Trafficking of Full-Length and N-Terminally Truncated Cathepsin B in Human Colorectal Carcinoma Cells
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Tissue Microarrays
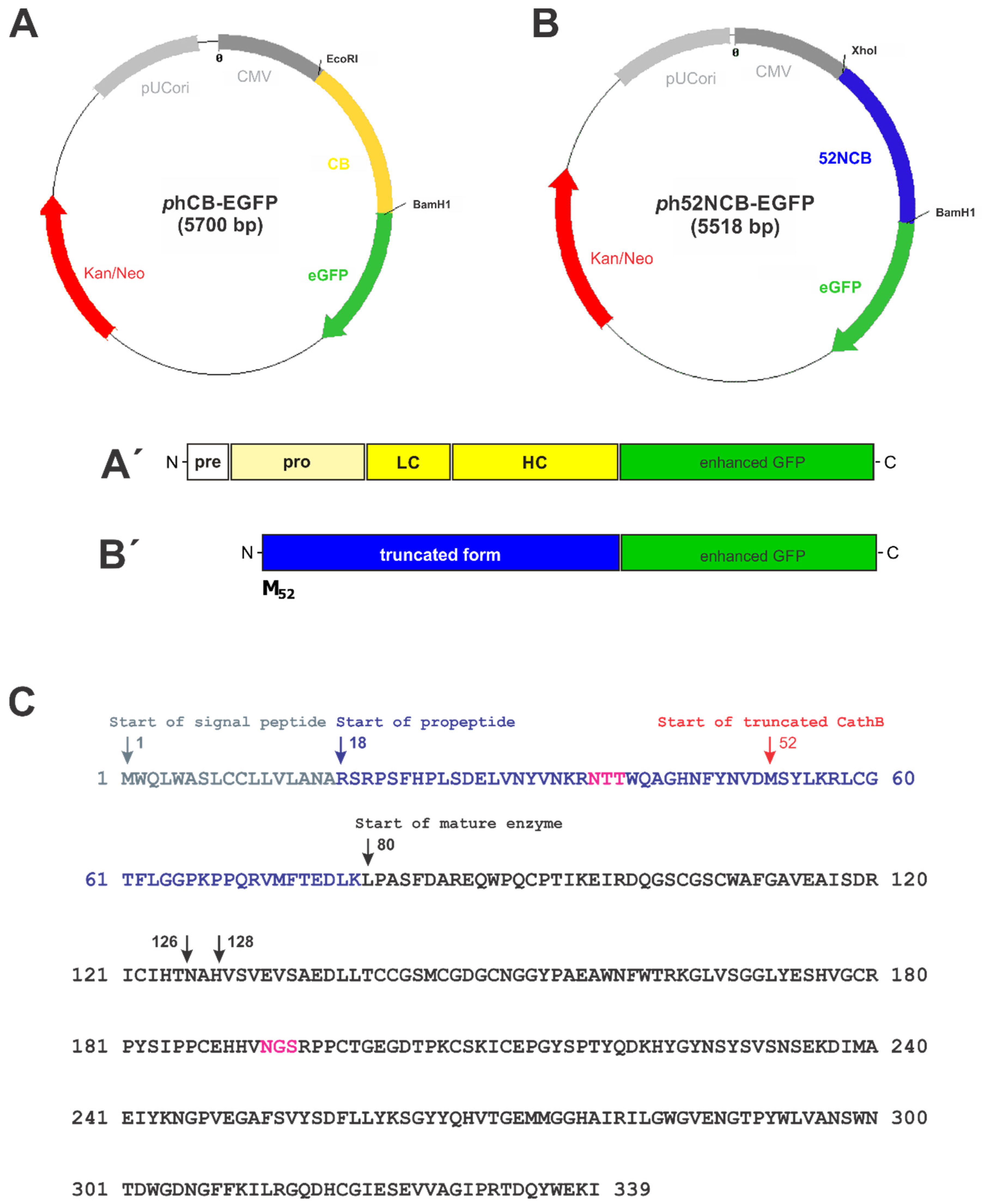
2.2. Plasmid Construction and Transfection
2.3. Cell Cycle Synchronization
2.4. Immunofluorescence Labeling
2.5. Aggresome Detection
2.6. Microscopy
2.7. Subcellular Fractionation, SDS-PAGE, and Immunoblotting
2.8. FACS Analysis
2.9. Densitometry Analysis and Statistical Evaluations
3. Results
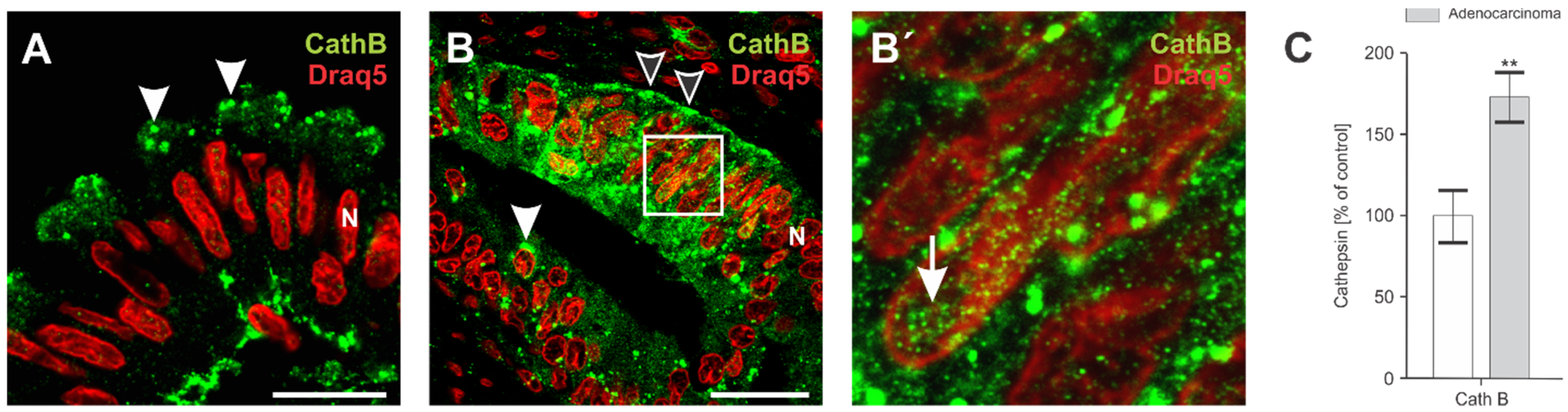
3.1. Elevated Levels and Dispersed Sub-Cellular Distribution of Cathepsin B in Adenocarcinoma of Colon Tissue
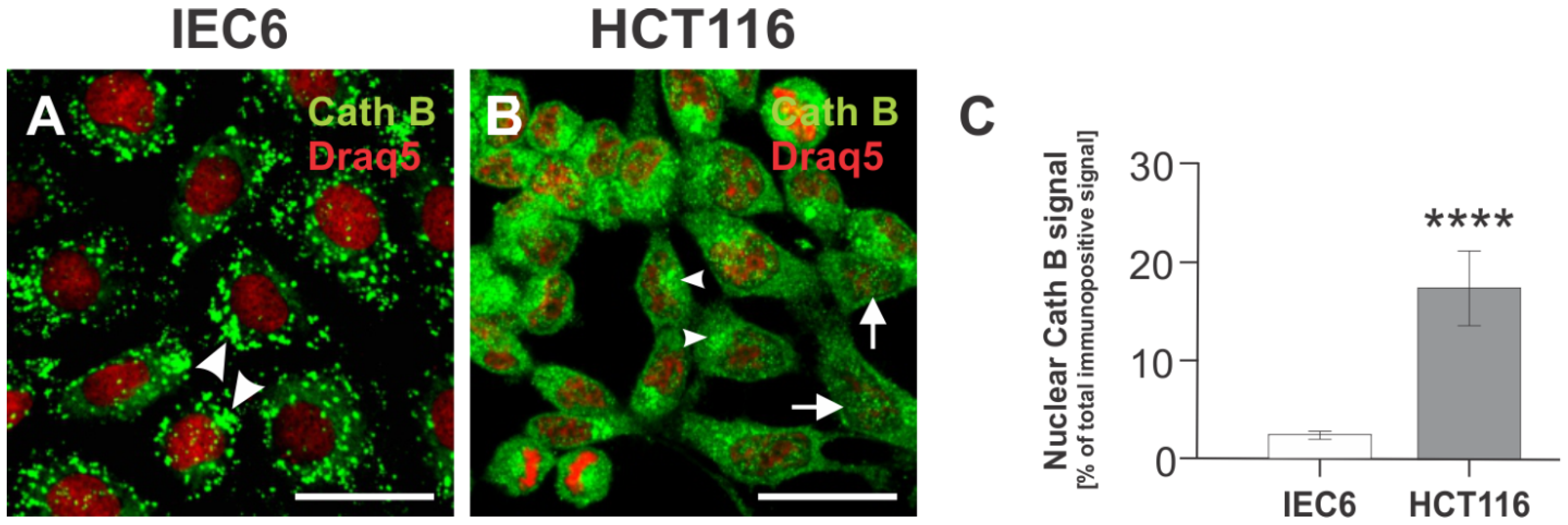
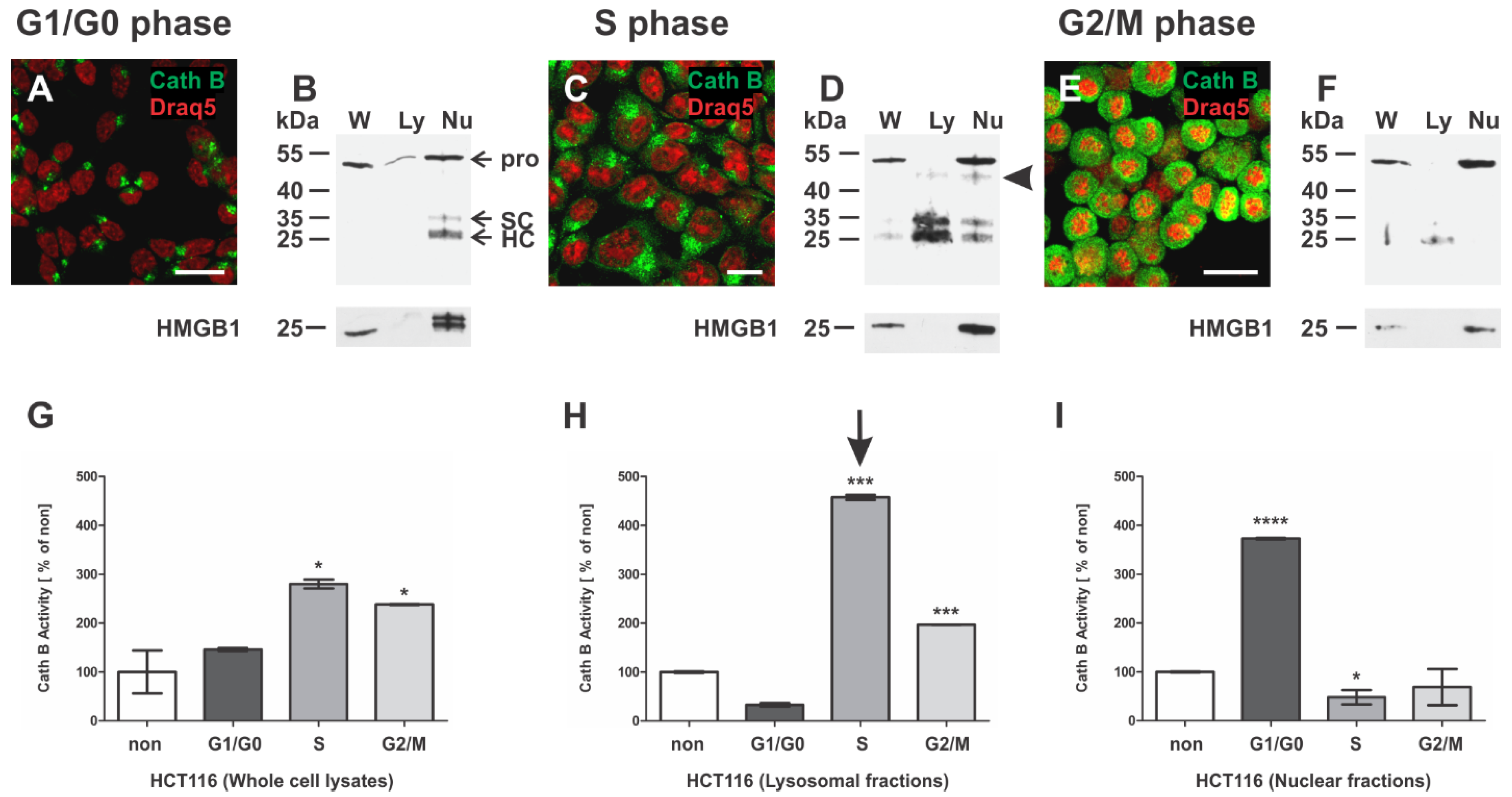
3.2. Nuclear Localization of Cathepsin B in the HCT116 Human Colorectal Carcinoma Cell Line
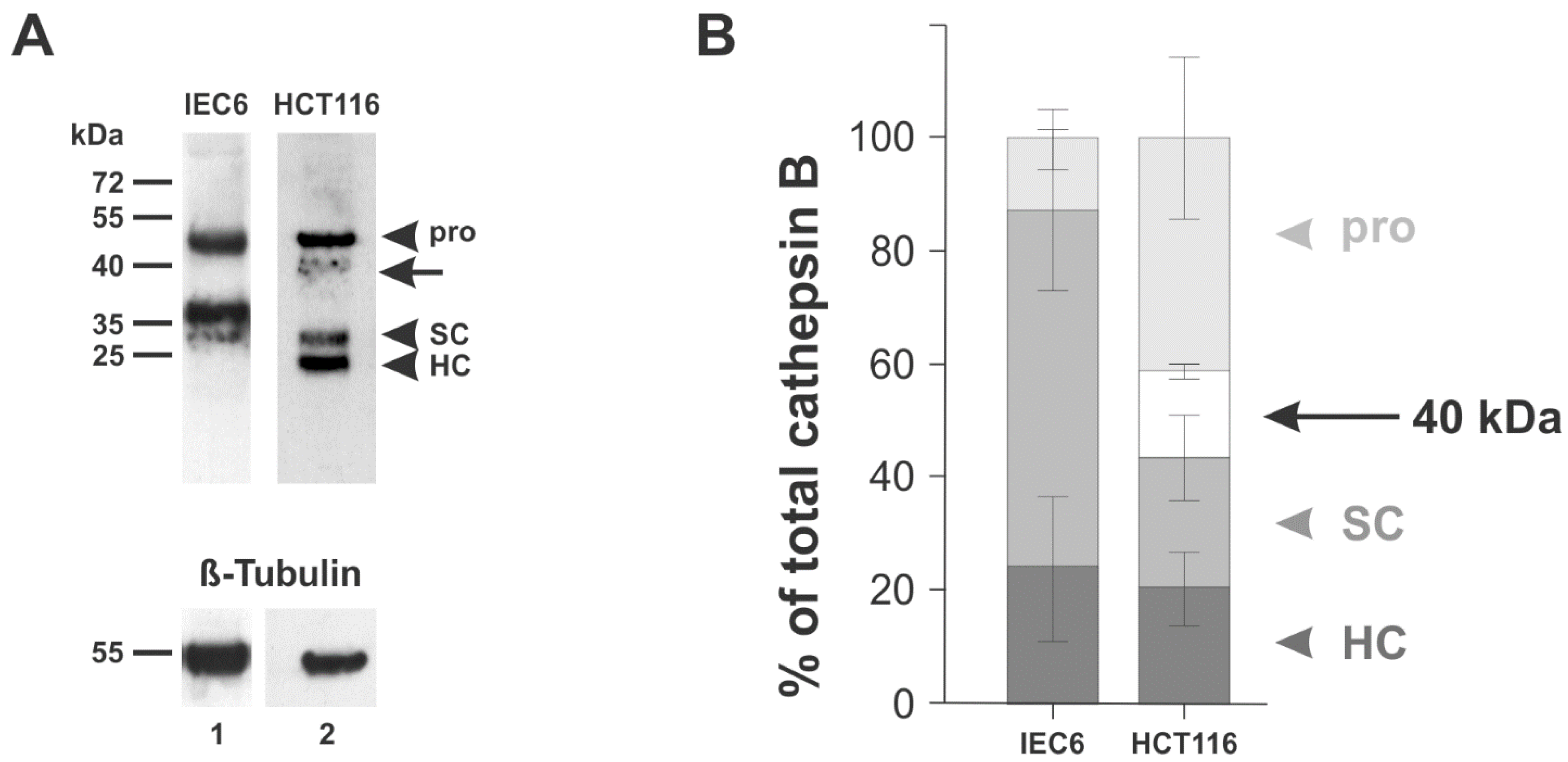
3.3. Expression of a 40-kDa Variant of Cathepsin B in CRC Cells
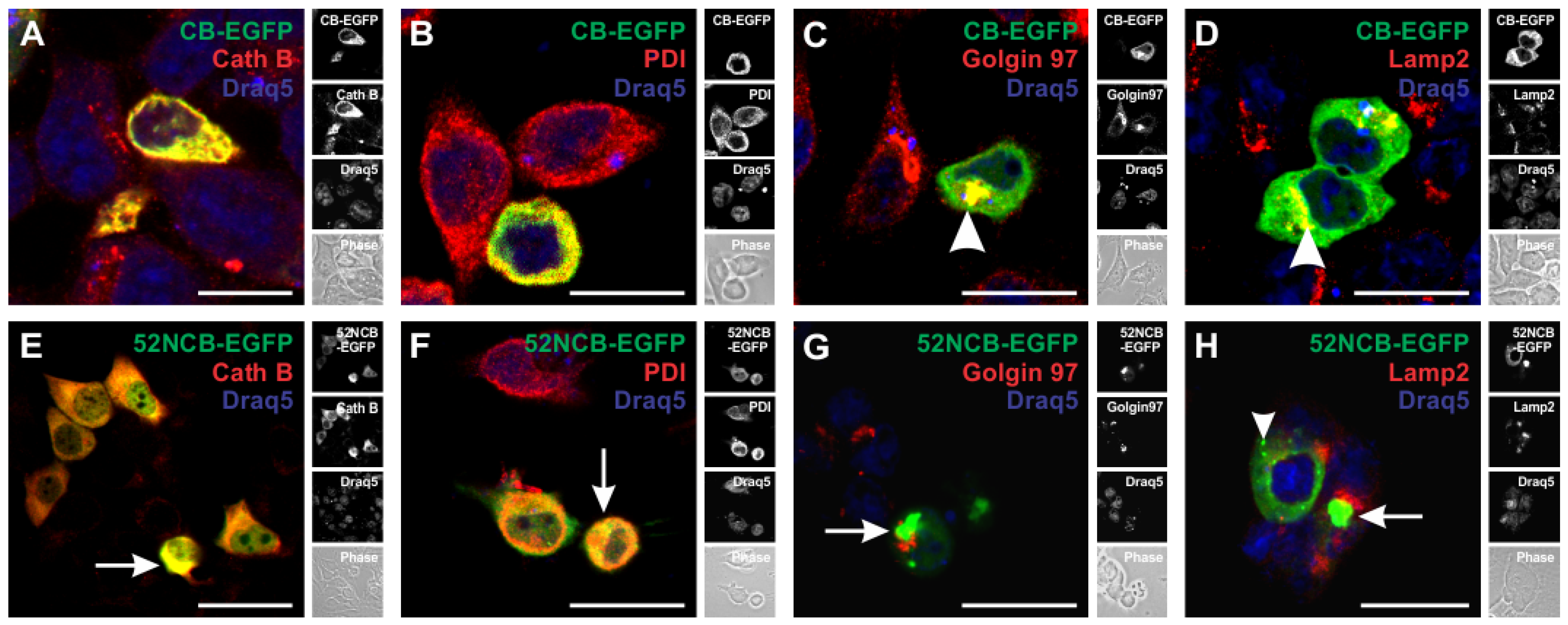
3.4. Trafficking of N-Terminally Truncated Human Cathepsin B-EGFP in HCT116 Cells
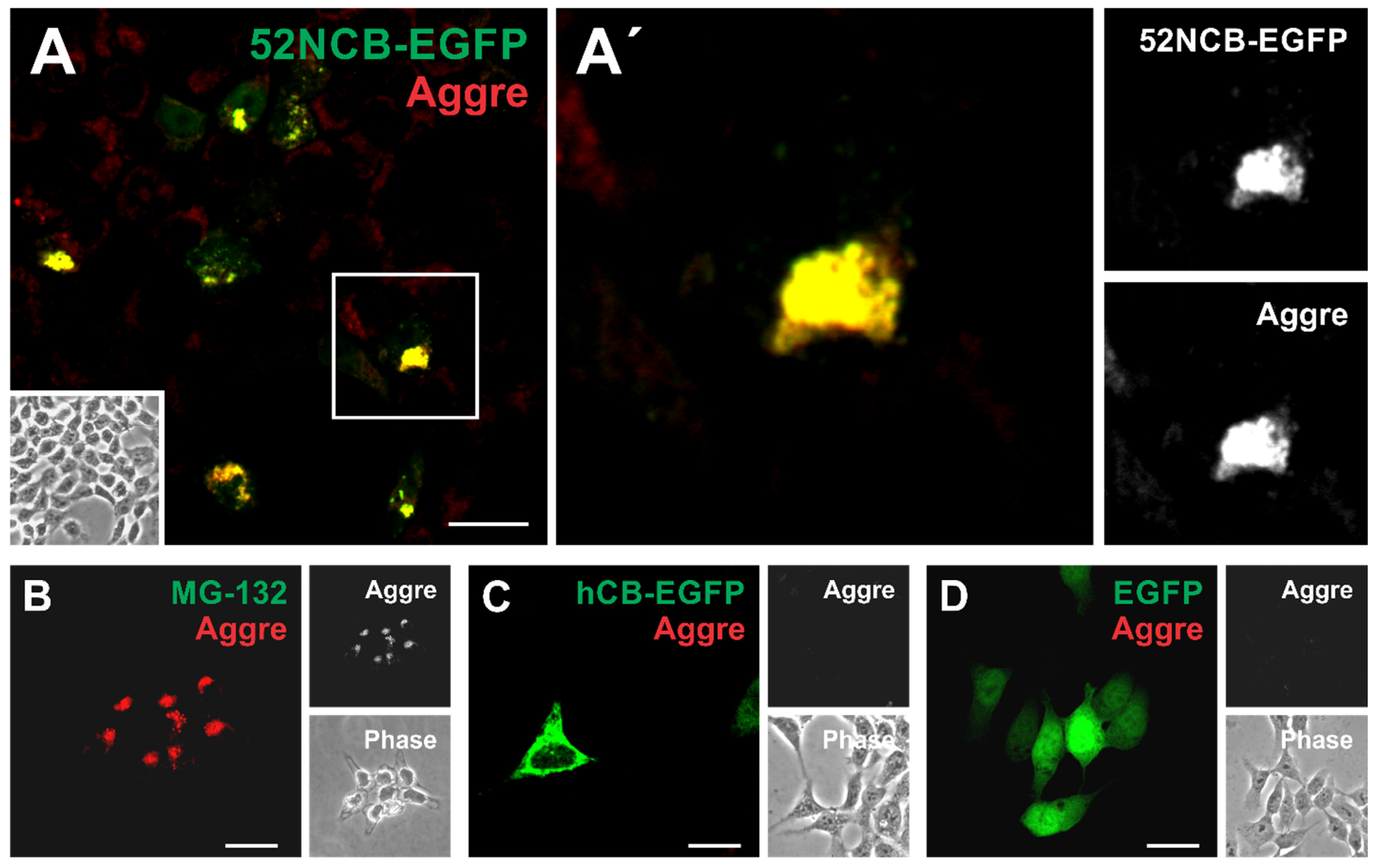
3.5. N-Terminally Truncated Human Cathepsin B-EGFP Chimeras Accumulate in Aggresome-like Inclusion Bodies of HCT116 Cells
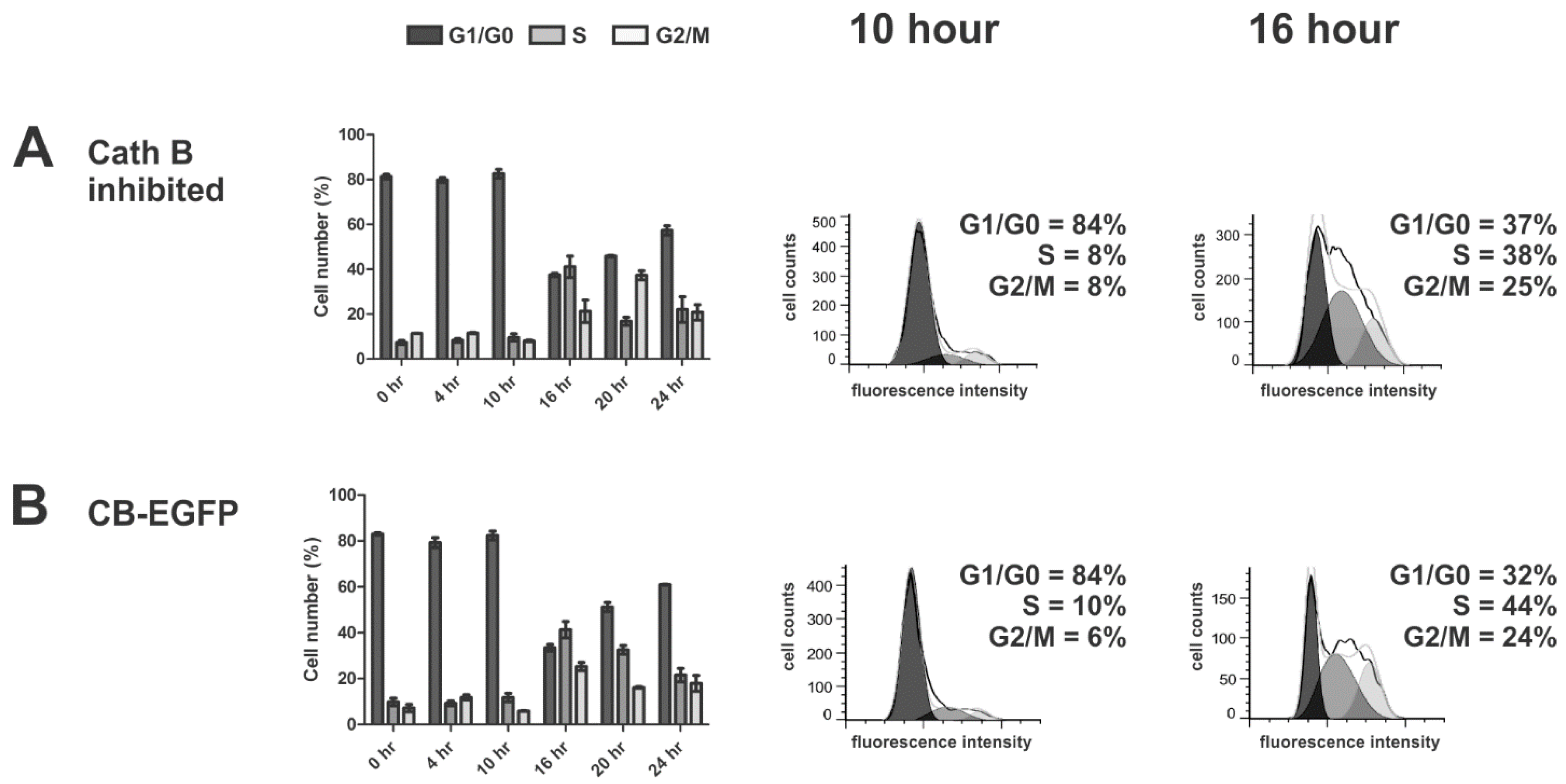
3.6. Cathepsin B of HCT116 Cells during Cell Cycle Progression
4. Discussion
4.1. Unusual Sub-Cellular Locations Reached by Specific Forms of Cathepsins
4.2. Cell Cycle Regulation of CRC Cells Is Independent of Cathepsin B
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Riese, R.J.; Chapman, H.A. Cathepsins and compartmentalization in antigen presentation. Curr. Opin. Immunol. 2000, 12, 107–113. [Google Scholar] [CrossRef]
- Turk, V.; Turk, B.; Turk, D. Lysosomal cysteine proteases: Facts and opportunities. EMBO J. 2001, 20, 4629–4633. [Google Scholar] [CrossRef]
- Büth, H.; Buttigieg, P.L.; Ostafe, R.; Rehders, M.; Dannenmann, S.R.; Schaschke, N.; Stark, H.J.; Boukamp, P.; Brix, K. Cathepsin B is essential for regeneration of scratch-wounded normal human epidermal keratinocytes. Eur. J. Cell Biol. 2007, 86, 747–761. [Google Scholar] [CrossRef]
- Brix, K.; Dunkhorst, A.; Mayer, K.; Jordans, S. Cysteine cathepsins: Cellular roadmap to different functions. Biochimie 2008, 90, 194–207. [Google Scholar] [CrossRef]
- Yadati, T.; Houben, T.; Bitorina, A.; Shiri-Sverdlov, R. The Ins and Outs of Cathepsins: Physiological Function and Role in Disease Management. Cells 2020, 9, 1679. [Google Scholar] [CrossRef] [PubMed]
- Berdowska, I. Cysteine proteases as disease markers. Clin. Chim. Acta 2004, 342, 41–69. [Google Scholar] [CrossRef] [PubMed]
- Bühling, F.; Peitz, U.; Krüger, S.; Küster, D.; Vieth, M.; Gebert, I.; Roessner, A.; Weber, E.; Malfertheiner, P.; Wex, T. Cathepsins K, L, B, X and W are differentially expressed in normal and chronically inflamed gastric mucosa. Biol. Chem. 2004, 385, 439–445. [Google Scholar] [CrossRef]
- Mohamed, M.M.; Sloane, B.F. Cysteine cathepsins: Multifunctional enzymes in cancer. Nat. Rev. Cancer 2006, 6, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Tedelind, S.; Poliakova, K.; Valeta, A.; Hunegnaw, R.; Yemanaberhan, E.L.; Heldin, N.E.; Kurebayashi, J.; Weber, E.; Kopitar-Jerala, N.; Turk, B.; et al. Nuclear cysteine cathepsin variants in thyroid carcinoma cells. Biol. Chem. 2010, 391, 923–935. [Google Scholar] [CrossRef] [PubMed]
- Gopinathan, A.; DeNicola, G.M.; Frese, K.K.; Cook, N.; Karreth, F.A.; Mayerle, J.; Lerch, M.M.; Reinheckel, T.; Tuveson, D.A. Cathepsin B promotes the progression of pancreatic ductal adenocarcinoma in mice. Gut 2012, 61, 877–884. [Google Scholar] [CrossRef] [PubMed]
- Gondi, C.S.; Rao, J.S. Cathepsin B as a cancer target. Expert Opin. Ther. Targets 2013, 17, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Brix, K.; McInnes, J.; Al-Hashimi, A.; Rehders, M.; Tamhane, T.; Haugen, M.H. Proteolysis mediated by cysteine cathepsins and legumain-recent advances and cell biological challenges. Protoplasma 2015, 252, 755–774. [Google Scholar] [CrossRef] [PubMed]
- Rudzińska, M.; Parodi, A.; Soond, S.M.; Vinarov, A.Z.; Korolev, D.O.; Morozov, A.O.; Daglioglu, C.; Tutar, Y.; Zamyatnin, A.A. The Role of Cysteine Cathepsins in Cancer Progression and Drug Resistance. Int. J. Mol. Sci. 2019, 20, 3602. [Google Scholar] [CrossRef]
- Soond, S.M.; Kozhevnikova, M.V.; Townsend, P.A.; Zamyatnin, A.A. Cysteine Cathepsin Protease Inhibition: An update on its Diagnostic, Prognostic and Therapeutic Potential in Cancer. Pharmaceuticals 2019, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Hämälistö, S.; Stahl-Meyer, J.; Jäättelä, M. They Might Cut It-Lysosomes and Autophagy in Mitotic Progression. Front. Cell Dev. Biol. 2021, 9, 727538. [Google Scholar] [CrossRef]
- Vasiljeva, O.; Sevenich, L.; Reinheckel, T. Analyzing the Role of Proteases in Breast Cancer Progression and Metastasis Using Primary Cells from Transgenic Oncomice. In Metastasis: Methods and Protocols; Stein, U.S., Ed.; Springer: New York, NY, USA, 2021; pp. 275–293. [Google Scholar]
- Olson, O.C.; Joyce, J.A. Cysteine cathepsin proteases: Regulators of cancer progression and therapeutic response. Nat. Rev. Cancer 2015, 15, 712–729. [Google Scholar] [CrossRef] [PubMed]
- Soond, S.M.; Kozhevnikova, M.V.; Frolova, A.S.; Savvateeva, L.V.; Plotnikov, E.Y.; Townsend, P.A.; Han, Y.P.; Zamyatnin, A.A., Jr. Lost or Forgotten: The nuclear cathepsin protein isoforms in cancer. Cancer Lett. 2019, 462, 43–50. [Google Scholar] [CrossRef]
- Linke, M.; Herzog, V.; Brix, K. Trafficking of lysosomal cathepsin B-green fluorescent protein to the surface of thyroid epithelial cells involves the endosomal/lysosomal compartment. J. Cell Sci. 2002, 115, 4877–4889. [Google Scholar] [CrossRef] [PubMed]
- Gong, Q.; Chan, S.J.; Bajkowski, A.S.; Steiner, D.F.; Frankfater, A. Characterization of the cathepsin B gene and multiple mRNAs in human tissues: Evidence for alternative splicing of cathepsin B pre-mRNA. DNA Cell Biol. 1993, 12, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Mehtani, S.; Gong, Q.; Panella, J.; Subbiah, S.; Peffley, D.M.; Frankfater, A. In Vivo Expression of an Alternatively Spliced Human Tumor Message That Encodes a Truncated Form of Cathepsin B: Subcellular Distribution of the Truncated Enzyme in Cos Cells. J. Biol. Chem. 1998, 273, 13236–13244. [Google Scholar] [CrossRef][Green Version]
- Müntener, K.; Willimann, A.; Zwicky, R.; Svoboda, B.; Mach, L.; Baici, A. Folding competence of N-terminally truncated forms of human procathepsin B. J. Biol. Chem. 2005, 280, 11973–11980. [Google Scholar] [CrossRef]
- Baici, A.; Müntener, K.; Willimann, A.; Zwicky, R. Regulation of human cathepsin B by alternative mRNA splicing: Homeostasis, fatal errors and cell death. Biol. Chem. 2006, 387, 1017–1021. [Google Scholar] [CrossRef] [PubMed]
- Berquin, I.M.; Cao, L.; Fong, D.; Sloane, B.F. Identification of two new exons and multiple transcription start points in the 5’-untranslated region of the human cathepsin-B-encoding gene. Gene 1995, 159, 143–149. [Google Scholar] [CrossRef]
- Müntener, K.; Zwicky, R.; Csucs, G.; Rohrer, J.; Baici, A. Exon skipping of cathepsin B: Mitochondrial targeting of a lysosomal peptidase provokes cell death. J. Biol. Chem. 2004, 279, 41012–41017. [Google Scholar] [CrossRef]
- Bestvater, F.; Dallner, C.; Spiess, E. The C-terminal subunit of artificially truncated human cathepsin B mediates its nuclear targeting and contributes to cell viability. BMC Cell Biol. 2005, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Hölzen, L.; Parigiani, M.A.; Reinheckel, T. Tumor cell- and microenvironment-specific roles of cysteine cathepsins in mouse models of human cancers. Biochim. Biophys. Acta-Proteins Proteom. 2020, 1868, 140423. [Google Scholar] [CrossRef] [PubMed]
- Müntener, K.; Zwicky, R.; Csucs, G.; Baici, A. The alternative use of exons 2 and 3 in cathepsin B mRNA controls enzyme trafficking and triggers nuclear fragmentation in human cells. Histochem. Cell Biol. 2003, 119, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Waghray, A.; Keppler, D.; Sloane, B.F.; Schuger, L.; Chen, Y.Q. Analysis of a truncated form of cathepsin H in human prostate tumor cells. J. Biol. Chem. 2002, 277, 11533–11538. [Google Scholar] [CrossRef]
- Goulet, B.; Baruch, A.; Moon, N.-S.; Poirier, M.; Sansregret, L.L.; Erickson, A.; Bogyo, M.; Nepveu, A. A Cathepsin L Isoform that is Devoid of a Signal Peptide Localizes to the Nucleus in S Phase and Processes the CDP/Cux Transcription Factor. Mol. Cell 2004, 14, 207–219. [Google Scholar] [CrossRef]
- Burton, L.J.; Henderson, V.; Liburd, L.; Odero-Marah, V.A. Snail transcription factor NLS and importin β1 regulate the subcellular localization of Cathepsin L and Cux1. Biochem. Biophys. Res. Comm. 2017, 491, 59–64. [Google Scholar] [CrossRef]
- Al-Hashimi, A.; Venugopalan, V.; Sereesongsaeng, N.; Tedelind, S.; Pinzaru, A.M.; Hein, Z.; Springer, S.; Weber, E.; Führer, D.; Scott, C.J.; et al. Significance of nuclear cathepsin V in normal thyroid epithelial and carcinoma cells. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118846. [Google Scholar] [CrossRef] [PubMed]
- Sloane, B.F.; List, K.; Fingleton, B.; Matrisian, L. Proteases in cancer: Significance for invasion and metastasis. In Proteases: Structure and Function; Brix, K., Stöcker, W., Eds.; Springer: Wien, Austria, 2013; pp. 491–550. [Google Scholar] [CrossRef]
- Zhang, Z.; Yue, P.; Lu, T.; Wang, Y.; Wei, Y.; Wei, X. Role of lysosomes in physiological activities, diseases, and therapy. J. Hematol. Oncol. 2021, 14, 79. [Google Scholar] [CrossRef]
- Tamhane, T.; Lllukkumbura, R.; Lu, S.; Maelandsmo, G.M.; Haugen, M.H.; Brix, K. Nuclear cathepsin L activity is required for cell cycle progression of colorectal carcinoma cells. Biochimie 2016, 122, 208–218. [Google Scholar] [CrossRef]
- Büth, H. Contribution of the Lysosomal Cysteine Protease Cathepsin B to Extracellular Matrix Remodeling during Keratinocyte Migration and Wound Healing. Ph.D. Thesis, International University Bremen, Bremen, Germany, 2006. [Google Scholar]
- Kamentsky, L.; Jones, T.R.; Fraser, A.; Bray, M.A.; Logan, D.J.; Madden, K.L.; Ljosa, V.; Rueden, C.; Eliceiri, K.W.; Carpenter, A.E. Improved structure, function and compatibility for CellProfiler: Modular high-throughput image analysis software. Bioinformatics 2011, 27, 1179–1180. [Google Scholar] [CrossRef] [PubMed]
- Haugen, M.H.; Johansen, H.T.; Pettersen, S.J.; Solberg, R.; Brix, K.; Flatmark, K.; Maelandsmo, G.M. Nuclear legumain activity in colorectal cancer. PLoS ONE 2013, 8, e52980. [Google Scholar] [CrossRef]
- Brix, K.; Lemansky, P.; Herzog, V. Evidence for extracellularly acting cathepsins mediating thyroid hormone liberation in thyroid epithelial cells. Endocrinology 1996, 137, 1963–1974. [Google Scholar] [CrossRef]
- Tamhane, T.; Arampatzidou, M.; Gerganova, V.; Tacke, M.; Illukkumbura, R.; Dauth, S.; Schaschke, N.; Peters, C.; Reinheckel, T.; Brix, K. The activity and localization patterns of cathepsins B and X in cells of the mouse gastrointestinal tract differ along its length. Biol. Chem. 2014, 395, 1201–1219. [Google Scholar] [CrossRef] [PubMed]
- Kopito, R.R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000, 10, 524–530. [Google Scholar] [CrossRef]
- Shen, D.; Coleman, J.; Chan, E.; Nicholson, T.P.; Dai, L.; Sheppard, P.W.; Patton, W.F. Novel cell- and tissue-based assays for detecting misfolded and aggregated protein accumulation within aggresomes and inclusion bodies. Cell Biochem. Biophys. 2011, 60, 173–185. [Google Scholar] [CrossRef]
- Jakoš, T.; Pišlar, A.; Jewett, A.; Kos, J. Cysteine Cathepsins in Tumor-Associated Immune Cells. Front. Immunol. 2019, 10, 2037. [Google Scholar] [CrossRef]
- Reiser, J.; Adair, B.; Reinheckel, T. Specialized roles for cysteine cathepsins in health and disease. J. Clin. Investig. 2010, 120, 3421–3431. [Google Scholar] [CrossRef]
- Brix, K.; Scott, C.J.; Heck, M.M.S. Compartmentalization of Proteolysis. In Proteases: Structure and Function; Brix, K., Stöcker, W., Eds.; Springer: Wien, Austria, 2013; pp. 85–125. [Google Scholar] [CrossRef]
- Tholen, M.; Hillebrand, L.E.; Tholen, S.; Sedelmeier, O.; Arnold, S.J.; Reinheckel, T. Out-of-frame start codons prevent translation of truncated nucleo-cytosolic cathepsin L in vivo. Nat. Commun. 2014, 5, 4931. [Google Scholar] [CrossRef]
- Haugen, M.H.; Boye, K.; Nesland, J.M.; Pettersen, S.J.; Egeland, E.V.; Tamhane, T.; Brix, K.; Maelandsmo, G.M.; Flatmark, K. High expression of the cysteine proteinase legumain in colorectal cancer—Implications for therapeutic targeting. Eur. J. Cancer 2015, 51, 9–17. [Google Scholar] [CrossRef]
- Hämälistö, S.; Stahl, J.L.; Favaro, E.; Yang, Q.; Liu, B.; Christoffersen, L.; Loos, B.; Guasch Boldú, C.; Joyce, J.A.; Reinheckel, T.; et al. Spatially and temporally defined lysosomal leakage facilitates mitotic chromosome segregation. Nat. Commun. 2020, 11, 229. [Google Scholar] [CrossRef] [PubMed]
- Shuja, S.; Cai, J.; Iacobuzio-Donahue, C.; Zacks, J.; Beazley, R.M.; Kasznica, J.M.; O’Hara, C.J.; Heimann, R.; Murnane, M.J. Cathepsin B activity and protein levels in thyroid carcinoma, Graves’ disease, and multinodular goiters. Thyroid 1999, 9, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Wiederanders, B.; Kaulmann, G.; Schilling, K. Functions of propeptide parts in cysteine proteases. Curr. Protein Pept. Sci. 2003, 4, 309–326. [Google Scholar] [CrossRef]
- Burden, R.E.; Snoddy, P.; Jefferies, C.A.; Walker, B.; Scott, C.J. Inhibition of cathepsin L-like proteases by cathepsin V propeptide. Biol. Chem. 2007, 388, 541–545. [Google Scholar] [CrossRef] [PubMed]
- Erickson, A.H.; Isidoro, C.; Mach, L.; Mort, J.S. Cathepsins: Getting in Shape for Lysosomal Proteolysis. In Proteases: Structure and Function; Brix, K., Stöcker, W., Eds.; Springer: Wien, Austria, 2013; pp. 127–173. [Google Scholar] [CrossRef]
- Jerič, B.; Dolenc, I.; Mihelič, M.; Klarić, M.; Zavašnik-Bergant, T.; Gunčar, G.; Turk, B.; Turk, V.; Stoka, V. N-terminally truncated forms of human cathepsin F accumulate in aggresome-like inclusions. Biochim. Biophys. Acta (BBA)-Mol. Cell Res. 2013, 1833, 2254–2266. [Google Scholar] [CrossRef] [PubMed]
- Johnston, J.A.; Ward, C.L.; Kopito, R.R. Aggresomes: A cellular response to misfolded proteins. J. Cell Biol. 1998, 143, 1883–1898. [Google Scholar] [CrossRef]
- García-Mata, R.; Bebök, Z.; Sorscher, E.J.; Sztul, E.S. Characterization and Dynamics of Aggresome Formation by a Cytosolic Gfp-Chimera. J. Cell Biol. 1999, 146, 1239–1254. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.; Jahreiss, L.; Sarkar, S.; Saiki, S.; Menzies, F.M.; Ravikumar, B.; Rubinsztein, D.C. Aggregate-Prone Proteins are Cleared from the Cytosol by Autophagy: Therapeutic Implications. In Current Topics in Developmental Biology; Academic Press: Cambridge, MA, USA, 2006; Volume 76, pp. 89–101. [Google Scholar]
- Beaudoin, S.; Goggin, K.; Bissonnette, C.; Grenier, C.; Roucou, X. Aggresomes do not represent a general cellular response to protein misfolding in mammalian cells. BMC Cell Biol. 2008, 9, 59. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.S.P.; Tan, J.M.M.; Soong, W.-E.; Hussein, K.; Nukina, N.; Dawson, V.L.; Dawson, T.M.; Cuervo, A.M.; Lim, K.-L. Autophagy-mediated clearance of aggresomes is not a universal phenomenon. Hum. Mol. Genet. 2008, 17, 2570–2582. [Google Scholar] [CrossRef] [PubMed]
- Bian, B.; Mongrain, S.; Cagnol, S.; Langlois, M.J.; Boulanger, J.; Bernatchez, G.; Carrier, J.C.; Boudreau, F.; Rivard, N. Cathepsin B promotes colorectal tumorigenesis, cell invasion, and metastasis. Mol. Carcinog. 2016, 55, 671–687. [Google Scholar] [CrossRef] [PubMed]
- Tedelind, S.; Jordans, S.; Resemann, H.; Blum, G.; Bogyo, M.; Führer, D.; Brix, K. Cathepsin B trafficking in thyroid carcinoma cells. Thyroid Res. 2011, 4 (Suppl. S1), 1–16. [Google Scholar] [CrossRef]
- Sevenich, L.; Bowman, R.L.; Mason, S.D.; Quail, D.F.; Rapaport, F.; Elie, B.T.; Brogi, E.; Brastianos, P.K.; Hahn, W.C.; Holsinger, L.J.; et al. Analysis of tumour- and stroma-supplied proteolytic networks reveals a brain-metastasis-promoting role for cathepsin S. Nat. Cell Biol. 2014, 16, 876–888. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamhane, T.; Njenga, R.W.; Burden, R.E.; Büth, H.; Maelandsmo, G.M.; Haugen, M.H.; Scott, C.J.; Brix, K. Trafficking of Full-Length and N-Terminally Truncated Cathepsin B in Human Colorectal Carcinoma Cells. Appl. Sci. 2021, 11, 11936. https://doi.org/10.3390/app112411936
Tamhane T, Njenga RW, Burden RE, Büth H, Maelandsmo GM, Haugen MH, Scott CJ, Brix K. Trafficking of Full-Length and N-Terminally Truncated Cathepsin B in Human Colorectal Carcinoma Cells. Applied Sciences. 2021; 11(24):11936. https://doi.org/10.3390/app112411936
Chicago/Turabian StyleTamhane, Tripti, Robin W. Njenga, Roberta E. Burden, Heiko Büth, Gunhild M. Maelandsmo, Mads H. Haugen, Christopher J. Scott, and Klaudia Brix. 2021. "Trafficking of Full-Length and N-Terminally Truncated Cathepsin B in Human Colorectal Carcinoma Cells" Applied Sciences 11, no. 24: 11936. https://doi.org/10.3390/app112411936
APA StyleTamhane, T., Njenga, R. W., Burden, R. E., Büth, H., Maelandsmo, G. M., Haugen, M. H., Scott, C. J., & Brix, K. (2021). Trafficking of Full-Length and N-Terminally Truncated Cathepsin B in Human Colorectal Carcinoma Cells. Applied Sciences, 11(24), 11936. https://doi.org/10.3390/app112411936





