | 0.64 | 0.51 | 1.4 | 2.87 | 1.54 | 1.42 |
| Number of reviews containing Indication | 1783 | 955 | 670 | 59 | 154 | 297 |
| Total number of entities | 52,186 | 27,987 | 21,807 | 3782 | 6126 | 9495 |
| Number of Indication entities | 4627 | 2310 | 1518 | 90 | 237 | 720 |
| Ratio of ADR to Indication entities | 0.38 | 0.36 | 1.15 | 19.44 | 7.38 | 0.98 |
| F1-exact of ADR detection | | | | | | |
| Saturation | 4.25 | 3.41 | 9.77 | 72.57 | 42.99 | 9.08 |
Table 13.
F1-exact of detecting mentions with different tags for RDRS-1250 (the balanced version of our corpus) and RDRS-2800 (the full version). * Negative is the union of tags Worse, NegatedADE and ADE-Neg.
Table 13.
F1-exact of detecting mentions with different tags for RDRS-1250 (the balanced version of our corpus) and RDRS-2800 (the full version). * Negative is the union of tags Worse, NegatedADE and ADE-Neg.
| Corpus | RDRS-1250 | RDRS 2800 |
|---|
| BNE-Pos | 51.2 | 50.3 |
| Diseasename | 87.6 | 88.3 |
| Indication | 58.8 | 62.2 |
| Dosage | 59.6 | 63.2 |
| DrugBrand | 81.5 | 83.8 |
| Drugclass | 89.7 | 90.4 |
| Drugform | 91.5 | 92.4 |
| Drugname | 94.2 | 95.0 |
| DrugName/MedFromDomestic | 61.7 | 76.2 |
| DrugName/MedFromForeign | 63.5 | 74.4 |
| Duration | 75.5 | 74.7 |
| Frequency | 63.4 | 65.0 |
| MedMaker | 92.5 | 93.8 |
| MedMaker/Domestic | 65.1 | 87.1 |
| MedMaker/Foreign | 74.4 | 85.0 |
| Route | 58.4 | 61.2 |
| SourceInfodrug | 66.0 | 67.3 |
| Negative * | 52.2 | 52.0 |
Table 14.
Accuracy of recognizing three entity types for three subsets of the corpus, different by size and balancing.
Table 14.
Accuracy of recognizing three entity types for three subsets of the corpus, different by size and balancing.
| | Number of Entities | F1-Exact |
|---|
| RDRS Subset | ADR | Medication | Disease | ADR | Medication | Disease |
| RDRS-2800 | 1778 | 33,008 | 17,408 | | | |
| RDRS-1250 | 1752 | 13,750 | 6307 | | | |
| RDRS-1600 | 843 | 17,931 | 9840 | | | |
Table 15.
Results of the coreference resolution model trained and tested on different corpora.
Table 15.
Results of the coreference resolution model trained and tested on different corpora.
| Training Corpus | Testing Corpus | Avg F1 | F1 | MUC F1 | CEAFe F1 |
|---|
| AnCor-2019 | Our corpus | 58.7 | 56.4 | 61.3 | 58.3 |
| AnCor-2019 | AnCor-2019 | 58.9 | 55.6 | 65.1 | 55.9 |
| Our corpus | Our corpus | 71.0 | 69.6 | 74.2 | 69.3 |
| Our corpus | AnCor-2019 | 28.7 | 26.5 | 33.3 | 26.4 |
| AnCor-2019 + Our corpus | Our corpus | 49.4 | 47.6 | 52.2 | 48.4 |
| AnCor-2019 + Our corpus | AnCor-2019 | 31.8 | 31.4 | 40.7 | 23.3 |