Featured Application
Our work is to better discover potential DTI and provide new options for drug redirection.
Abstract
The prediction of drug–target interactions is always a key task in the field of drug redirection. However, traditional methods of predicting drug–target interactions are either mediocre or rely heavily on data stacking. In this work, we proposed our model named GraphMS. We merged heterogeneous graph information and obtained effective node information and substructure information based on mutual information in graph embeddings. We then learned high quality representations for downstream tasks, and proposed an end–to–end auto–encoder model to complete the task of link prediction. Experimental results show that our method outperforms several state–of–the–art models. The model can achieve the area under the receiver operating characteristics (AUROC) curve of 0.959 and area under the precise recall curve (AUPR) of 0.847. We found that the mutual information between the substructure and graph–level representations contributes most to the mutual information index in a relatively sparse network. And the mutual information between the node–level and graph–level representations contributes most in a relatively dense network.
1. Introduction
The prediction of drug target interactions(DTIs) is a key task in the field of drug redirection [1]. Since biochemical experimental assays are extremely costly and time–consuming, efficient methods for identifying new DTIs are essential and valuable. Two main methods for DTI prediction have been studied: molecular docking and machine learning. Molecular docking technology is widely used due to its reasonable accuracy. However, the performance of molecular docking is limited when large scale simulations take time [2]. Compared with the traditional molecular docking technology, machine learning method can conduct large scale testing of drug and protein data in a relatively short time. Several computing strategies have been introduced into machine learning methods to obtain high quality embedding for predicting DTIs.
Since the progress of deep learning, researchers have been able to develop deep neural network models. Deep learning methods are also widely used in feature mapping [3], classification task [4] and disease prediction [5]. Moreover, differentiable representation learning methods can be directly applied on low–level representations to enable the potential of interpretable DTI predictions. In particular, Graph Representation Learning (GRL) can effectively combine feature information and structural information to obtain low-dimensional embedding [6]. Such method includes the random walk method [7] and Graph Convolutional Network (GCN) model [8]. Ordinary graph embedding tends to make the entire input graph smoother. The substructural information about the part of the input graph will be ignored. However, there have been few studies on preserving substructure information in GRL. A substructure is a set of subgraphs represented by a subset of vertices and edges, which is often able to express the unique semantics and fundamental expressions of the graph. More precisely, neighbor nodes in the graph (such as first-order neighbor nodes) are usually trained to obtain similar embedding representations [9]. However, nodes that are far apart in the graph have no similar representations, even if they are similar in structure. Preserving substructure information could effectively prevent such a situation from occurring.
Substructure in graph is generally used to solve three types of problems. One type can be utilized to accelerate large–scale graph training. Cluster–GCN [10] is an example of this. The core idea of Cluster–GCN is to apply the clustering algorithm to divide the large graph into multiple clusters. The division follows the principle of fewer connections between clusters and more connections within clusters. The simple method effectively reduces the consumption of memory and computing resources. At the same time, good prediction accuracy can be achieved. One type can be used for self-supervised learning. SUBG–CON [11] exploits the strong correlation between the central graph node and its sampled subgraphs to capture regional structure information. SUBG–CON is a self–supervised representation learning method based on subgraph contrast. The other type can be applied on denoising in a network, such as a graph. For instance, there are only three nodes around a node. The substructure embedding will select the most representative neighbor node, which can eliminate unnecessary confusion of neighbor nodes. Mutual Information (MI) is a measure of quantifying the relationship between two random variables. Inspired by the MI-based learning algorithms [12,13], we combine substructure embedding with mutual information, ie, adversarial learning, and apply it to DTI prediction to obtain more accurate embedding. In order to make the embedding pay more attention to the contribution of the substructure in the graph, the representation of the substructure should be highly relevant to the graph-level representation. That is, maximizing the correlation between the graph-level and substructure representations helps to retain substructure information. To a certain extent, the application of substructures can improve the embedding effect of the sparse graph network.
In this paper, we propose an end–to–end network model that predicts DTIs from low level representations, called GraphMS. As shown in Figure 1, we apply to guarantee accountability in the node–level representation by maximizing mutual information between the node–level and graph–level representations to guide the encoding step. And then, we propose to preserve the substructure information in the graph–level representation by maximizing mutual information between the graph–level and substructure representations. The high quality embedded information learned by the model is useful for downstream tasks. Finally, combined with learning interpretable feature embedding from heterogeneous information, we use an auto–encoder model to achieve the task of link prediction.
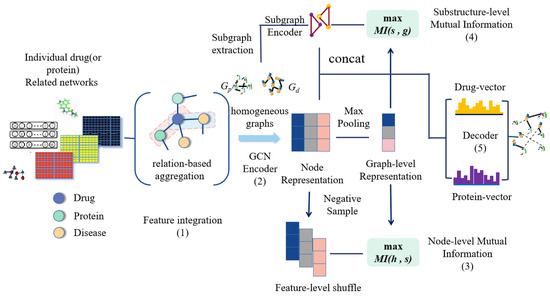
Figure 1.
Framework overview. The model contains five parts: (1) Perform feature integration on heterogeneous graph. (2) GCN encoder is designed to obtain the node-level representation. (3) Feature-level shuffle is taken on the node representation to generate negative samples. Then maximize the mutual information between the node–level representation and graph–level representation. (4) Partition the homogeneous graphs into k subgraphs. Shuffle nodes except the current subgraph and select k nodes randomly to obtain the corresponding negative subgraph. Then maximize the mutual information between the substructure–level representation and graph–level representation. (5) A decoder takes latent vectors as input and output the reconstructed drug-protein matrix.
To summarize, our major contributions include:
- We apply the substructure embedding to DTI prediction, and remove certain noise in the graph network. The subgraph comparison strengthens the correlation between graph-level representation and subgraph representation to capture substructure information;
- We maximize the mutual information of node representation and graph–level representation. This allows the graph–level representation to contain more information about the node itself, and it will be more concentrated on the representative nodes in the embedded representation;
- Case study and comparison method experiments also show that our model is effective.
2. Related Work
2.1. DTI Prediction
In recent years, drug–protein targeting prediction has been widely investigated. The molecular docking method, which takes the 3D structure of a given drug molecule and the target as input, is widely used to predict binding patterns and scores. Although molecular docking can provide visual interpretability, it takes time and is limited by the need to obtain the 3D structure of protein targets [14].
Much effort has been devoted to developing machine learning methods for computational DTI prediction. Wan et al. [15] extracted hidden characteristics of drugs and targets by integrating heterogeneous information and neighbor information. Faulon et al. [16] applied an SVM model to predict DTIs, based on chemical structure and enzyme reactions. Bleakley et al. [2] developed an SVM framework for predicting DTIs, based on a bipartite local model, named BLM. Mei et al. [17] extended this framework by combining BLM with a neighbor–based interaction–profile–inferring (NII) procedure (named BLM–NII), which is able to learn DTI features from neighbors.
As the amount of data on drugs and protein targets has increased, algorithms from the field of deep learning have been used to predict DTIs. Wen et al. [18] developed the deep belief network model, whose input is the fingerprint of the drug and composition of the protein sequence. Chan et al. [19] used a stacked auto–encoder for representing learning, and developed other machine learning methods to predict DTIs.
Recently, GRL has also been applied as an advanced method for identifying potential DTIs. The purpose of GRL is to encode the structural information into low-dimensional vectors and then quantify the graph. Gao et al. [20] and Duvenaud et al. [21] proposed graph convolutional networks with attention mechanisms to model chemical structures and demonstrated good interpretability. Che et al. [22] developed Att-GCN to predict drugs for both ordinary diseases and COVID-19.
Our work solves the problem of retaining the substructure information of a graph. We also obtain an explanatory DTI prediction from low-dimensional representations.
2.2. Graph Representation Learning
In recent years, graph embedding emerged as a promising approach in network data analysis and application. DeepWalk [23] is the first network embedding method proposed to use technology that represents a learning (or deep learning) community. DeepWalk treats nodes as words and generates short random walks. Random walk paths are used as sentences to bridge the gap between network embedding and word embedding. Node2Vec [24] is an extension of DeepWalk. It introduces a biased random walk program that combines BFS style and DFS style neighborhood exploration. LINE [25] generates context nodes with a breadth-first search strategy. Only nodes that are at most two hops away from a given node are considered neighbors. In addition, compared to the hierarchical softmax used in DeepWalk, it uses negative sampling to optimize the Skip-gram model. GCN can capture the global information of graph, so as to well represent the characteristics of node. However, GCN needs all nodes to participate in training to get node embedding. There are many nodes in the graph and the structure of the graph is very complicated. The training cost is very high and it is difficult to quickly adapt to changes in the graph structure. Graph Attention Network (GAT) [26] uses the attention mechanism to perform weighted summation on neighbor nodes.
In traditional graph embedding learning, nodes are adjacent to each other in the input diagram, and embedded represents are similar. Although these methods claim that the snap nodes are close, they still suffer from some limitations. Most notably, they place too much emphasis on proximity similarity, making it difficult to capture inherent graphical structure information. Our work solves the problem of retaining the substructure information of a graph. We also obtain an explanatory DTI prediction from low-dimensional representations.
3. Our Approach
In this section, we introduce the framework of our proposed model. As shown in Figure 1, the framework consists of five parts, including the feature integration, a GCN encoder, a Node-level mutual information estimator, a Substructure-level mutual information estimator, and a decoder for network reconstruction. We also list the steps of our method GraphMS in Algorithm 1 and the algorithm describes as follows:
3.1. Problem Formulation
GraphMS predicts unknown DTIs through a heterogeneous graph associated with drugs and targets.
Definition 1.
(Heterogeneous graph) A heterogeneous graph is defined as a directed or undirected graph , where each node and each edge . Each node v in the node set V belongs to the object type N, and each edge e in the relationship set E belongs to the object type R. The type of node N includes drugs, targets and diseases. The type of relation R includes protein-disease-interaction, drug-protein-interaction, drug-disease-interaction, drug-drug-interaction, protein-protein-interaction, drug-structure-similarity and protein-sequence-similarity.
In our current model, each node only belongs to a single type and all edges are undirected and non–negatively weighted. We adopted the heterogeneous graph that was built in our team previous study. The data example of the heterogeneous graph is shown in the Figure 2.
| Algorithm 1 GraphMS |
| Input: A Heterogeneous Graph |
| Output: Drug–Protein Reconstruction Matrix M |
| 1: Perform feature integration on nodes whose node type according to Equation (1); |
| 2: Select the relationship type and convert to homogeneous graphs ,; |
| 3: while not Convergence do |
| 4: Shuffle X on row dimension to obtain ; |
| 5: for node type do |
| 6: ; //Obtain Node-level Representation (Positive) |
| 7: ; //Obtain Node-level Representation (Negative) |
| 8: ; //Obtain Graph-level Representation |
| 9: Partition , nodes into k subgraphs , , ⋯, by METIS separately; |
| 10: for all each subgraph do |
| 11: Form the subgraph with nodes and edges into ; |
| 12: Shuffle other nodes except nodes of the current subgraph and select k nodes randomly to obtain the corresponding negative subgraph ; |
| 13: ; //Obtain Substructure-level Representation (Positive) |
| 14: //Obtain Substructure-level Representation (Negative); |
| 15: end for |
| 16: ; |
| 17: end for |
| 18: ; |
| 19: Compute the final loss and update parameters according to Equation (14); |
| 20: ; |
| 21: end while |
| 22: return M |
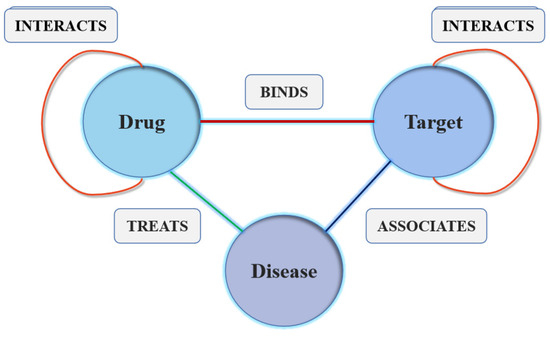
Figure 2.
The data example of the heterogeneous graph.
3.2. Information Fusion on Heterogeneous Graph
The first step is to integrate and transform different types of relationships. The edges whose relationship object type is drug-drug interaction and protein-protein interaction will be converted into isomorphic matrix, that is, homogeneous graph. The edges whose relationship object type is drug structure similarity and protein sequence similarity will be transformed into the initial feature matrix , which is the embedding representation of the node itself. In order to obtain better versatility, we follow the idea of NeoDTI [27] and integrate the neighborhood information of each node (not including the same type of node) with its own embedded integration into a richer feature representation. Given a heterogeneous network graph, we embed the node whose type into a d-dimensional vector, namely . And we embed the edge whose relation type is protein-disease interaction, drug-protein interaction and drug-disease interaction into the real number space through the weight function, namely . The information aggregation of node v is defined as:
where is the set of adjacent nodes connected to through edges of type . f stands for a nonlinear activation function over a single-layer neural network parameterized by weights and bias . represents the edge weight. indicates a normalization term for . Next, we combine the two parts: the aggregated neighbor information and the initial features in the same dimension to obtain the final feature representation.
where .
3.3. GCN Encoder
After obtaining the feature representation, we apply the GCN encoder to calculate the node-level representation. For the drug representation, the drug adjacency matrix, that is, isomorphic matrix is added to one of the identity matrix, and then Laplace decomposition is used to obtain the network matrix. Similarly, the protein representation vector is processed through the same steps.
where , I is the identity matrix, and D is the degree matrix. The corresponding degree matrix is . Following the structure of Graph Convolutional Network, we apply one GCN layer to encode the nodes in our graph, the node representation of the drug or protein view is expressed as:
where is the feature matrix, is the trainable weight matrix, is the network matrix obtained by Laplace decomposition, and is the node representation of the drug or protein view.
3.4. Mutual Information between Node–Level and Graph–Level Representation
Although the graph is compressed and effectively quantified, the learned node representation should be consistent with the level representing the graph that contains global information. Relevance can be quantified by correlation. Similarly, the learned node representation should be highly relevant to the graph-level representation. This prompts us to use mutual information as a measure of quantifying the relationship between two random variables [12]. High mutual information corresponds to a significant reduction in uncertainty, whereas zero mutual information means that the variables are independent [13].
To ensure the reliability represented by the node, we use mutual information to measure the correlation between node–level and graph–level representations. Taking the drug or protein representation vector as an example, we calculate the graph–level global representation of the drug view through the aggregation function:
where is the i-th row vector of the node representation , n is the number of row vectors, is max pooling function, and is the graph-level global representation.
We simply shuffle the feature matrix X on the row-wise dimensions and generate negative example, i.e., . Similarly, we encode the disturbed feature matrix according to the above formula.
where represents the negative node-level Representation. We then apply the bilinear function as the discriminator, namely
where D is the discriminator function, is the trainable weight matrix, is the bilinear function, is the transpose of the node–level representation, and s is the graph–level global representation.
We calculate the cross–entropy loss between the node–level representation h and the graph-level global representation s. In the process of optimizing the loss function, the mutual information between the node–level representation of the drug view and the graph–level representation is captured.
where is the number of positive pairs, is the number of negative pairs, is the node representation of a positive example pair, is the node representation of a negative example pair.
3.5. Mutual Information Between Graph–Level Representation and Substructure Representation
Substructure, a subset of graph structure, is uniquely representative in distinguishing graphs. Therefore, to quantify the common goal of the graph, the representation of the substructure should be highly relevant to the graph-level representation. That is, maximizing the correlation between the graph-level and substructure representations helps to retain substructure information.
As Figure 3 shows, we use the Metis [28] algorithm to extract k subgraphs with nodes and edges formed as from homogeneous graphs ,. Where , represent drug–drug relationship matrix and protein–protein relationship matrix. Then we obtain the substructure-level representation of positive samples. Since the decomposed subgraph is sparser than the original graph, we enhance the node embedding of the subgraph by enlarging the diagonal part of the adjacency matrix. It also means that there are some subgraphs containing distant nodes in the graph, which can make this type of subgraph contributes more to the embedding that is finally used to reconstruct the drug–protein matrix. is formed as:
where represents a learnable parameter. is to diagonalize the matrix. Then we obtain the substructure-level representation of positive samples .
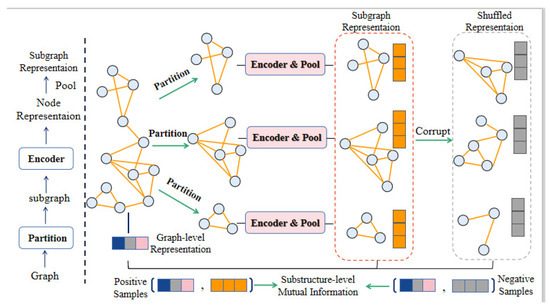
Figure 3.
The process of retaining the sub-structure mutual information. Specifically, on the left is the process of obtaining the subgraph embedding. On the right is the process of generating negative subgraphs. We shuffle nodes except the current subgraph and select k nodes randomly to obtain the corresponding negative subgraph.
The goal is to construct subgraphs on the vertices of the graph so that there are more links between subgraphs than links within subgraphs. This can better capture the clustering structure of the graph, and can effectively focus on the sparse structure. Intuitively speaking, each node and its neighbors are usually located in the same subgraph. After a few hops, the neighbor nodes are still in the same subgraph with a high probability. For the k-th subgraph, we obtain the graph–level representation s, and generate the substructure representation with the nodes related to the substructure.
For the k-th subgraph, we take as a positive sample and as a negative sample, where is a subgraph representation in which nodes are randomly selected from other graphs. The detailed process of retaining the sub-structure mutual information is shown in Figure 3. In detail, for the k-th subgraph representation, our corruption function shuffles the other nodes and select k nodes randomly to obtain the negative subgraphs . Then we obtain the substructure-level representation of negative samples.
In this way, the graph-level representation is closely related to the subgraph representation, while the correlation with other random negative subgraphs is weaker.
A neural network is used to maximize the mutual information between the graph–level representation s and the substructure representation g, to ensure a high correlation. Using cross-entropy to calculate the loss function, the mutual information between the graph–level representation and the substructure representation of the drug view is captured by
where is the substructure representation of the k-th subgraph in a positive example pair, and is the substructure representation of the k-th subgraph in a negative example pair.
3.6. Automatic Decoder for Prediction
We connect the above subgraph embeddings and perform transposition operations. Then we combine the embeddings of the entire graph by concating on the same dimension to obtain the final embedding, i.e., .
According to the final learned drug–embedding and protein–embedding representations, i.e., , , the drug–protein relationship matrix is reconstructed by performing inverse matrix decomposition. We obtain the predicted drug–protein matrix, which is compared with the known drug–protein relationship matrix. We then integrate the loss function of the reconstructed drug–protein matrix and capture the cross–entropy loss function in the mutual information. Finally, we perform gradient update, and optimize the loss function:
Reconstructing the final drug–protein matrix, we obtain
where G is the original matrix, U is the final learned drug-embedding representation, V is the final learned protein-embedding representation, and are the learnable weight matrix, and M is the final predicted drug–protein matrix.
4. Experiments and Results
4.1. Datasets
We used datasets that were compiled in previous studies. We give the source of the data used in the section Data Availability Statement in the article. We describe the data in detail. The drug-drug relational network, i.e., mentioned in the model and protein sequence similarity are shown in Figure 4. 1 means known interaction, 0 means unknown interaction.
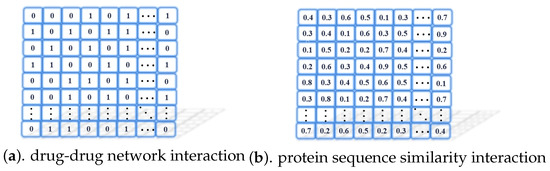
Figure 4.
Some data examples used in model.
We also perform basic statistical analysis on the data. Table 1 and Table 2. show the statistics of node and edge of the heterogeneous graph. These datasets include five individual drug-related and protein-related networks: drug–protein interaction and drug–drug interaction, protein–protein interaction, and drug–disease and protein–disease association networks. We also used two feature networks: the drug–structure similarity network and the protein–sequence similarity network.

Table 1.
Statistics of nodes.

Table 2.
Statistics of edges.
4.2. Experimental Settings
We applied the Adam gradient descent optimization algorithm, with initial learning rate set to 0.001, to train the parameters. During training, the parameters were initialized randomly from a uniform distribution . We trained the model for 10 epochs, where each epoch contained 100 steps. The model used a 10–fold cross–validation procedure after each epoch. Our method shows the best performance among these comparative methods. Performance was measured by the area under the Receiver Operating Characteristics (ROC) curve and the area under the Precision Recall (PR) curve. Then we calculated the mean and standard deviation of the indicator.
4.3. Baselines
We compared our model with four baseline methods. Two of the comparison methods are DTI prediction methods including NeoDTI, DTINet, and two other graph embedding methods including LightGCN and GAT.
- NeoDTI [27] integrates the neighborhood information constructed by different data sources through a large number of information transmission and aggregation operations.
- DTINet [29] aggregates information on heterogeneous data sources, and can tolerate large amounts of noise and incompleteness by learning low–dimensional vector representations of drugs and proteins.
- LightGCN [30] simplified the design of GCN to make it more concise. This model only contains the most important part of GCN–neighborhood aggregation for collaborative filtering.
- GAT [26] proposes to use the attention mechanism to weight and sum the features of neighboring nodes. The weight of features of neighboring nodes depends entirely on the features of the nodes and is independent of the graph structure. GAT uses the attention mechanism to replace the fixed standardized operations in GCN. In essence, GAT just replaces the original GCN standardization function with a neighbor node feature aggregation function using attention weights.
4.4. Comparative Experiment
In the chart, 1:10 means that the ratio of positive samples to negative samples was 1:10. 1:all means that all unknown drug–target interaction pairs were considered. Specially, the ratio between positive and negative samples was around 1:500. The whole network is sparse against the network with the ratio of 1:10. Single–view means that we only use the drug-structure-similarity network and protein-sequence-similarity network. Multi–view means that we use all networks mentioned above. The results are shown in Table 3 and Table 4. In particular, the numbers reported for the NeoDTI method differ from our work with the same dataset. On the one hand, we have reproduced NeoDTI without evaluation strategies. Experiments under various conditions have been carried out more than 5 times. The average level of our reproduced results is lower than the results reported in NeoDTI. NeoDTI followed the evaluation strategies by removing DTIs with similar drugs or similar proteins and so on while we didn’t follow the evaluation strategies. Also our comparison procedure extends to the other methods. We conduct comparative experiments without evaluation strategies in this paper. The calculation strategy improves the performance of NeoDTI to a certain extent, which should be the main reason for the difference in results from our work. On the other hand, we did not use the drug-side-effect network in the comparative experiment, while the network was used in NeoDTI.

Table 3.
Comparison of AUROC scores for various methods under different experimental settings. The best results in each column are in bold.

Table 4.
Comparison of AUPR scores for various methods under different experimental settings. The best results in each column are in bold.
The AUPR and AUROC metrics were used to evaluate the performance of the above prediction methods. Among the existing methods, LightGCN showed the best performance. Our model improved AUROC by nearly 3% and AUPR by nearly 5% against NeoDTI. In highly skewed datasets, AUPR is usually more informative than AUROC. Since drug discovery is usually a problem like finding a needle in a haystack, the high AUPR score also truly proves the superior performance of GraphMS, compared to other methods.
The multi-view model is better than the single-view model experimental results. This is because the multi-view model integrates more feature information of drug targets. Higher quality features are extracted through the framework to provide good results for subsequent reconstruction of the matrix. The graph convolutional neural network indicates that the feature prediction result of the node vector extracted by learning is higher than that of the ordinary auto–encoder. This also proves from the side that the graph convolutional neural network has stronger feature expression ability for non-Euclidean data. Compared with the GAT network index, LightGCN has little change, which proves that the graph embedding makes the input graph smoother. The indicators of our model perform well. On the one hand, the addition of mutual information allows the model to consider the strong correlation between the graph-level representation and its subgraph representation. On the other hand, the subgraph embedding can eliminate certain noise in the network.
4.5. Ablation Experiment
We visualized the embedding learned by Graph-M and Graph-S to see if Graph-S could capture the structure of an interactive network better than Graph-M. Graph-M uses only mutual information between node-level and graph-level representations, whereas Graph-S uses only mutual information between substructure and graph-level representations. Then we visualize the learned embedding in Figure 5. We could observe that there are potentially linked targets near some relatively marginal drug points in the embedded space which Graph-S learns. And the distribution of drug targets in the embedded space of Graph-M learning is more concentrated. We further corroboration with AUROC and AUPR indicators.
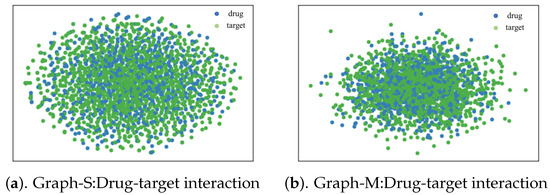
Figure 5.
Visualizations of drug-target interaction network. We use Graph-M and Graph-S on the drug-target interaction dataset to learn drug/target embedding, which are visualized using t-SNE. Graph-M means the model uses only mutual information between node-level and graph-level representations. Graph-S means the model uses only mutual information between substructure and graph-level representations. The detail could be refered to the specific part of the above framework.
Our method uses the mutual information between the substructure and graph-level representations and the mutual information between the node–level and graph–level representations. It can be observed that the mutual information between the substructure and graph-level representations contributes more to the mutual information in a relatively sparse network. In a relatively dense network, the node–level and graph–level representations contribute more to the mutual information (see Figure 6).
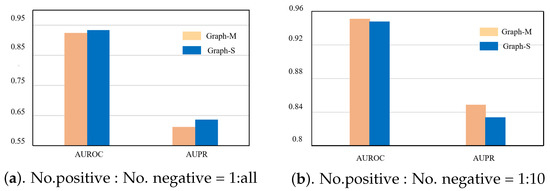
Figure 6.
(a) Diagram to compare ablation experiments under the condition in which all unknown drug–target interaction pairs were considered. (b) Diagram to compare ablation experiments under the condition in which the ratio of positive to negative samples was 1:10.
4.6. Case Study for Interpretability
The network visualization of the top 30 novel DTIs predicted by GraphMS can be found in Figure 7. Ethoxzolamide (EZA) interacts with only two types of drugs and EZA (drug) has a high link probability with DNA methyltransferases (DNMT1). EZA is an FDA-approved diuretic as a human carbonic anhydrase inhibitor. After consulting relevant medical literature, it was found that EZA has potential to treat duodenal ulcer and will be developed into a new anti-Helicobacter drug [31]. Chronic inflammation is closely related to various human diseases, such as cancer, neurodegenerative diseases, and metabolic diseases [32]. Among these, abnormal DNA methylation occurs to some extent, and the enzymatic activity of DNMTs increases. This also shows that there is a nonzero probability of a link between EZA and DNMT1.
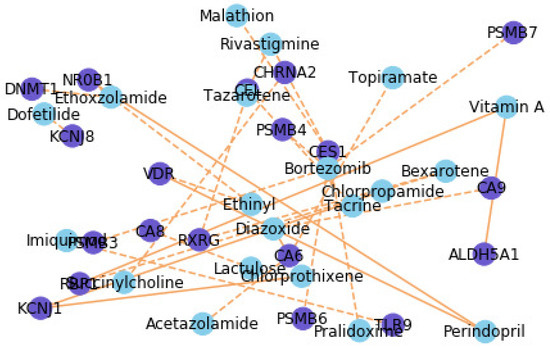
Figure 7.
Network visualization of the top 30 novel drug–target interactions predicted by GraphMS. Purple and blue nodes represent proteins and drugs, respectively. solid and dashed lines represent the known and predicted drug–target interactions.
5. Discussion
This paper merges heterogeneous graph information and obtains effective node information and substructure information based on mutual information in heterogeneous graph. We apply the subgraph embedding to DTI prediction, and remove certain noise in the graph network. Then we present an end-to-end auto–encoder model to predict the interaction of drug targets. The overall experimental evaluation showed that the method was superior to all baselines and at a better level in sparse networks, which was essential for drug discovery. In the ablation experiment, the substructure representation is more important in a relatively sparse network, and some unnecessary noise information in the network can be eliminated. In addition, our model shows top30 DTI pairs and we have shown through a case study that our approach can understand the nature of predictive interactions from a biological perspective.
Our work provides solutions for drug redirection. At the same time, this work can also help medical staff provide new drug ideas for protein targets corresponding to some special diseases. Our work also has some flaws. Due to the large number of training parameters, when it comes to using GCN embedding to calculate the graph-level representation, the nodes of the entire graph will participate in the calculation. This also leads to a longer training time for the entire model. Therefore, in future work, we will refer to some graph network acceleration algorithms such as Cluster-GCN and some new deep learning algorithms, such as meta-learning, to improve the computational efficiency of our model.
6. Patents
Part of the work in this manuscript has been applied for China’s national invention patents. Patents have been made public. The patent application number is 202011275141.6. The application publication number is CN112382411A.
Author Contributions
Conceptualization, S.C. and B.J.; methodology, S.C. and X.L.; validation, S.C.; data collation, L.Z.; writing—original draft preparation, S.C. and Q.Z.; writing—review and editing, L.Z., M.Y.; visualization, S.C.; project administration, B.J., M.Y., X.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Key Program of China (2018YFC0116800) and National Natural Science Foundation of China (No. 61772110).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data is avaiable at https://github.com/FangpingWan/NeoDTI (accessed on 2 April 2021).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Altae-Tran, H.; Ramsundar, B.; Pappu, A.S.; Pande, V. Low data drug discovery with one-shot learning. ACS Cent. Sci. 2017, 3, 283–293. [Google Scholar] [CrossRef]
- Bleakley, K.; Yamanishi, Y. Supervised prediction of drug–target interactions using bipartite local models. Bioinformatics 2009, 25, 2397–2403. [Google Scholar] [CrossRef]
- Ali, F.; El-Sappagh, S.; Islam, S.; Ali, A.; Attique, M.; Imran, M.; Kwak, K. An intelligent healthcare monitoring framework using wearable sensors and social networking data. Future Gener. Comput. Syst. 2021, 114, 23–43. [Google Scholar] [CrossRef]
- Xie, Y.; Yao, C.; Gong, M.; Chen, C.; Qin, A. Graph convolutional networks with multi-level coarsening for graph classification. Knowl. Based Syst. 2020, 194, 105578. [Google Scholar] [CrossRef]
- Ali, F.; El-Sappagh, S.; Islam, S.; Kwak, D.; Ali, A.; Imran, M.; Kwak, K. A smart healthcare monitoring system for heart disease prediction based on ensemble deep learning and feature fusion. Inf. Fusion 2020, 63, 208–222. [Google Scholar] [CrossRef]
- Zhao, T.; Hu, Y.; Valsdottir, L.R.; Zang, T.; Peng, J. Identifying drug-target interactions based on graph convolutional network and deep neural network. Briefings Bioinform. 2020, 22, 2141–2150. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Cao, S.; Lu, W.; Xu, Q. Grarep: Learning graph representations with global structural information. In Proceedings of the 24th ACM International on Conference on Information and Knowledge Management, Melbourne, VIC, Australia, 19–23 October 2015; pp. 891–900. [Google Scholar]
- Chiang, W.L.; Liu, X.; Si, S.; Li, Y.; Bengio, S.; Hsieh, C.J. Cluster-gcn: An efficient algorithm for training deep and large graph convolutional networks. In Proceedings of the 25th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining, Anchorage, AK, USA, 4–8 August 2019; pp. 257–266. [Google Scholar]
- Jiao, Y.; Xiong, Y.; Zhang, J.; Zhang, Y.; Zhang, T.; Zhu, Y. Sub-graph Contrast for Scalable Self-Supervised Graph Representation Learning. arXiv 2020, arXiv:2009.10273. [Google Scholar]
- Velickovic, P.; Fedus, W.; Hamilton, W.L.; Liò, P.; Bengio, Y.; Hjelm, R.D. Deep Graph Infomax. arXiv 2019, arXiv:1809.10341. [Google Scholar]
- Park, C.; Han, J.; Yu, H. Deep multiplex graph infomax: Attentive multiplex network embedding using global information. Knowl. Based Syst. 2020, 197, 105861. [Google Scholar] [CrossRef]
- Zhu, Y.; Che, C.; Jin, B.; Zhang, N.; Su, C.; Wang, F. Knowledge-driven drug repurposing using a comprehensive drug knowledge graph. Health Inform. J. 2020, 26, 2737–2750. [Google Scholar] [CrossRef] [PubMed]
- Wan, F.; Zeng, J.M. Deep learning with feature embedding for compound-protein interaction prediction. bioRxiv 2016, 086033. [Google Scholar] [CrossRef]
- Faulon, J.L.; Misra, M.; Martin, S.; Sale, K.; Sapra, R. Genome scale enzyme–metabolite and drug–target interaction predictions using the signature molecular descriptor. Bioinformatics 2008, 24, 225–233. [Google Scholar] [CrossRef]
- Mei, J.P.; Kwoh, C.K.; Yang, P.; Li, X.L.; Zheng, J. Drug–target interaction prediction by learning from local information and neighbors. Bioinformatics 2013, 29, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Zhang, Z.; Niu, S.; Sha, H.; Yang, R.; Yun, Y.; Lu, H. Deep-learning-based drug–target interaction prediction. J. Proteome Res. 2017, 16, 1401–1409. [Google Scholar] [CrossRef]
- Hu, P.W.; Chan, K.C.; You, Z.H. Large-scale prediction of drug-target interactions from deep representations. In Proceedings of the 2016 International Joint Conference on Neural Networks (IJCNN), Vancouver, BC, Canada, 24–29 July 2016; pp. 1236–1243. [Google Scholar]
- Gao, K.Y.; Fokoue, A.; Luo, H.; Iyengar, A.; Dey, S.; Zhang, P. Interpretable Drug Target Prediction Using Deep Neural Representation. IJCAI 2018, 2018, 3371–3377. [Google Scholar]
- Duvenaud, D.; Maclaurin, D.; Aguilera-Iparraguirre, J.; Gómez-Bombarelli, R.; Hirzel, T.; Aspuru-Guzik, A.; Adams, R.P. Convolutional networks on graphs for learning molecular fingerprints. arXiv 2015, arXiv:1509.09292. [Google Scholar]
- Che, M.; Yao, K.; Che, C.; Cao, Z.; Kong, F. Knowledge-Graph-Based Drug Repositioning against COVID-19 by Graph Convolutional Network with Attention Mechanism. Future Internet 2021, 13, 13. [Google Scholar] [CrossRef]
- Perozzi, B.; Al-Rfou, R.; Skiena, S. Deepwalk: Online learning of social representations. In Proceedings of the 20th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 24–27 August 2014; pp. 701–710. [Google Scholar]
- Grover, A.; Leskovec, J. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 855–864. [Google Scholar]
- Tang, J.; Qu, M.; Wang, M.; Zhang, M.; Yan, J.; Mei, Q. Line: Large-scale information network embedding. In Proceedings of the 24th International Conference on World Wide Web, Florence, Italy, 18–22 May 2015; pp. 1067–1077. [Google Scholar]
- Veličković, P.; Cucurull, G.; Casanova, A.; Romero, A.; Lio, P.; Bengio, Y. Graph attention networks. arXiv 2017, arXiv:1710.10903. [Google Scholar]
- Wan, F.; Hong, L.; Xiao, A.; Jiang, T.; Zeng, J. NeoDTI: Neural integration of neighbor information from a heterogeneous network for discovering new drug–target interactions. Bioinformatics 2019, 35, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Karypis, G.; Kumar, V. A Fast and High Quality Multilevel Scheme for Partitioning Irregular Graphs. SIAM J. Sci. Comput. 1998, 20, 359–392. [Google Scholar] [CrossRef]
- Luo, Y.; Zhao, X.; Zhou, J.; Yang, J.; Zhang, Y.; Kuang, W.; Peng, J.; Chen, L.; Zeng, J. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Deng, K.; Wang, X.; Li, Y.; Zhang, Y.; Wang, M. Lightgcn: Simplifying and powering graph convolution network for recommendation. In Proceedings of the 43rd International ACM SIGIR Conference on Research and Development in Information Retrieval, Xi’an, China, 25–30 July 2020; pp. 639–648. [Google Scholar]
- Takeshima, H.; Niwa, T.; Yamashita, S.; Takamura-Enya, T.; Iida, N.; Wakabayashi, M.; Nanjo, S.; Abe, M.; Sugiyama, T.; Kim, Y.J.; et al. TET repression and increased DNMT activity synergistically induce aberrant DNA methylation. J. Clin. Investig. 2020, 130, 10. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M.; Tikhomirova, A.; Modak, J.K.; Hutton, M.L.; Supuran, C.T.; Roujeinikova, A. Antibacterial activity of ethoxzolamide against Helicobacter pylori strains SS1 and 26695. Gut Pathog. 2020, 12, 1–7. [Google Scholar] [CrossRef]
Short Biography of Authors
 | Bo Jin Professor, PhD supervisor, innovative talents in colleges and universities in Liaoning Province, special consultant of Baidu Research Institute, outstanding member of CCF of China Computer Society, senior member of American ACM and IEEE. He graduated from Dalian University of Technology with a Ph.D., and visited Rutgers, the State University of New Jersey in the United States under the tutelage of Professor Xiong Hui, an authoritative scholar in the field of international big data. Served as Chair at ICDM, a top conference in the field of data mining for two consecutive years, and only two domestic experts hold relevant positions each year. The main research direction is deep learning, big data mining, artificial intelligence, and is committed to the analysis and mining methods of multi-source heterogeneous networked and serialized data. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).