1. Introduction
Functional irregularities of the heart and blood circulatory system are referred to as cardiovascular diseases (CVDs). CVDs cause significant degeneration of patients’ life quality, while severe cases result in death. Research statistics provided by the World Health Organization show that CVDs account for 17.9 million deaths per year, which makes them the leading cause of death globally [
1]. Early diagnosis of CVDs enables timely and appropriate treatment and prevention of patients’ death. The diagnostic process includes obtaining images of unhealthy or weakened heart structures using imaging devices such as echocardiography, computed tomography (CT), or magnetic resonance (MRI). After that, collected images are observed, interpreted, and analyzed by clinical experts using specialized medical software built with advanced image processing methods.
Various clinical applications require insights into different cardiovascular system structures. For example, whole heart segmentation is crucial for pathology localization and hearts’ anatomy and function analysis. The construction of patient-specific three-dimensional (3D) heart models and surgical implants greatly benefits pre-surgical planning for patients with atherosclerosis, congenital heart defects, cardiomyopathy, or even inspecting different heart infections in post-surgical treatment [
2]. Whole heart segmentation includes delineation of four heart chambers, the entire blood pool, and the great vessels, which makes manual segmentation by clinical experts time-consuming and prone to observer variability. There is an increasing focus on developing accurate and robust automatic image processing methods. The development of accurate, efficient, and automatic image processing and analysis methods is a complex task, especially in the medical field. The main reason is in high variability in image intensities distribution and dynamic properties of cardiac structures. Nevertheless, the advancements and rapid development in image processing, computer vision, and artificial intelligence fields significantly facilitate this challenging task.
A commonly used approach for medical image segmentation includes encoder–decoder-based architectures such as the U-Net architecture [
3]. The U-Net architecture and its’ corresponding three-dimensional counterpart, 3D U-Net [
4], consist of contracting and expanding pathways. Throughout the contracting pathway, the network learns low-level features and reduces its numbers using sets of pooling and convolutional layers. In an expanding pathway, the network learns high-level features and recovers the original image resolution using deconvolutional layers. Features from contracting and expanding pathways are concatenated with skip connections to retrieve lost image information that occurs during the down-sampling process. Intuitively, this indicates that the part of the information is lost during the encoding process and can not be recovered when decoding. Variational autoencoders [
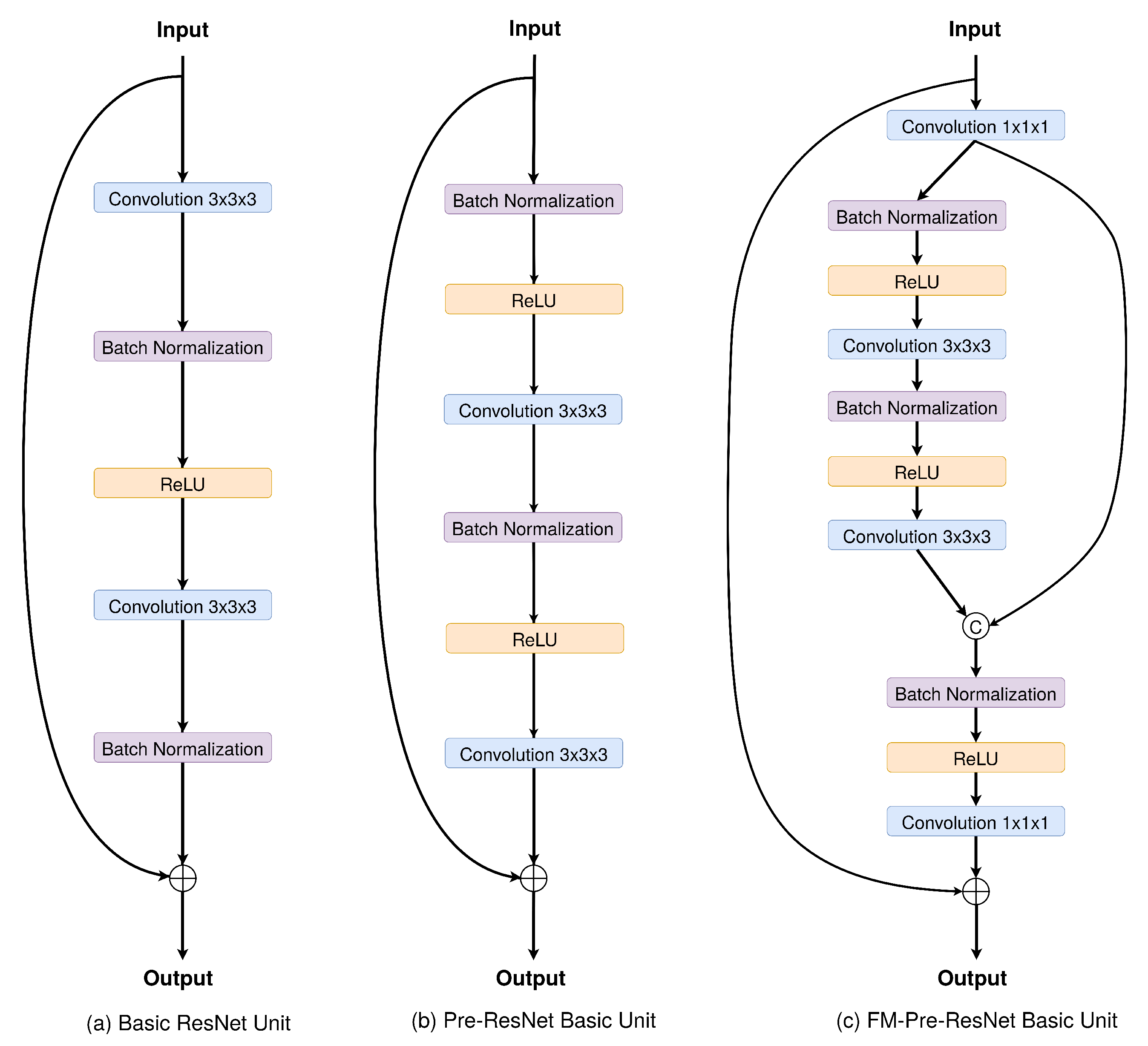
5] enable regularization during the training to ensure that the latent space, i.e., encoded space, keeps the maximum of information when encoding, which results in the minimum reconstruction error during the decoding. Furthermore, since the number of features in the contracting pathway is significantly lower than the number in the expanding pathway, direct concatenation of these features may not produce the most optimal results. The increment in the number of layers provides larger parameter space enabling learning of more abstract features. Therefore, deeper architectures could provide more abstract learning that results in better performance and higher accuracy in medical segmentation tasks. Common obstacles in training deeper neural network architectures are the appearance of vanishing gradients, accuracy degradations, and extensive parameter growth that lead to computationally expensive models. Recent advancements have shown that convolutional networks can be significantly deeper and still preserve high efficiency and accuracy if they contain shorter and direct connections in between each layer. The introduction of skip connections in ResNets [
6] allows for copying of the activations from layer to layer. Since some features are best constructed in shallow networks and others require more depth, skip connections increase the network’s capability, flexibility, and performance.
1.1. Research Contributions
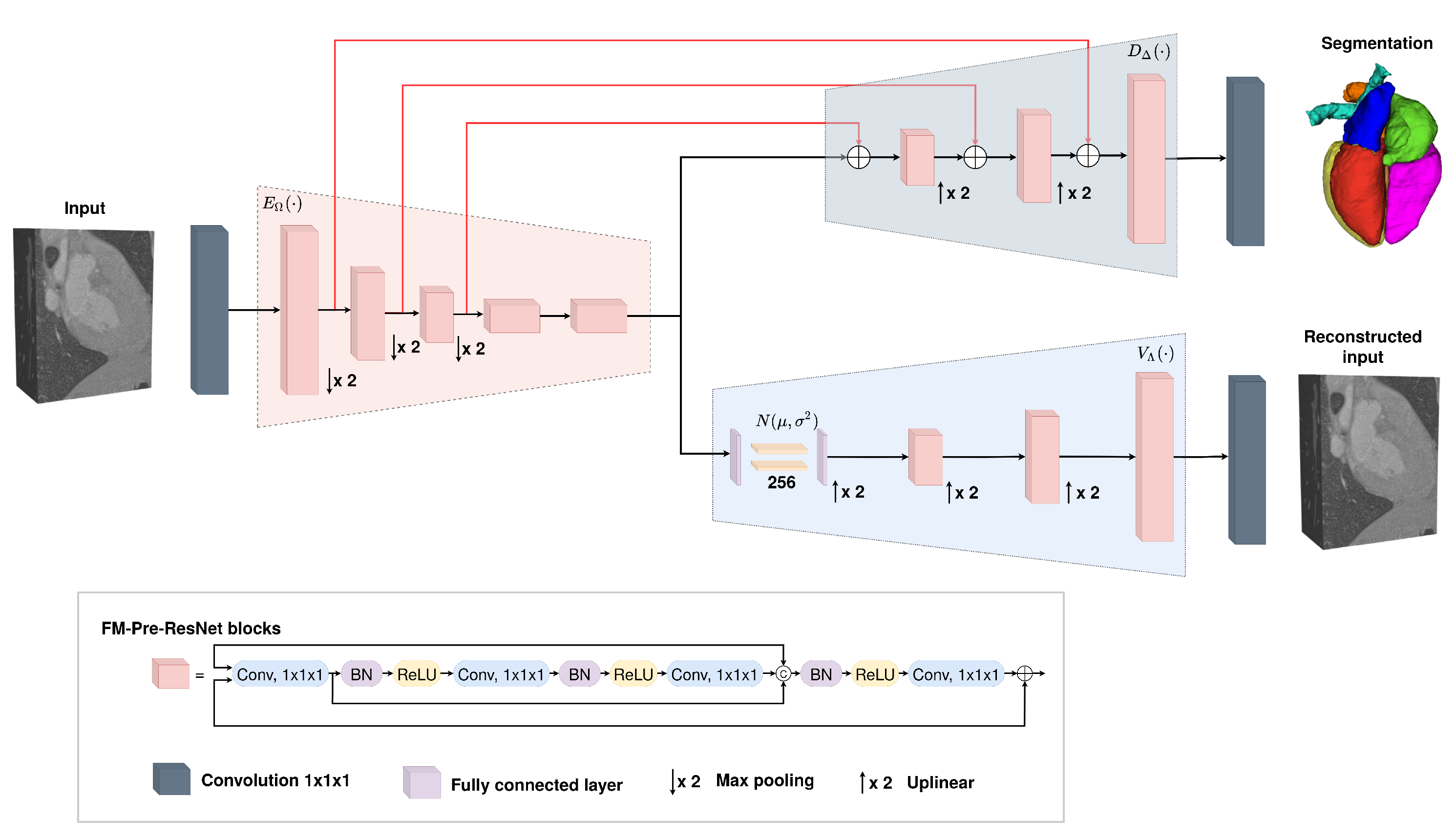
Motivated by previously described advancements, in this paper, we present a novel 3D encoder–decoder based architecture with variational autoencoder regularization. Our intention is to achieve maximum optimality in training performance, efficiency, and final segmentation result accuracy for the whole heart segmentation task. The work contributions can be summarized as:
We propose a new connectivity structure of residual unit that we refer to as feature merge residual units (FM-Pre-ResNet). The FM-Pre-ResNet unit attaches two convolution layers at the top and at the bottom of the pre-activation residual block. The top layer balances the parameters of the two branches, while the bottom layer reduces the channel dimension. This allows the construction of a deeper model with a similar number of parameters compared to the original pre-activation residual unit.
We present a 3D encoder–decoder based architecture that efficiently incorporates FM-Pre-ResNet units and is additionally guided with variational autoencoders (VAE). The architecture includes three stages. First, in an encoding stage, FM-Pre-ResNet units learn a low-dimensional representation of the input. Second, in the VAE stage, an input image is reduced to a low-dimensional latent space and reconstructs itself to provide a strong regularization of all model weights and is only used during the network training. Finally, the decoding stage creates the final whole heart segmentation.
The proposed approach obtains highly comparable dice scores to the state-of-the-art for whole heart segmentation tasks on CT images while outperforming the current state-of-the-art on the MRI images.
1.2. Paper Overview
The remainder of the paper is structured as follows. In
Section 2, we give an overview of related research. First, we briefly review previous research regarding whole heart segmentation. After that, we provide a background of the most successful ResNet variants and feature reuse mechanisms.
Section 3 gives details about our proposed method. First, we introduce an overall architecture design. After that, we present the encoder and decoder stages as well as a theoretical explanation of the new connectivity structure of the residual unit. We introduce the variational autoencoders and their role in the proposed architecture as well.
Section 4 provides dataset description, implementation details, and presents conducted experiments and obtained results for the whole heart segmentation. Finally, the concluding remarks are given in
Section 5.
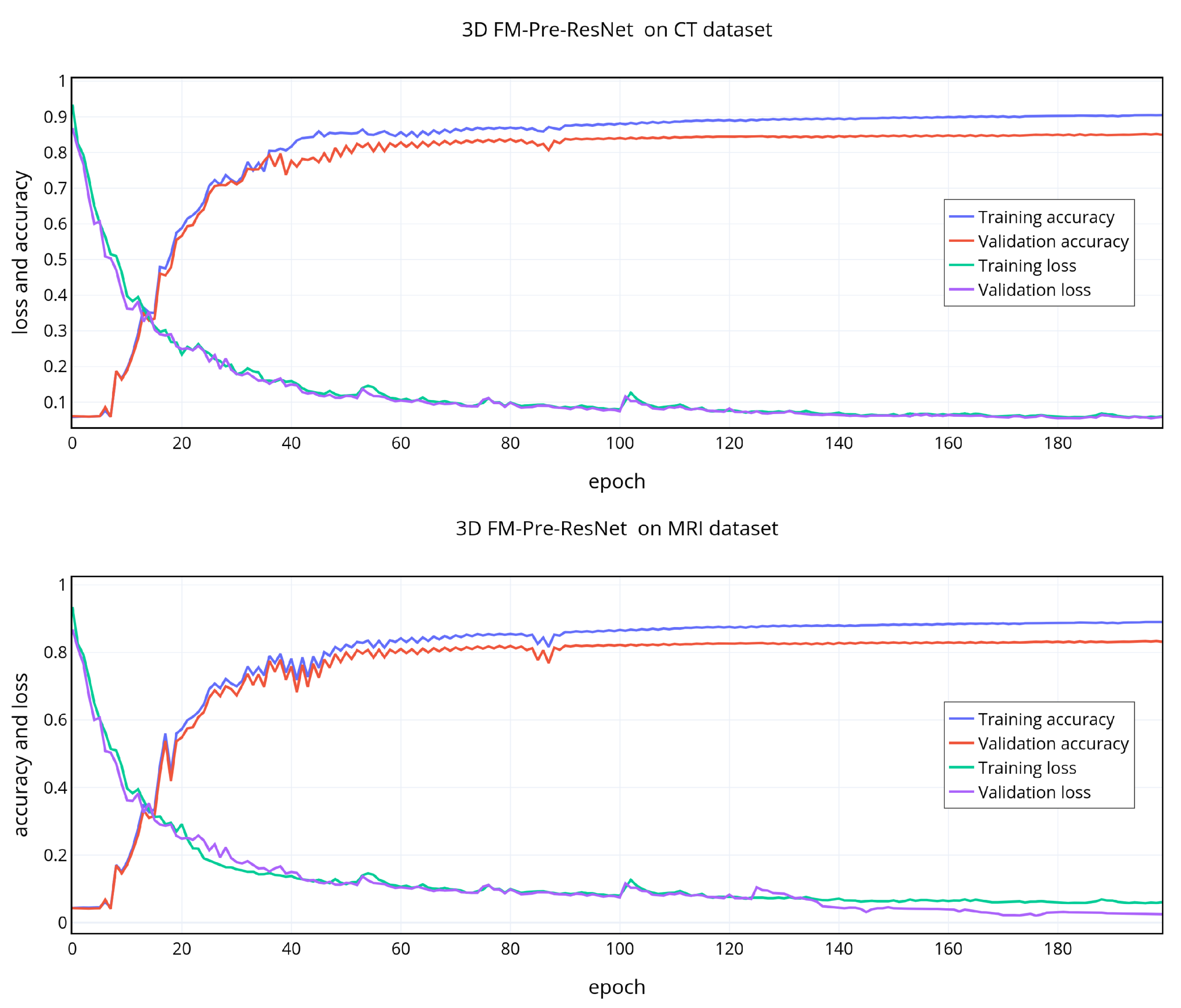
5. Experiments and Results
To demonstrate the effectiveness of the proposed approach and our design choice for the new FM-ResNet unit, we train encoder–decoder based architecture using 3D Pre-ResNet without and with VAE regularization as well as the proposed FM-Pre-ResNet without and with VAE regularization.
Table 2 summarizes an average whole heart segmentation results on CT and MRI images. On CT images, the 3D Pre-ResNet network achieves average WHS segmentation results for DSC, JI, SD, and HD of
,
,
mm, and
mm, respectively. The addition of VAE at Pre-ResNet segmentation encoders’ endpoint improve DSC, JI, SD and HD values for
,
,
mm, and
mm, respectively.
The 3D FM-Pre-ResNet network achieves DSC, JI, SD, and HD values of , , mm, and mm, respectively. Compared to the 3D Pre-ResNet, it achieves improvement in DSC, JI, SD, and HD values of , , mm, and 56,307 mm, which means that the proposed FM-PreResNet unit significantly improves segmentation accuracy. Moreover, the highest DSC, JI, SD, and HD are achieved using 3D FM-Pre-ResNet + VAE network and report values of , , mm, and mm, respectively.
Similarly, on MRI images, the 3D Pre-ResNet network achieves average WHS segmentation results for DSC, JI, SD, and HD of , , mm, and mm, respectively. The addition of VAE at Pre-ResNet segmentation encoders’ endpoint improve DSC, JI, SD, and HD values for , , mm mm.
The 3D FM-Pre-ResNet network achieves average DSC, JI, SD, and HD values of , , mm, and mm, respectively. Compared to 3D Pre-ResNet, it achieves improvement in DSC, JI, SD, and HD values of , , mm, and mm, which means that the proposed FM-PreResNet unit significantly improves segmentation accuracy. Moreover, the highest DSC, JI, SD, and HD are achieved using 3D FM-Pre-ResNet + VAE network and report values of , , mm and mm, respectively. These results highlight the improvement in segmentation accuracy afforded by the introduction of FM-Pre-ResNet units and VAE.
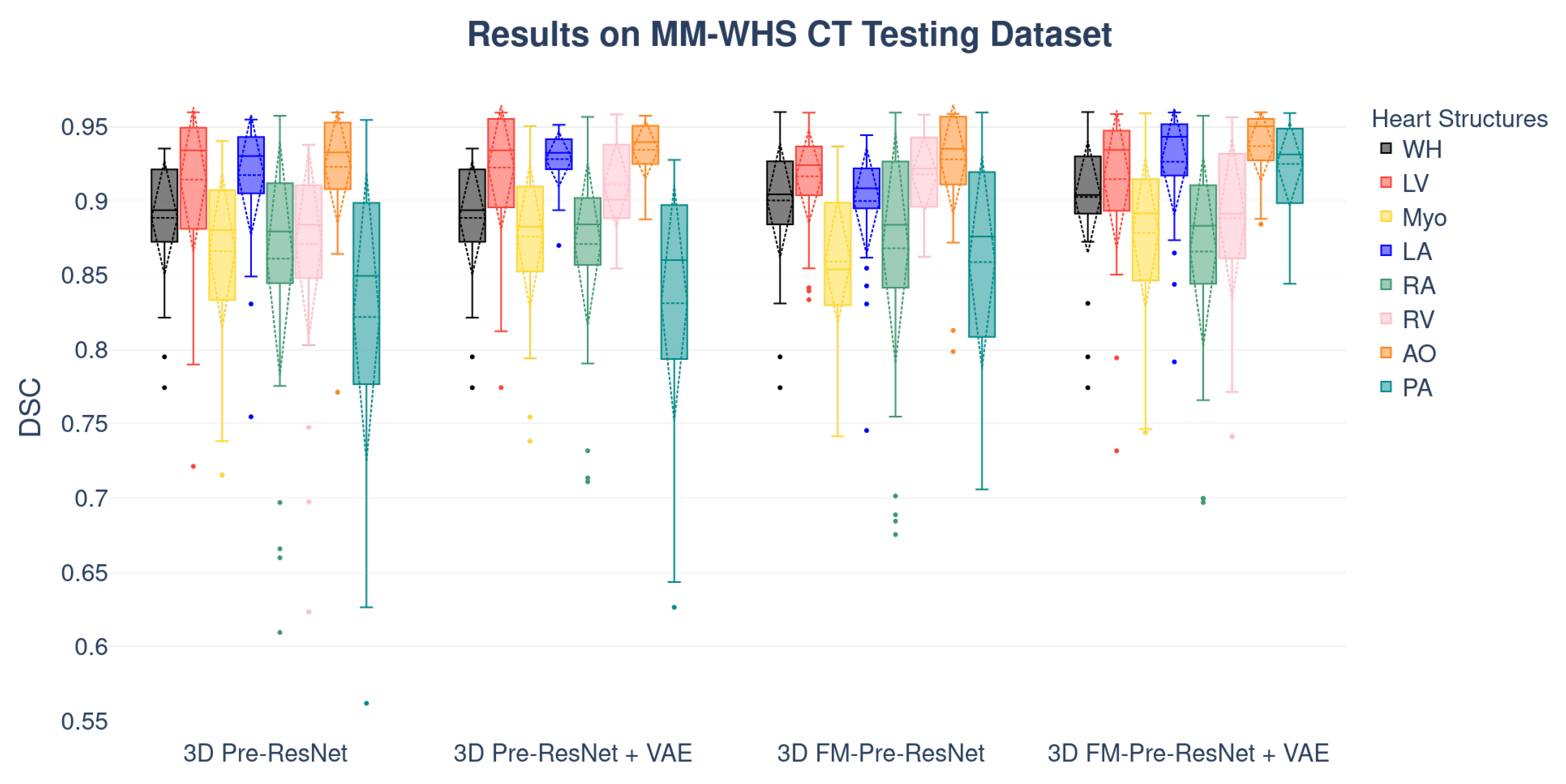
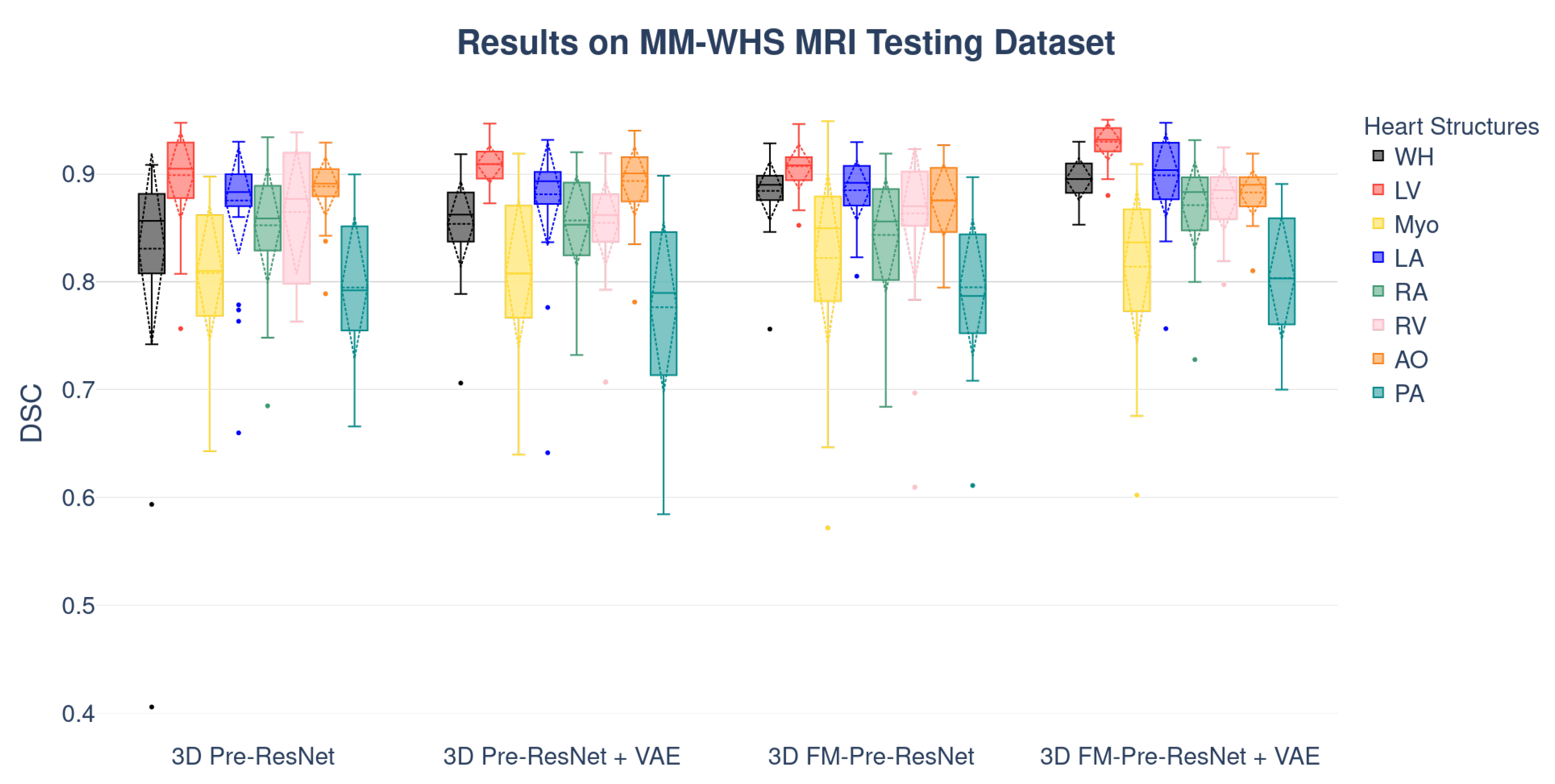
Boxplots showing the distribution of the DSC for WH, LV, Myo, LA, RA, RV, AO, and PA using different segmentation networks on MMWHS CT and MRI testing datasets are presented in
Figure 6 and
Figure 7, respectively. Additional, structure-wise segmentation accuracies for the LV, RV, LA, RA, Myo, Ao, and PA, for both CT and MRI images, are summarized in
Table 3 and
Table 4.
The
p-values have been calculated using a Wilcoxon rank-sum test to show the significant difference of used architectures. Bonferroni correction was used for controlling the family-wise error rate.
Figure 8 and
Figure 9 show the comparisons and
p-values for CT and MRI testing datasets, respectively.
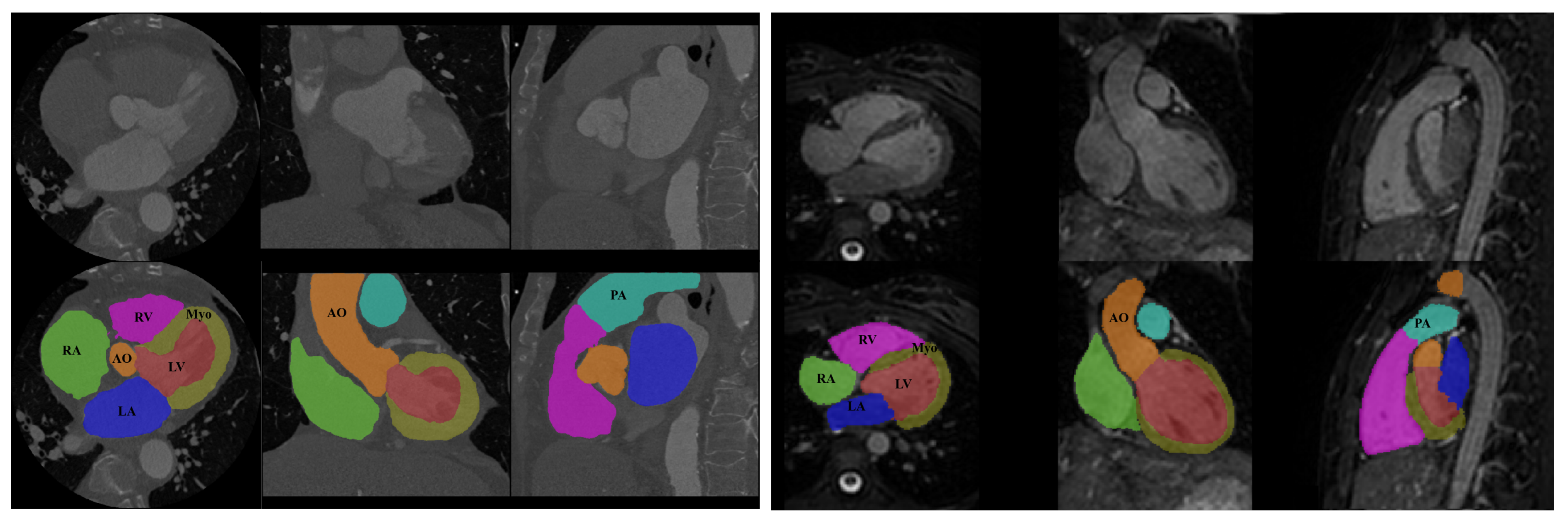
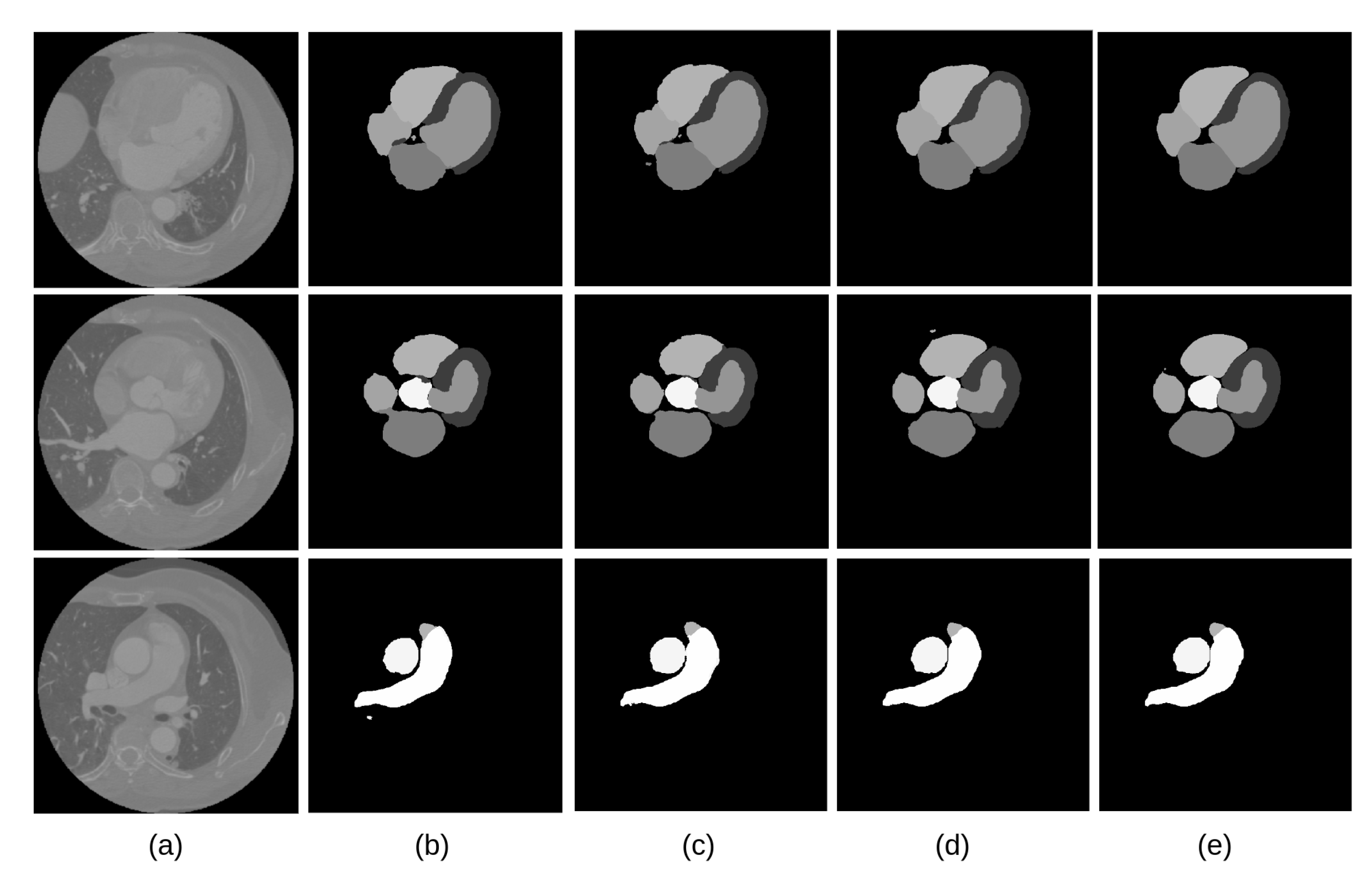
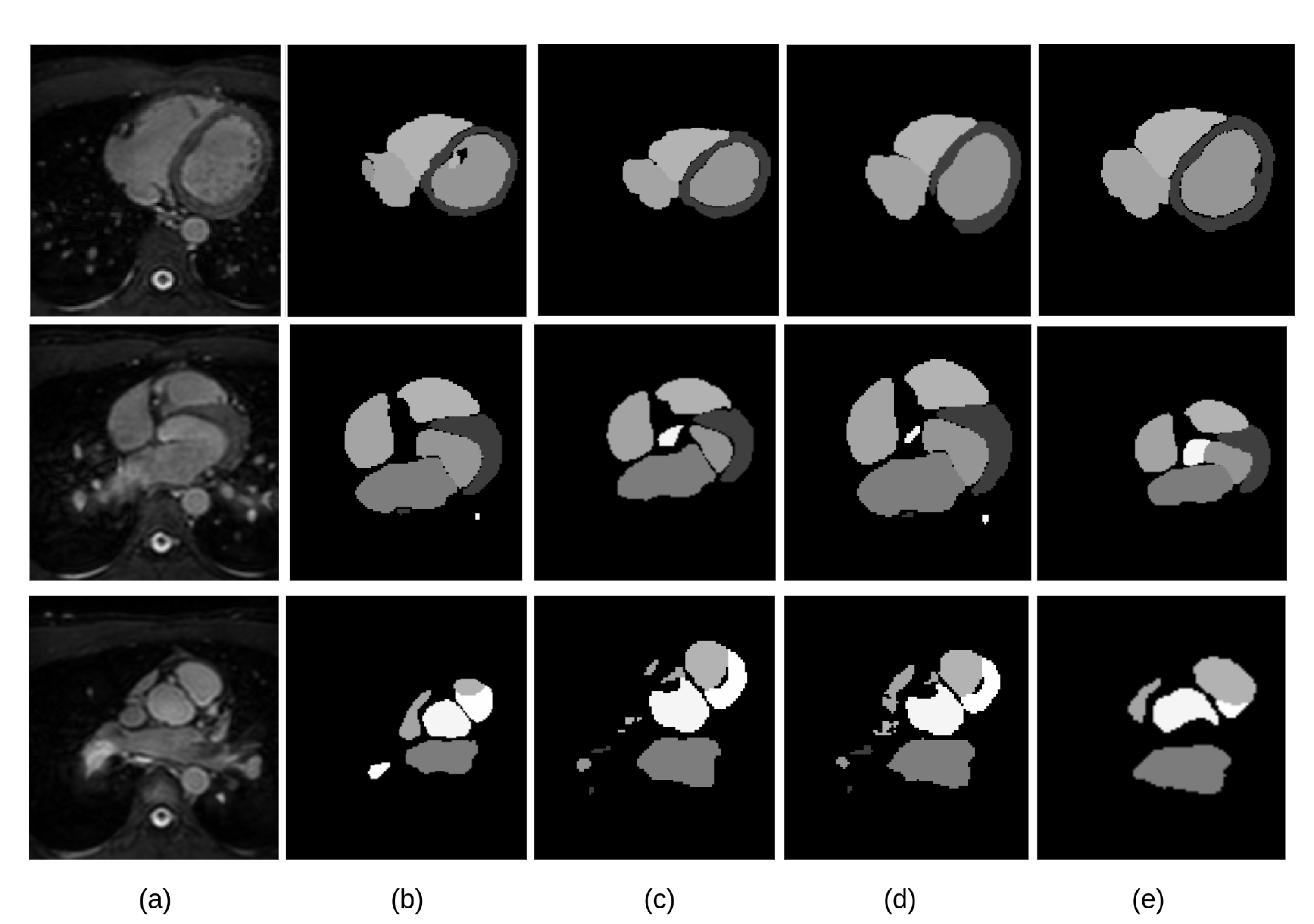
The visual inspection of the obtained segmentations using each network investigated in this work is presented in
Figure 10 for the CT dataset, and
Figure 11 for the MRI dataset. For example,
Figure 11d shows clear improvements regarding LV segmentation that is obtained using FM-Pre-ResNet compared to missed segmentation of LV parts while using Pre-ResNet without a proposed feature merge residual unit as shown in
Figure 11b. Moreover,
Figure 11e shows a significant reduction in segmentation error compared to all other presented networks. This further highlights the benefits of the proposed FM-Pre-ResNet + VAE approach. Nonetheless, in both modalities, PA and Myo’s segmentation results are significantly lower than other substructures due to high shape variations and heterogeneous intensity of blood fluctuations.
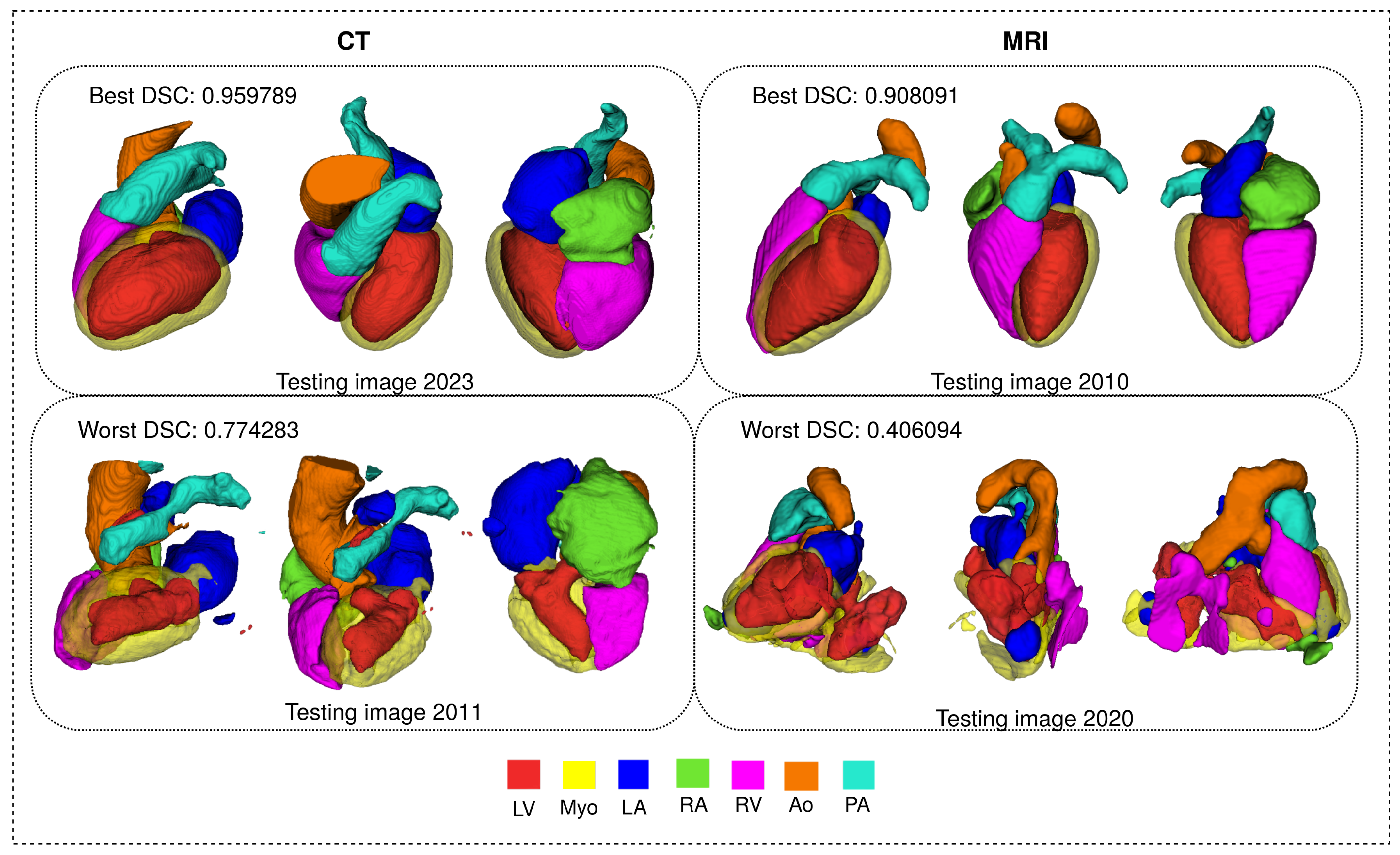
Figure 12 shows 3D visualization of the best and the worse segmentation cases on the CT and MRI test dataset obtained using the proposed FM-Pre-ResNet +VAE approach.
Furthermore, Pre-ResNet has demonstrated that increasing the depth of the network improves model performance significantly. The addition of two convolutional layers at the top and bottom of the pre-activation residual block introduced in our FM-Pre-ResNet unit allows for the feature fusion block to reach the same depth with fewer parameters which benefits model performance. Therefore, the proposed type of connectivity of the FM-Pre-ResNet unit in terms of depth and number of parameters regarding Pre-ResNet implies no increase in the number of parameters compared to the Pre-ResNet. Time-wise, each training epoch (200 cases) and prediction times on two GPU-s (NVIDIA Titan V) are significantly reduced with architectures with VAE. This shows the computational efficiency of our choice for VAE introduction. Comparison of depth, number of parameters, training times per epoch, and prediction time of one volume for different architectures is shown in
Table 5.
Comparison with State-of-the-Art Methods
The proposed approach was compared with other similar deep learning approaches in terms of image segmentation accuracy as shown in
Table 6 and
Table 7. An approach that combines atlas registration with CNNs’ [
12] provides an incremental segmentation that allows user interaction, which can be beneficial in a clinical setting. Nevertheless, the challenges of accurate atlas registration resulted in low accuracy on MRI images. The deep supervision mechanism [
23] and use of transfer learning [
21] result in an increase of trainable parameters and overall network complexity. In contrast, we aim to introduce a light-weight network that results in a significantly deep network without increasing the parameter number. Moreover, the authors in [
21] report an average WHS DSC of
on CT images and
on MRI images using a hold-out set of 10% of training data and evaluate their method with 10-fold cross-validation. Our results report
on CT images and
on MRI images and are evaluated on all unseen 40 subjects, which shows that the VAE stage’s introduction significantly helps in overcoming overfitting problems. Therefore, these results highlight the advantages of our proposed method.
6. Conclusions
This paper introduced an efficient encoder–decoder-based architecture for whole heart segmentation on CT and MRI images. Accurate heart and its substructures segmentation enable faster visualization of target structures and data navigation, which benefits clinical practice by reducing diagnosis and prognosis times. Our proposed method introduces a novel connectivity structure of residual unit that we refer to as feature merge residual unit (FM-Pre-ResNet). The proposed connectivity allows the creation of distinctly deep models without an increase in the number of parameters compared to the Pre-ResNet units. Furthermore, we construct an encoder–decoder-based architecture that incorporates the VAE encoder at the segmentation encoder output to have a regularizing effect on the encoder layers. The segmentation encoder learns a low-dimensional representation of the input, after which VAE reduces the input to a low-dimensional space of 256 (128 to represent std, and 128 to represent mean). A sample is then drawn from the Gaussian distribution with the given std and mean and reconstructed into the input image dimensions following the same architecture as the decoder but without inter-level skip connections. Therefore, VAE acts as a regulator of model weights, adds additional guidance, and exploits the encoder endpoint features. In the end, the segmentation decoder learns high-level features and creates the final segmentations. We evaluate the proposed approach on MMWHS CT and MRI testing datasets and obtain average WHS DSC, JI, SD, and HD values of , , , for CT images, and , , , for MRI images, respectively. Results for both datasets are highly comparable to the state-of-the-art.