Anomaly Analysis of Alzheimer’s Disease in PET Images Using an Unsupervised Adversarial Deep Learning Model
Abstract
:1. Introduction
- Novelty—the Alzheimer’s disease anomaly analysis of PET images using a proposed unsupervised adversarially trained model with a unique feature extractor model. To the authors’ knowledge, there are no anomaly detection studies in Alzheimer’s disease cases using adversarial deep learning models.
- Effectiveness—the proposed model quantitatively and qualitatively outperforms the state-of-the-art models.
2. Related Anomaly Detection Works
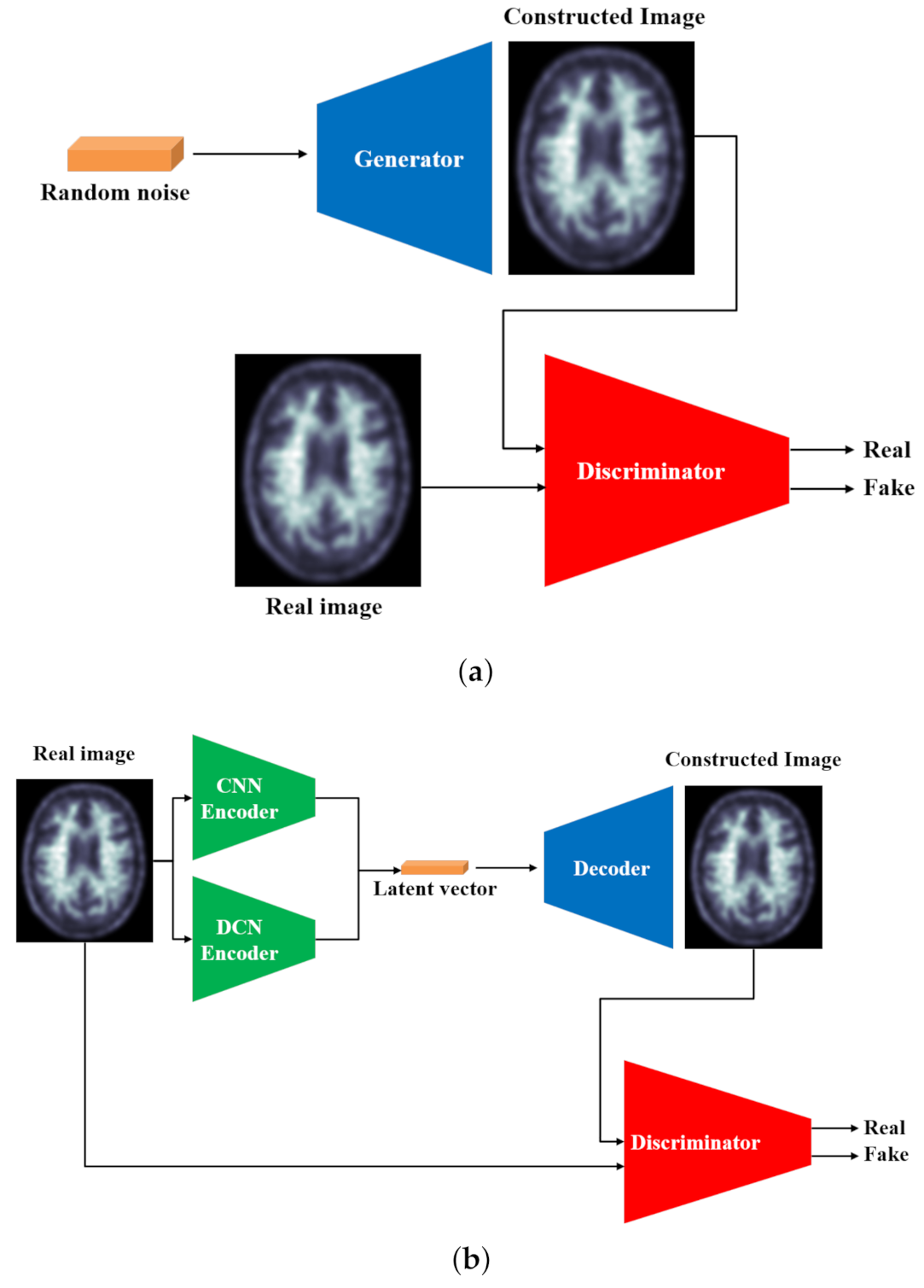
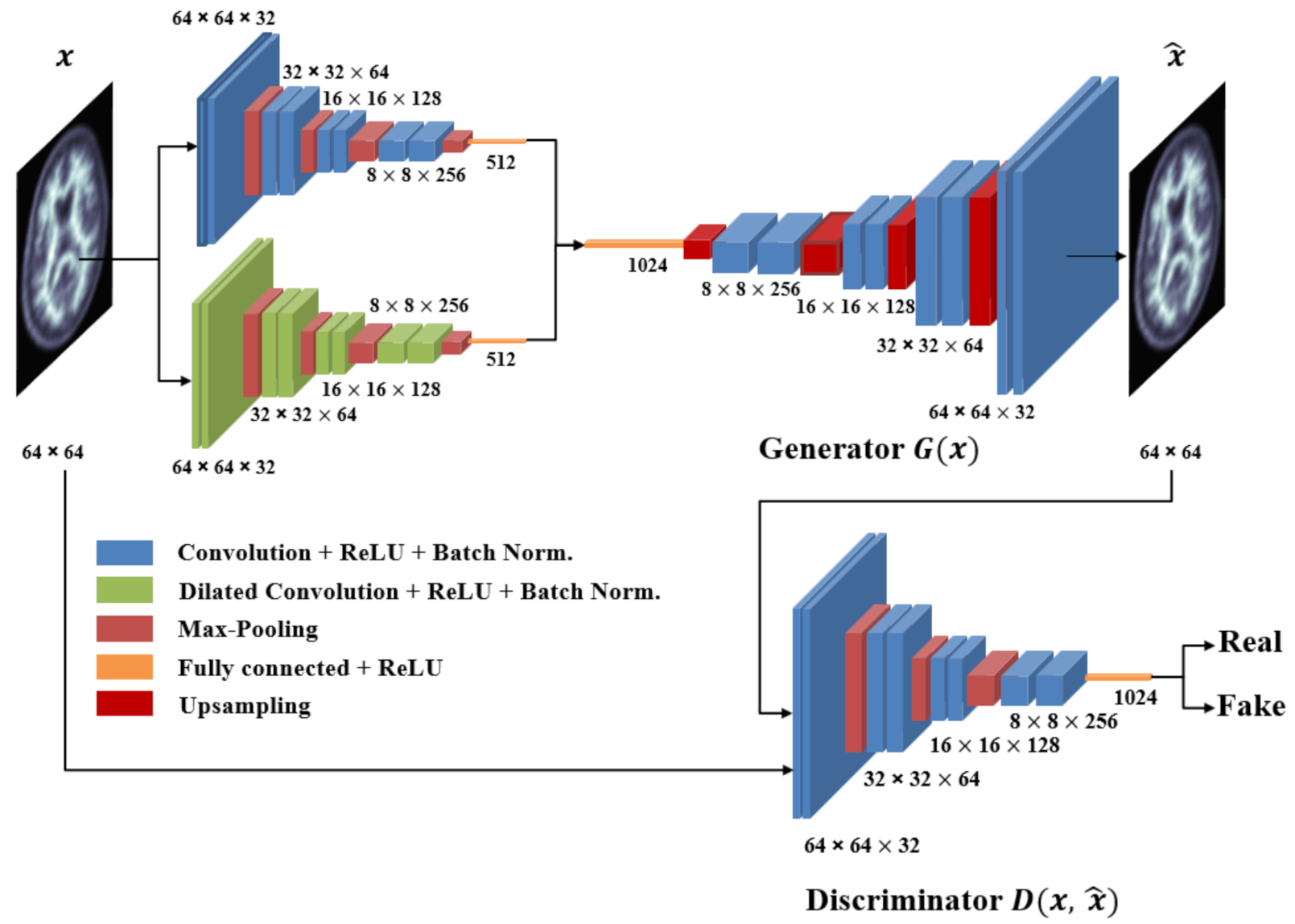
3. Proposed Model
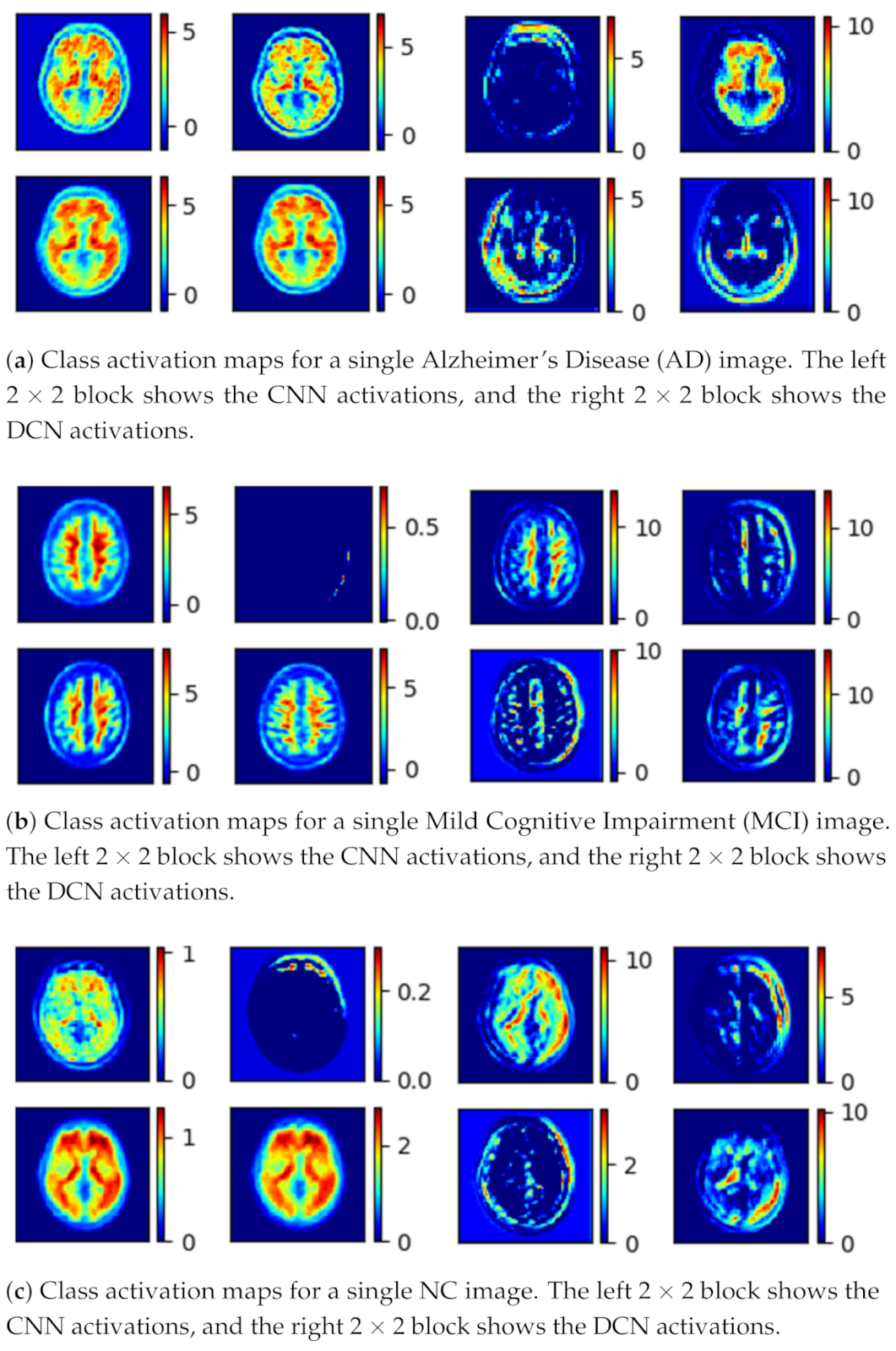
- The generator (G) learns the dataset distribution from the input image, encodes it into a latent vector, and reconstructs the image by upsampling. The uniqueness of the generator is that the encoder uses a parallel model that is comprised of a convolutional pipeline (CNN) and a dilated convolutional end network (DCN) that is 8 layers deep, each layer uses convolutional filters, a Rectified linear unit (ReLU) activation function, and a batch normalization operation. After two identical layers, a max-pooling operation is used for spatial dimension reduction and doubling the depth of the tensor. A latent vector of the input image is then generated.The DCN is eight layers deep, each layer uses convolutional filters with a dilation factor of 2, a ReLU (Rectified linear units) activation function, and a batch normalization operation. After 2 identical layers, a max-pooling operation is used for spatial dimension reduction and increasing the depth of the tensor. A latent vector of the input image is then generated. Concatenation of these features gives the optimal feature vector of the input image [58]. The class activation map for the given input image is shown in Figure 3.
- The discriminator (D) predicts the class of the input (whether it is fake or not) based on learned features. The discriminator generally uses an encoder-type architecture.
4. Experimental Environment and Results
4.1. The Dataset
4.2. Training the Model
- Contextual Loss: To learn the distribution of the dataset, normalization is applied to the input x and the output x̂. This helps the generation of contextually similar images from the normal samples and is proven to produce less blurry images than normalization [54]. The loss formula is given below as:
- Adversarial Loss: Taken from [28], this loss ensures that the generator G can reconstruct an image x as realistically as possible while the discriminator D can differentiate between the normal and fake images. The task is to minimize this objective for G and maximize it for D to achieve the min-max equilibrium where it is defined as:
- Latent Loss: This loss is used in obtaining the latent representations of the input x and the generated output x̂ as similar as possible. This ensures that the network can produce similar latent representations for sampling. Using the concatenated features (x) and the fully connected layer of the discriminator (x̂). The loss becomes:
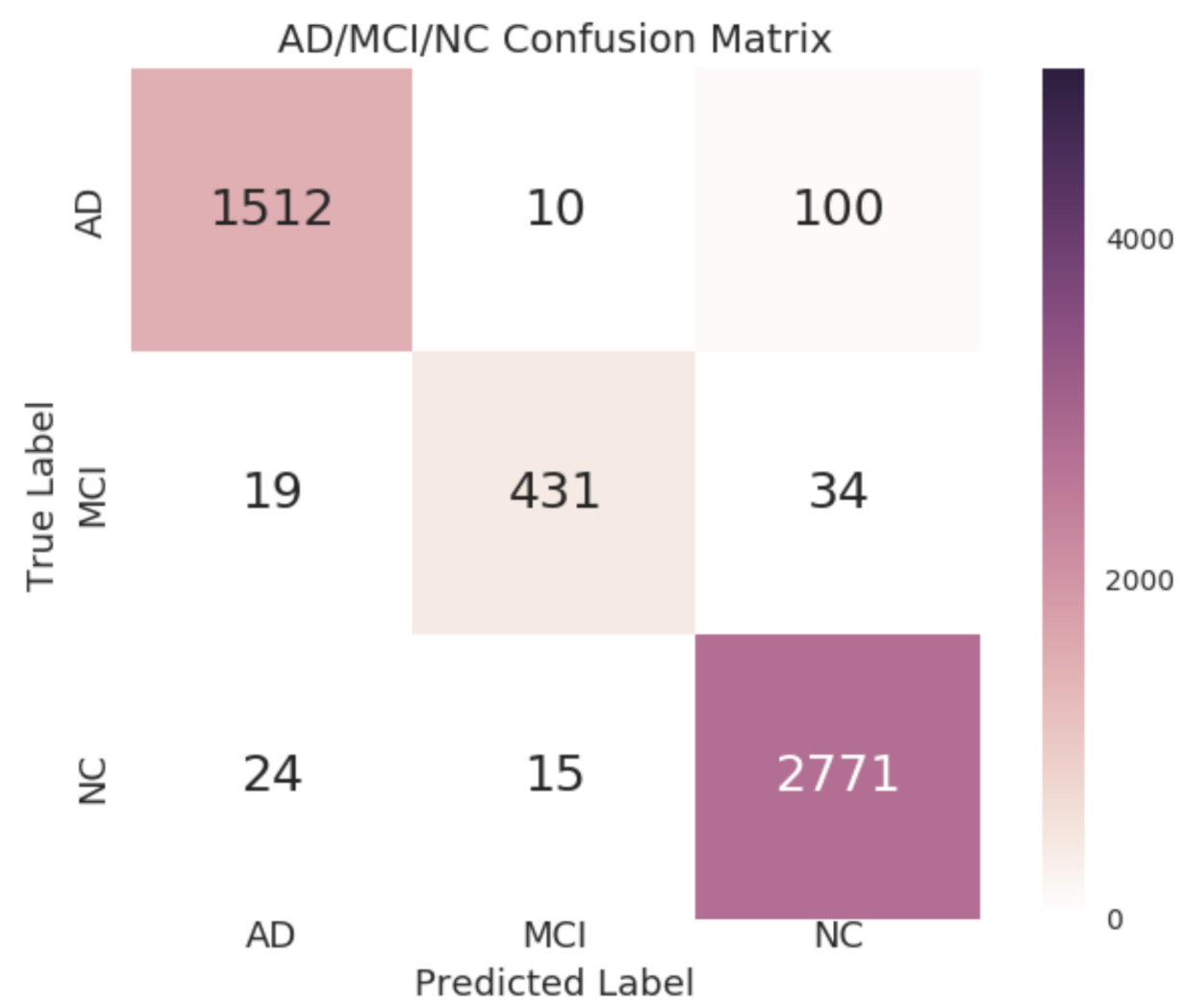
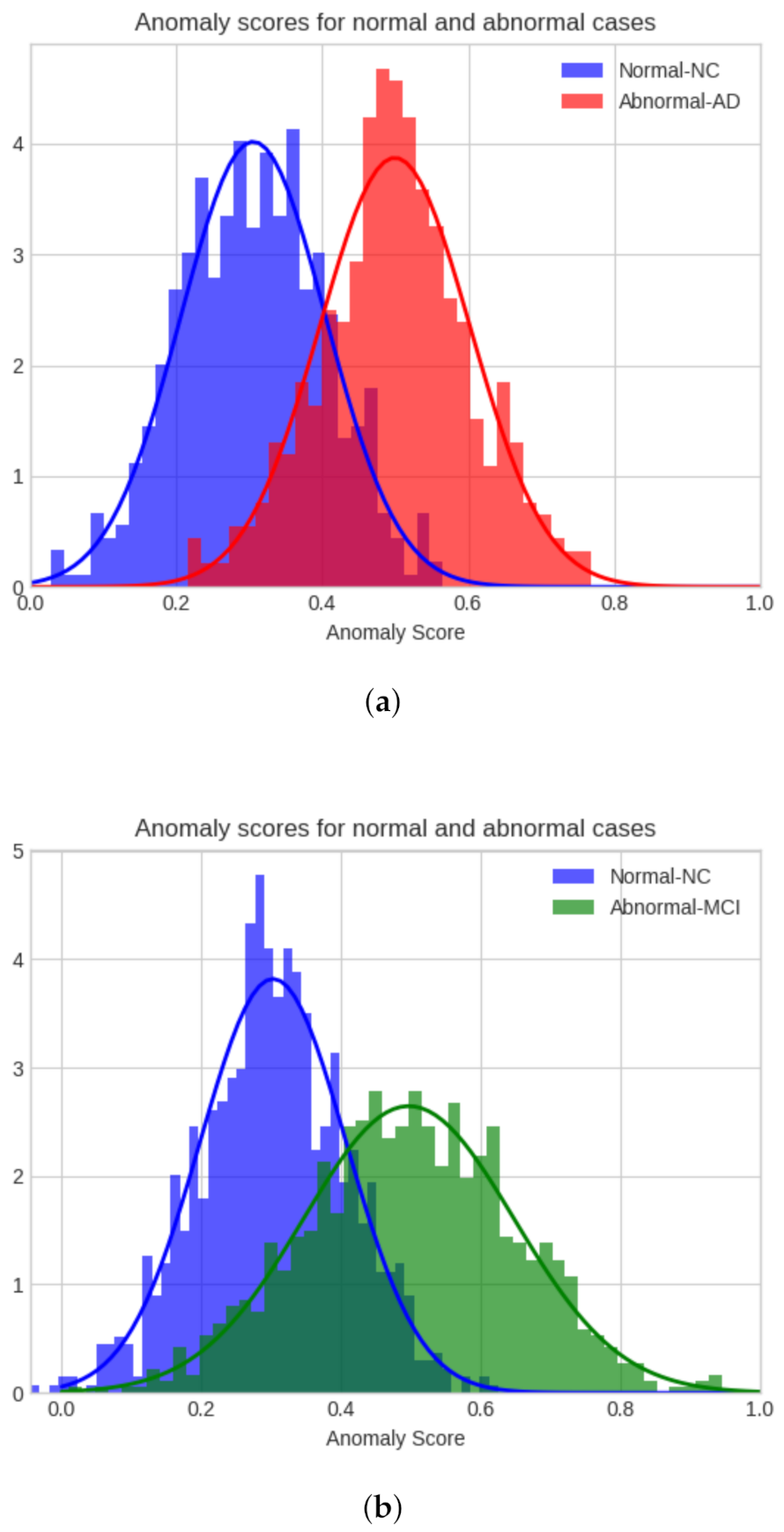
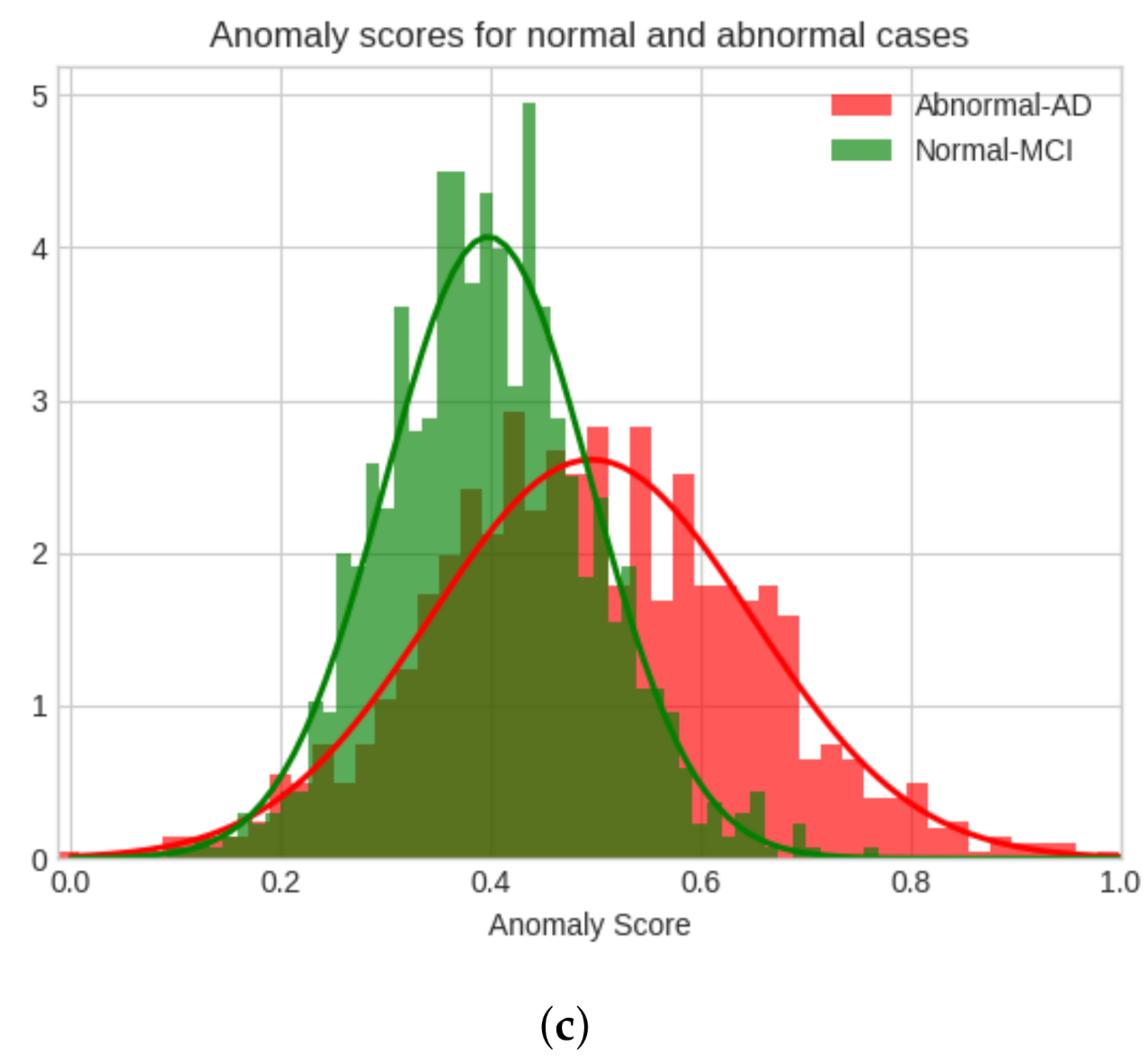
4.3. Model Evaluation
5. Conclusions
- Among the three loss functions, the importance of each loss function can be evaluated. A genetic algorithm or a grid search can be used to assign weights for each loss function to observe its effect on the model.
- The depth of the parallel model can be altered by using other parameter search algorithms. A deeper model may increase the computational cost while improving the accuracy.
- The skip connections used in autoencoders have been showing promising results [57]. The possibility and feasibility of skip connections will be investigated to further improve the performance of the model.
- Probable ways to improve the AUC score further will be investigated.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AD | Alzheimer’s Disease |
| MCI | Mild Cognitive Impairment |
| NC | Normal Control |
| A | Amyloid beta |
| PET | Positron emission tomography |
| MRI | Magnetic Resonance Imaging |
| MMSE | Mini Mental State Examination |
| CDR | Clinical Dementia Rating |
| GANs | Generative Adversarial Networks |
| DCGAN | Deep Convolutional Generative Adversarial Networks |
| AnoGAN | Anomaly GAN |
| EGBAD | Efficient GAN-Based Anomaly Detection |
| BiGAN | Bidirectional GAN |
| sMRI | structural Magnetic Resonance Imaging |
| fMRI | functional Magnetic Resonance Imaging |
| CNN | Convolutional Neural Network |
| ReLU | Rectified Linear Units |
| ROI | Region of Interest |
| DCN | Dilated Convolutional Network |
| AUC | Area Under the Curve |
| FID | Fréchet Inception Distance |
| FPR | False Positive Rate |
| TPR | True Positive Rate |
| SVM | Support Vector Machine |
| SGD | Stochastic Gradient Descent |
| ADNI | Alzheimer’s Disease Neuroimaging Initiative |
References
- Johns Hopkins Bloomberg School of Public Health. Alzheimer’s Disease to Quadruple Worldwide by 2050. Available online: https://www.jhsph.edu/news/news-releases/2007/brookmeyer-alzheimers-2050.html (accessed on 17 January 2021).
- Yangling, M.; Gage, F.H. Adult hippocampal neurogenesis and its role in Alzheimer’s disease. Mol. Neurodegener. 2011, 6, 85. [Google Scholar] [CrossRef] [Green Version]
- Villemagne, V.L.; Rowe, C.C.; Macfarlane, S.; Novakovic, K.; Masters, C.L. Imaginem oblivionis: The prospects of neuroimaging for early detection of Alzheimer’s disease. J. Clin. Neurosci. 2005, 12, 221–230. [Google Scholar] [CrossRef]
- Haass, C.; Selkoe, D.J. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007, 8, 101. [Google Scholar] [CrossRef]
- Chen, X.; Yan, S.D. Mitochondrial Abeta: A potential cause of metabolic dysfunction in Alzheimer’s disease. IUBMB Life 2006, 52, 686–694. [Google Scholar] [CrossRef]
- Alzheimer’s Association. Mild Cognitive Impairment (MCI). Available online: https://www.alz.org/alzheimers-dementia/what-is-dementia/related_conditions/mild-cognitive-impairment (accessed on 17 January 2021).
- Sabri, O.; Sabbagh, M.N.; Seibyl, J.; Barthel, H.; Akatsu, H.; Ouchi, Y.; Senda, K.; Murayama, S.; Ishii, K.; Takao, M.; et al. Florbetaben PET imaging to detect amyloid beta plaques in Alzheimer’s disease: Phase 3 study. Alzheimer’s Dement. 2015, 11, 964–974. [Google Scholar] [CrossRef]
- Johnson, K.A.; Fox, N.C.; Sperling, R.A.; Klunk, W.E. Brain Imaging in Alzheimer Disease. Cold Spring Harb. Perspect. Med. 2012, 4. [Google Scholar] [CrossRef]
- Catana, C.; Guimaraes, A.R.; Rosen, B.R. PET and MRI: The Odd Couple or a Match Made in Heaven? J. Nucl. Med. 2013, 54, 815–824. [Google Scholar] [CrossRef]
- Desai, B.; Sheth, S.; Conti, P. Review of novel radiotracers for positron emission tomography (PET) imaging. J. Nucl. Med. 2013, 54, 815–824. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.; Van Ginneken, B.; Sanchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Yanase, J.; Triantaphyllou, E. A systematic survey of computer-aided diagnosis in medicine: Past and present developments. Expert Syst. Appl. 2019, 138, 112821. [Google Scholar] [CrossRef]
- Jo, T.; Nho, K.; Saykin, A.J. Deep Learning in Alzheimer’s Disease: Diagnostic Classification and Prognostic Prediction Using Neuroimaging Data. Front. Aging Neurosci. 2019, 11, 220. [Google Scholar] [CrossRef] [Green Version]
- Samper-Gonzalez, J.; Burgos, N.; Bottani, S.; Fontanella, S.; Lu, P.; Marcoux, A.; Routier, A.; Guillon, J.; Bacci, M.; Wen, J.; et al. Reproducible evaluation of classification methods in Alzheimer’s disease: Framework and application to MRI and PET data. Neuroimage 2018, 183, 504–521. [Google Scholar] [CrossRef] [Green Version]
- Riedel, B.C.; Daianu, M.; Ver Steeg, G.; Mezher, A.; Salminen, L.E.; Galstyan, A.; Thompson, P.M. Uncovering biologically coherent peripheral signatures of health and risk for Alzheimer’s disease in the aging brain. Front. Aging Neurosci. 2019, 10, 390. [Google Scholar] [CrossRef] [Green Version]
- Shen, S.; Sandham, W.A.; Granat, M.H. Preprocessing and segmentation of brain magnetic resonance images. In Proceedings of the IEEE EMBS International Conference on Information Technology Applications in Biomedicine, Birmingham, UK, 24–26 April 2003. [Google Scholar]
- Dachena, C.; Casu, S.; Fanti, A.; Lodi, M.B.; Mazzarella, G. Combined Use of MRI, fMRIand Cognitive Data for Alzheimer’s Disease: Preliminary Results. Appl. Sci. 2019, 9, 3156. [Google Scholar] [CrossRef] [Green Version]
- Tufail, J.A.; Rudisill, C.; Egan, C.; Kapetanakis, V.V.; Vega-Salas, S.; Owen, C.G.; Lee, A.; Louw, V.; Anderson, J.; Liew, G.; et al. Automated Diabetic Retinopathy Image Assessment Software: Diagnostic Accuracy and Cost-Effectiveness Compared with Human Graders. Ophthalmology 2017, 124, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Johnson, J.M.; Khoshgoftaar, T.M. Survey on deep learning with class imbalance. J. Big Data 2019, 6, 1. [Google Scholar] [CrossRef]
- Willemink, M.J.; Koszek, W.A.; Hardell, C.; Wu, J.; Fleischmann, D.; Harvey, H.; Folio, L.R.; Summers, R.M.; Rubin, D.L.; Lungren, M.P. Preparing Medical Imaging Data for Machine Learning. Radiology 2020, 295, 4–15. [Google Scholar] [CrossRef]
- Ahmed, M.; Naser Mahmood, A.; Hu, J. A survey of network anomaly detection techniques. J. Netw. Comput. Appl. 2016, 60, 19–31. [Google Scholar] [CrossRef]
- Schlegl, T.; Seeböck, P.; Waldstein, S.M.; Schmidt-Erfurth, U.; Langs, G. Unsupervised anomaly detection with generative adversarial networks to guide marker discovery. In Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2017; Volume 10265, pp. 146–147. [Google Scholar] [CrossRef] [Green Version]
- Kiran, B.R.; Thomas, D.M.; Parakkal, R. An overview of deep learning based methods for unsupervised and semi-supervised anomaly detection in videos. J. Imaging 2018, 4, 36. [Google Scholar] [CrossRef] [Green Version]
- Hodge, V.; Austin, J. A Survey of Outlier Detection Methodologies. Artif. Intell. Rev. 2004, 22, 85–126. [Google Scholar] [CrossRef] [Green Version]
- Pimentel, M.A.; Clifton, D.A.; Clifton, L.; Tarassenko, L. A review of novelty detection. Signal Process. 2014, 99, 215–249. [Google Scholar] [CrossRef]
- Chandola, V.; Banerjee, A.; Kumar, V. Anomaly detection: A Survey. ACM Comput. Surv. 2009, 41, 1–58. [Google Scholar] [CrossRef]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the Conference on Advances in Neural Information Processing Systems, Montreal, QC, Canada, 8–13 December 2014; pp. 2672–2680. [Google Scholar]
- Akcay, S.; Atapour-Abarghouei, A.; Breckon, T.P. GANomaly: Semisupervised anomaly detection via adversarial training. In Proceedings of the Asian Conference on Computer Vision, Perth, Australia, 2–6 December 2018; pp. 622–637. [Google Scholar]
- Shin, H.C.; Ihsani, A.; Xu, Z.; Mandava, S.; Sreenivas, S.T.; Forster, C.; Cha, J.; Alzheimer’s Disease Neuroimaging Initiative. GANDALF: Generative Adversarial Networks with Discriminator-Adaptive Loss Fine-Tuning for Alzheimer’s Disease Diagnosis from MRI. In Proceedings of the Conference on Medical Image Computing and Computer Assisted Intervention, Lima, Peru, 4–8 October 2020; pp. 688–697.
- Islam, J.; Zhang, Y. GAN-based synthetic brain PET image generation. Brain Inform. 2020, 7. [Google Scholar] [CrossRef]
- Zenati, H.; Foo, C.S.; Lecouat, B.; Manek, G.; Chandrasekhar, V.R. Efficient gan-based anomaly detection. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Sabokrou, M.; Khalooei, M.; Fathy, M.; Adeli, E. Adversarially learned one-class classifier for novelty detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 30 April–3 May 2018; pp. 3379–3388. [Google Scholar]
- Fan, Y.; Resnick, S.M.; Wu, X.; Davatzikos, C. Structural and functional biomarkers of prodromal Alzheimer’s disease: A high-dimensional pattern classification study. Neuroimage 2008, 41, 277–285. [Google Scholar] [CrossRef] [Green Version]
- Hu, K.; Wang, Y.; Chen, K.; Hou, L.; Zhang, X. Multi-scale features extraction from baseline structure MRI for MCI patient classification and AD early diagnosis. Neurocomputing 2016, 175, 132–145. [Google Scholar] [CrossRef] [Green Version]
- Filiphovych, R.; Davatzikos, C.; Alzheimer’s Disease Neuroimaging Initiative. Semi-supervised pattern classification of medical images: Application to mild cognitive impairment (MCI). Neuroimage 2011, 55, 1109–1119. [Google Scholar] [CrossRef] [Green Version]
- Gray, K.R.; Wolz, R.; Heckermann, R.A.; Aljabar, P.; Hammers, A.; Rueckert, D.; The Alzheimer’s Disease Neuroimaging Initiative. Multi-region analysis of longitudinal FDG-PET for classification for the Alzheimer’s disease. Neuroimage 2012, 60, 221–229. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Cheng, D.; Yan, W.; The Alzheimer’s Disease Neuroimaging Initiative. Classification of Alzheimer’s Disease by Combination of Convolutional and Recurrent Neural Networks Using FDG-PET Images. Neuroimage 2012, 60, 221–229. [Google Scholar] [CrossRef] [Green Version]
- Davatzikos, C.; Bhatt, P.; Shaw, L.M.; Batmangehelich, K.N.; Trojanowski, J.Q. Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol. Aging 2011, 32, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Aderghal, K.; Benois-Pineau, J.; Afdel, K.; Gwenaëlle, C. FuseMe: Classification of sMRI images by fusion of Deep CNNs in 2D + ε projections. In Proceedings of the 15th International Workshop on Content-Based Multimedia Indexing, Firenze, Italy, 19–21 June 2017; pp. 1–7. [Google Scholar] [CrossRef]
- Suk, H.; Wee, C.Y.; Shen, D. Discriminative group sparse representation for mild cognitive impairment classification. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Nagoya, Japan, 22 September 2013; pp. 131–138. [Google Scholar]
- Ramzan, F.; Khan, M.U.G.; Rehmat, A.; Iqbal, S.; Saba, T.; Rehman, A.; Mehmood, Z. A Deep Learning Approach for Automated Diagnosis and Multi-Class Classification of Alzheimer’s Disease Stages Using Resting-State fMRI and Residual Neural Networks. J. Med. Syst. 2020, 44, 916–922. [Google Scholar] [CrossRef]
- Perrin, R.J.; Fagan, A.M.; Holtzman, D.M. Multimodal techniques for diagnosis and prognosis of Alzheimer’s disease. Nature 2009, 461, 916–922. [Google Scholar] [CrossRef]
- Liu, M.; Cheng, D.; Wang, K.; Wang, Y.; The Alzheimer’s Disease Neuroimaging Initiative. Multi-Modality Cascaded Convolutional Neural Networks for Alzheimer’s Disease Diagnosis. Neuroinformatics 2009, 16, 295–308. [Google Scholar] [CrossRef]
- Singh, S.; Srivastava, A.; Mi, L.; Chen, K. Deep Learning based Classification of FDG-PET Data for Alzheimers Disease Categories. In Proceedings of the International Conference on Medical Information Processing and Analysis, San Andres Island, Colombia, 17 November 2017. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Xu, T.; Zhang, H.; Long, L.R.; Huang, X. SegAN: Adversarial Network with Multi-scale L1 Loss for Medical Image Segmentation. Neuroinformatics 2017, 16, 383–392. [Google Scholar] [CrossRef] [Green Version]
- Armanious, K.; Jiang, C.; Fischer, M.; Kustner, T.; Hepp, T.; Nikolau, K.; Gatidis, S.; Yang, B. MedGAN: Medical image translation using GANs. Comput. Med. Imaging Graph. 2019, 79. [Google Scholar] [CrossRef]
- Creswell, A.; White, T.; Dumoulin, V.; Arulkumaran, K.; Sengupta, B.; Bharath, A.A. Generative adversarial networks: An overview. IEEE Signal Process. Mag. 2018, 35, 53–65. [Google Scholar]
- Salimans, T.; Goodfellow, I.; Zaremba, W.; Cheung, V.; Radford, A.; Chen, X. Improved techniques for training gans. In Advances in Neural Information Processing; MIT Press: Cambridge, MA, USA, 2016; pp. 2234–2242. [Google Scholar]
- Radford, A.; Metz, L.; Chintala, S. Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. In Proceedings of the International Conference on Learning Representations, San Juan, Puerto Rico, 2–4 May 2016. [Google Scholar]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 7–9 July 2015; pp. 448–456. [Google Scholar]
- Arjovsky, M.; Chintala, S.; Bottou, L. Wasserstein generative adversarial networks. In Proceedings of the International Conference on Machine Learning, Sydney, Australia, 6–11 August 2017; pp. 214–223. [Google Scholar]
- Ravanbakhsh, M.; Sangineto, E.; Nabi, M.; Sebe, N. Training Adversarial Discriminators for Cross-channel Abnormal Event Detection in Crowds. In Proceedings of the IEEE Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 7–11 January 2019. [Google Scholar] [CrossRef] [Green Version]
- Isola, P.; Zhu, J.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 5967–5976. [Google Scholar] [CrossRef] [Green Version]
- Donahue, J.; Krahenbuhl, P.; Darrell, T. Adversarial Feature Learning. In Proceedings of the International Conference on Learning Representations, Toulon, France, 24–26 April 2017. [Google Scholar]
- Akcay, S.; Atapour-Abarghouei, A.; Breckon, T.P. Skip-GANomaly: Skip Connected and Adversarially Trained Encoder-Decoder Anomaly Detection. In Proceedings of the International Joint Conference on Neural Networks, Budapest, Hungary, 14–19 July 2019. [Google Scholar] [CrossRef] [Green Version]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2017; pp. 234–241. [Google Scholar] [CrossRef]
- Baydargil, H.B.; Park, J.S.; Kang, D.Y.; Kang, H.; Cho, K. A Parallel Deep Convolutional Neural Network for Alzheimer’s disease classification on PET/CT brain images. KSII Trans. Internet Inf. Syst. 2020, 14. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.J.; van der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017. [Google Scholar] [CrossRef] [Green Version]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Sizzerman, A. Very deep convolutional networks for large-scale image recognition. In Proceedings of the International Conference on Machine Learning, Lille, France, 6–11 July 2016. [Google Scholar] [CrossRef] [Green Version]
- Ling, C.X.; Huang, J.; Zhang, H. Auc: A statistically consistent and more discriminating measure than accuracy. In Proceedings of the International Joint Conferences on Artificial Intelligence, Acapulco, Mexico, 9–15 August 2003; pp. 519–524. [Google Scholar]
- Heusel, M.; Ramsauer, H.; Unterthinter, T.; Nessler, B. VGANs Trained by a Two Time-Scale Update Rule Converge to a Local Nash Equilibrium. In Proceedings of the International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 6629–6640. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar] [CrossRef] [Green Version]
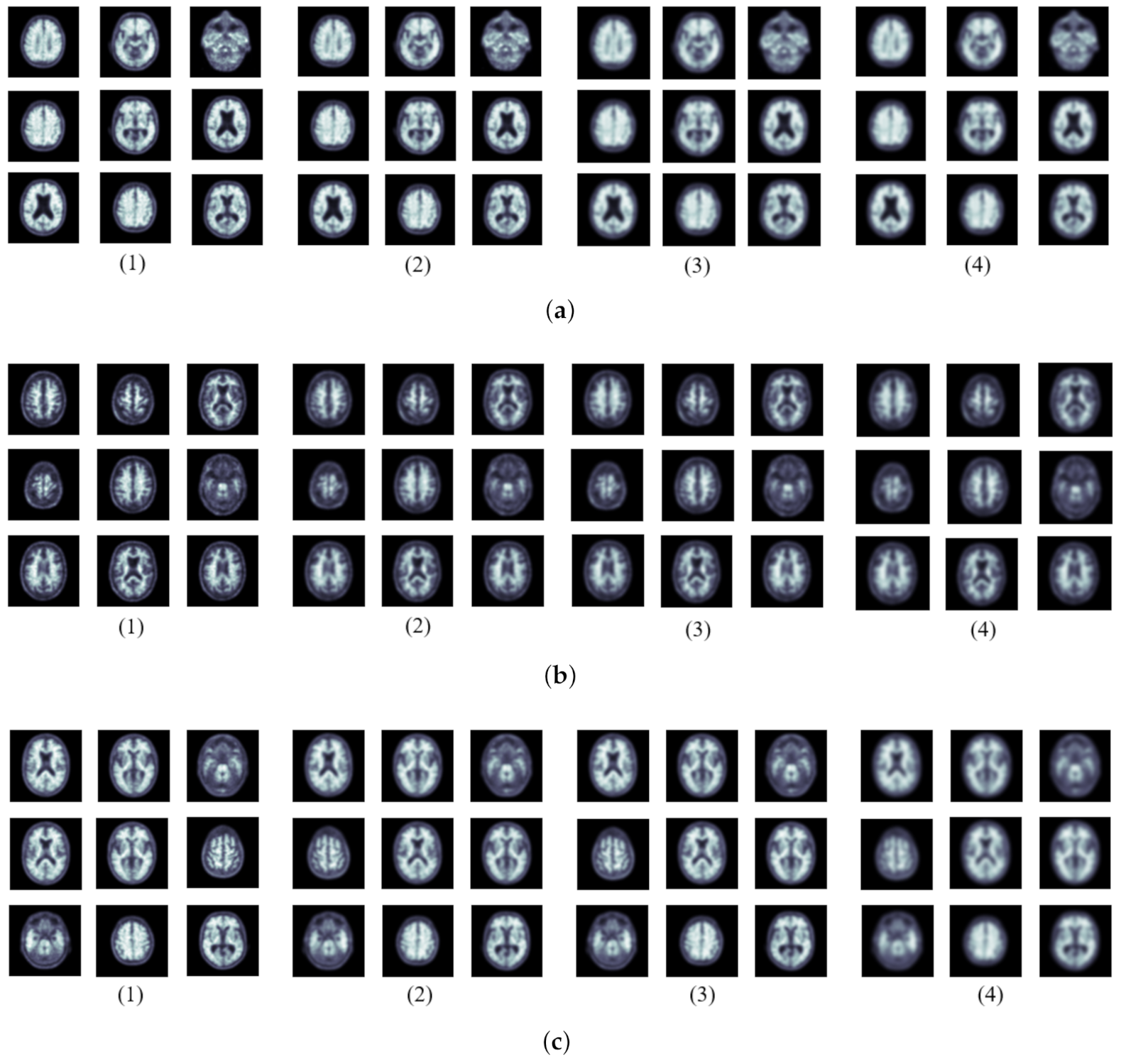
| Group | NC | MCI | AD | Total |
|---|---|---|---|---|
| No. of subjects | 148 | 83 | 25 | 256 |
| Gender (M/F) | 82/66 | 45/38 | 15/10 | |
| No. of Images | 14,208 | 7968 | 2400 | 24,576 |
| Age, mean ± SD | 75.9 ± 4.5 | 74.8 ± 7.1 | 76.5 ± 6.7 | |
| CDR, mean ± SD | 0.0 ± 0.0 | 0.5 ± 0.0 | 0.8 ± 0.2 | |
| MMSE, mean ± SD | 29.1 ± 1.0 | 27.2 ± 1.6 | 23.4 ± 2.0 |
| Training | Testing | Total | |||||
|---|---|---|---|---|---|---|---|
| Cases | AD | MCI | NC | AD | MCI | NC | |
| AD-NC | 11,366 | 2400 | 2842 | 16,608 | |||
| MCI-NC | 11,366 | 2842 | 2842 | 17,050 | |||
| AD-MCI | 6374 | 2400 | 1594 | 10,368 | |||
| Performance | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Parallel Model | 96.03 | 95.70 | 93.73 | 94.67 |
| DenseNet169 [59] | 95.44 | 93.98 | 92.31 | 94.20 |
| Inception-V3 [60] | 90.06 | 88.73 | 87.12 | 89.44 |
| ResNet50 [61] | 89.41 | 86.81 | 87.29 | 88.52 |
| VGG16 [62] | 85.13 | 83.98 | 82.31 | 85.01 |
| Model | AD-NC | MCI-NC | AD-MCI |
|---|---|---|---|
| GANomaly [29] | 59.31 | 57.33 | 48.34 |
| EGBAD [32] | 58.46 | 60.31 | 54.71 |
| AnoGAN [23] | 55.71 | 52.93 | 48.11 |
| Skip-GANomaly [56] | 71.42 | 67.02 | 68.56 |
| Ablation-Conv. | 68.71 | 65.34 | 61.25 |
| Ablation-Dilated Conv. | 60.82 | 54.02 | 52.40 |
| Proposed Model | 75.21 | 70.84 | 71.85 |
| Models | AD | MCI | NC |
|---|---|---|---|
| GANomaly [29] | 18.216 | 17.353 | 17.885 |
| EGBAD [32] | 20.815 | 21.621 | 19.134 |
| AnoGan [23] | 19.908 | 17.991 | 21.759 |
| Skip-GANomaly [56] | 11.892 | 12.516 | 11.908 |
| Ablation Conv. | 16.175 | 18.023 | 17.659 |
| Ablation Dilated Conv. | 21.762 | 18.895 | 21.342 |
| Proposed Model | 9.481 | 10.024 | 10.858 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baydargil, H.B.; Park, J.-S.; Kang, D.-Y. Anomaly Analysis of Alzheimer’s Disease in PET Images Using an Unsupervised Adversarial Deep Learning Model. Appl. Sci. 2021, 11, 2187. https://doi.org/10.3390/app11052187
Baydargil HB, Park J-S, Kang D-Y. Anomaly Analysis of Alzheimer’s Disease in PET Images Using an Unsupervised Adversarial Deep Learning Model. Applied Sciences. 2021; 11(5):2187. https://doi.org/10.3390/app11052187
Chicago/Turabian StyleBaydargil, Husnu Baris, Jang-Sik Park, and Do-Young Kang. 2021. "Anomaly Analysis of Alzheimer’s Disease in PET Images Using an Unsupervised Adversarial Deep Learning Model" Applied Sciences 11, no. 5: 2187. https://doi.org/10.3390/app11052187
APA StyleBaydargil, H. B., Park, J.-S., & Kang, D.-Y. (2021). Anomaly Analysis of Alzheimer’s Disease in PET Images Using an Unsupervised Adversarial Deep Learning Model. Applied Sciences, 11(5), 2187. https://doi.org/10.3390/app11052187







