Abstract
The modification and degradation of pectin in cell walls are necessary for the fruit softening process, which involves a series of pectin-modifying enzymes. Polygalacturonases (PGs) are a major group of pectin-hydrolyzing enzymes, which participate in fruit maturation, organ shedding, pollen development, and other processes by catalyzing the degradation of polygalacturonic acid. However, their function in plants has not yet been fully elucidated. In this paper, a full-length cDNA encoding SlPG49 was cloned from a tomato. Sequence alignment and phylogenetic analysis demonstrated that SlPG49 contains four typical conserved domains and belongs to clade E in PG classification. Quantitative real-time PCR analysis showed that SlPG49 was highly expressed in fruits during the softening stage, indicating that SlPG49 may be involved in fruit softening. Subcellular localization results revealed that SlPG49 was located in the cell membrane and the cell wall. In addition, an in vitro enzymatic activity assay confirmed that SlPG49 does have the ability to catalyze the hydrolysis of polygalacturonic acid. These results indicate that SlPG49 is a newly discovered PG gene involved in tomato fruit softening, and provide an experimental basis for elucidating the biological functions of plant PGs during fruit softening.
1. Introduction
Fruit ripening is a highly coordinated, genetically programmed, and irreversible process, accompanied by a series of physiological, biochemical, and sensory changes [1,2]. Fruit softening has an important impact on the changes in the flavor, aroma, color, and texture of the fruits, as well as their storage, transportation, and shelf life [3,4]. Previous studies have shown that the softening of fruits is due to enzyme-mediated changes in the structure and composition of the cell wall, especially the partial or complete degradation of cell wall polysaccharides, such as pectin and cellulose [5,6,7]. Among cell wall hydrolases, pectin-hydrolytic enzymes are a group of key cell wall hydrolases leading to fruit softening, which can promote the dissolution of pectin substances, resulting in a gradual decrease in fruit firmness [8,9,10].
Four pectin-hydrolytic enzymes involved in pectin modification and degradation have been identified, namely pectin methylesterases (PMEs), pectin acetylesterases (PAEs), polygalacturonases (PGs), and pectate lyases-like (PLLs) [11,12,13,14], which participate in different processes of pectin degradation. PMEs are responsible for the removal of methyl ester groups from pectin [15]; PAEs can cleave the acetylester bond from pectin [16,17]; PGs cleave the α-(1,4)-galacturonosyl linkages in pectin [18]; PLLs can hydrolyze d-galacturonic acid in pectin through a β-elimination reaction [19,20]. The PG family of pectin-hydrolytic enzymes has been known to function in plant growth and development for three decades [21]. PGs are one of the largest hydrolase families in plants [22], and contain four common conserved domains (domains I–IV) [23]. Based on their modes of action, PGs can be further divided into three different types: endo-PGs, exo-PGs, and rhamnose-PGs [24,25].
PGs are needed to degrade polygalacturonic acid and promote pectin degradation, and are of great significance to plant growth and development [6]. PGs are involved in plant morphogenesis [26,27], pollen development [28,29], organ shedding [30], fruit ripening [1], and other processes in plants [31]. Arabidopsis ATPG45 regulates leaf shape by affecting the proliferation and expansion of epidermal cells [26,27]. The absence of BcMF16 in Brassica campestris leads to the loss of pollen inner wall and inclusions [32]. PGZAT is a PG gene expressed in leaf abscission zones and functions in the cell separation process [33].
The relationship between PGs and fruit development has always been a research hotspot in this field, mainly focusing on species that use fruits as edible organs, such as Citrus sinensis, apples, and strawberries [34,35,36]. Strawberry lines that silence FaPG1 have soluble pectin of a higher molecular weight and show more supramolecular pectin aggregates than controls, indicating that FaPG1 plays a role in promoting the degradation of pectin, thereby accelerating fruit ripening [37]. Studies have found that mature fruits with inhibited MDPG1 expression in apples have higher intercellular adhesion, weakened subcutaneous cell expansion, and denser cells [38]. In addition, it was also verified that PGs could promote fruit softening by hydrolyzing pectin in cell walls of bananas, kiwis, and pears [39,40,41]. A transformed tomato with an antisense PG gene resulted in hardened fruit texture and lengthened shelf life [42]. Interestingly, although the mutation of tomato PG2A affects fruit color and weight, it has no effect on fruit softening [8], which may be due to the redundancy of the PGs in tomato plants. In a recent study, we characterized 54 PG members from tomato plants and found that multiple PG genes were highly expressed during fruit ripening [18]. Intriguingly, Solanum lycopersicum POLYGALACTURONASE49 (SlPG49) was highly expressed in the pericarp, suggesting that it may be critical to fruit softening. However, its biological functions remain unclear.
Here, the full length of a tomato PG gene (SlPG49) cDNA was cloned and bioinformatically analyzed. The expression pattern of SlPG49 was further confirmed by qRT-PCR. Moreover, the bio-functions of SlPG49 were further identified through subcellular localization and enzymatic activity analysis in vitro. This study could be valuable for providing a reliable experimental basis for future PG research and developing new tomato varieties with enhanced tolerance to fruit softening or long shelf life.
2. Materials and Methods
2.1. Plant Material and Growth Conditions
Tomato plants (Solanum lycopersicum L. cv. Micro-Tom) were grown in an artificial climate chamber. The growth conditions were set as described by Ke et al. [18], with minor modification.
2.2. SlPG49 Cloning and Bioinformatic Analysis
The full-length open reading frame (ORF) of SlPG49 (Solyc08g082170) was obtained from the Sol Genomic Network database (https://solgenomics.net, accessed on 5 March 2021). Molecular weight and isoelectric point of SlPG49 were predicted by the ProtParam tool (http://web.expasy.org/protparam, accessed on 5 March 2021). Clustal W was used to align the amino acid sequences of SlPG49 and PGs in other selected species. A neighbor-joining phylogenetic tree was constructed on the basis of the amino acid sequence by using MEGA 6.0 software.
2.3. RNA Extraction and qRT-PCR Analysis
The roots, stems, leaves, flowers, and fruits (mature green, breaker, and red ripening stage) used for RNA extraction were collected. All the materials were frozen in liquid nitrogen immediately and stored at −80 °C [43]. RNA extraction and cDNA synthesis were conducted using a MiniBEST Plant RNA Extraction Kit and a PrimeScript RT reagent Kit with gDNA Eraser (Takara, Japan). The tomato gene Ubi [44] was used as the internal reference gene. QRT-PCR analysis was performed using Takara’s SYBR Premix Ex Taq kit on a Real-Time PCR Thermocycler (Jena, Germany). Three biological replicates for each sample were performed. All primers used in this study were listed in Table S1.
2.4. Transient Expression Analysis of SlPG49
The coding sequence (CDS) of SlPG49 was amplified and cloned into pFGC-GFP to form the CaMV35S::GFP-SlPG49 fusion construct. The recombinant product was transformed into E. coli DH5α competent cells and verified by sequencing. Transient expression of the GFP-SlPG49 fusion protein in onion epidermal cells was carried out by a gene bombardment transformation method. The validated plasmids harboring CaMV35S::SlPG49-GFP were used to prepare the particles for bombardment. The inner epidermis of the onion (1–2 cm) was pre-incubated on Murashige and Skoog (MS) medium. The bombardment was performed using a gene gun at 1100 p.s.i. with DNA-coated gold particles from a distance of 9 cm. The bombarded samples were cultured on MS medium for 16–24 h at 25 °C in the dark. The images were taken with a laser scanning confocal microscope (Olympus, Japan).
2.5. Prokaryotic Expression and Purification of SlPG49
SlPG49 CDS (without the sequence encoding a signal peptide) was cloned into pMAL-c2x to construct the pMAL-MBP-SlPG49 bacterial expression vector. The generated pMAL-MBP-SlPG49 vector and pMAL-MBP empty vector were transformed into BL21-DE3. The extracted plasmid was verified by sequencing and transferred into E. coli Rosetta strain. Expression of pMAL-MBP-SlPG49 fusion protein and pMAL-MBP protein were induced by 0.05 mM isopropyl b-D-thiogalactopyranoside (IPTG, Sangon Biotech, Shanghai, China) at 37 °C for 2 h. Cells were harvested by centrifugation (8000× g, 8 min, 4 °C), then resuspended in protein extraction buffer (10 mmol·L−1 disodium phosphate, 1.75 mmol·L−1 potassium dihydrogen phosphate, 137 mmol·L−1 sodium chloride, and 2.65 mmol·L−1 potassium chloride, pH 7.4; Sangon Biotech, Shanghai, China). The resuspended cells were sonicated for 30 min (10 s/10 s) and then centrifuged (8000× g, 10 min, 4 °C). For protein purification, supernatants were incubated with amylose resin and then loaded into a purification column (Sangon Biotech, Shanghai, China). The purification column was washed with extraction and washing buffer (1 mmol·L−1 ethylene diamine tetraacetic acid, 200 mmol·L−1 sodium chloride, and 20 mmol·L−1 tris-hydrochloride, pH 8.0). The bound proteins were eluted with elution buffer (10 mmol·L−1 maltose) and stored at 4 °C for later use.
2.6. SDS-PAGE and Western Blot Analysis
Denatured proteins (100 °C, 8 min) were separated by 12% Biofuraw Precast Gel (Tanon, Shanghai, China). Then, the gel was stained with Coomassie Brilliant Blue R-250 (Sangon Biotech, Shanghai, China) for 1 h and washed off with decolorizing solution (5% ethanol and 10% acetic acid) for imaging.
For Western blot, purified proteins were denatured (100 °C, 8 min) before loading onto Biofuraw Precast Gel. After electrophoresis, the separated proteins were electroblotted onto NC membranes (ShineGene, Shanghai, China). After blocking in 5% skim milk for 1 h, Maltose Binding Protein Rabbit Monoclonal Antibody and HRP-labeled Goat Anti-Rabbit IgG (H+L, Beyotime, Shanghai, China) were bound sequentially to the target proteins. Signals on nitrocellulose membranes were captured with High-sig ECL Western Blotting Substrate (Tanon, Shanghai, China).
2.7. Analysis of SlPG49 Enzymatic Activity In Vitro
SlPG49 activity assays of purified protein were performed by using the Modified Bradford Protein Assay Kit (Sangon Biotech, Shanghai, China) and polygalacturonic acid activity assay kit (Solarbio, Beijing, China). In brief, one unit of PG activity was defined as the amount of enzyme releasing 1 μmol of galacturonic acid per minute per gram of purified protein at 40 °C and pH = 6.0. The absorbance at 520 nm was measured using a UV spectrophotometer (Shimadzu, Japan).
2.8. Statistical Analysis
Data were presented as mean ± SE with at least three biological replicates. One-way ANOVA was used for statistical analysis. A value of p < 0.05 was considered statistically significant and indicated by different letters in all the figures in this research.
3. Results
3.1. Sequence and Phylogenetic Analysis of SlPG49
A PG gene (Solyc08g082170.2.1), named SlPG49, was isolated and cloned from Micro-Tom on the basis of the sequence available in Sol Genomics Network (solgenomics.net). SlPG49 contains seven exons and six introns (Figure 1A), and encodes a protein of 452 amino acid residues (49.03 kD). Transmembrane domain analysis indicated that SlPG49 contains a transmembrane domain and belongs to the category of membrane localization proteins (Figure 1B). To determine the evolutionary position of SlPG49, a phylogenetic tree containing SlPG49 and 37 PGs (Table S2) from other species was constructed. The results showed that SlPG49 belongs to clade E, and clustered with the other eight PGs, including AtPG5 in Arabidopsis thaliana and BcMF23 in Brassica rapa (Figure 1C). Then, sequence alignment was performed on the nine PGs from clade E and the other four PGs from different clades. As expected, the nine PGs that belong to the clade E showed higher amino acid sequence similarity. Further analysis revealed that the PGs in clade E all contained conserved domains Ⅰ, Ⅱ, and Ⅳ, but domain Ⅲ was less conserved (Figure 1D).
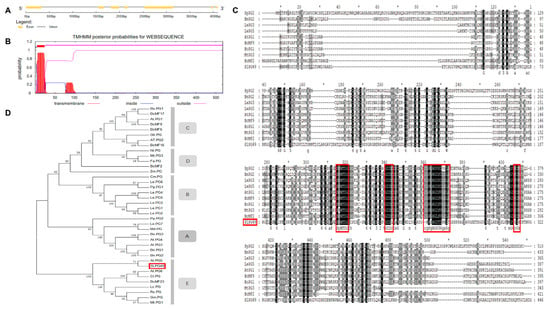
Figure 1.
Gene structure, transmembrane region, multiple alignment, and phylogenetic analysis of SlPG49: (A) gene structure of SlPG49; (B) transmembrane region of SlPG49; (C) phylogenetic analysis of SlPG49; (D) multiple alignment analysis of SlPG49.
3.2. Expression Pattern Analysis of SlPG49
To further understand the role of SlPG49, qRT-PCR was first conducted to study the expression pattern of SlPG49 in tomato plants. The results showed that SlPG49 transcripts were detectable in all organs examined except roots. SlPG49 was highly expressed both in fruits of red ripening stage and leaves, but moderately accumulated in flowers, and weakly expressed in mature green fruits, breaker fruits, and stems (Figure 2). These findings suggested that SlPG49 may be crucial for tomato fruit softening, as well as leaf and flower development.
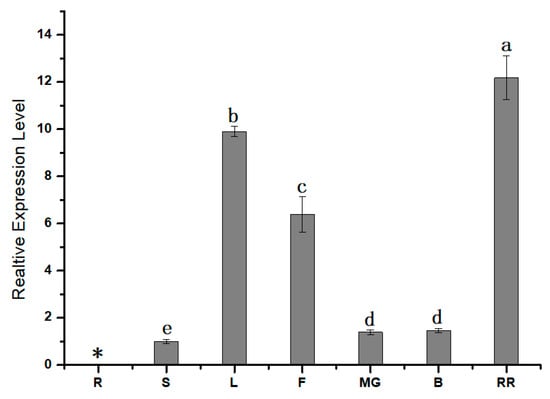
Figure 2.
Expression pattern analysis of SlPG49 by qRT-PCR. R: roots; S: stems; L: leaves; F: flowers; MG: mature green fruit; B: breaker fruit; RR: red ripening fruit. Data are presented as mean ± SE with at least three biological replicates. Significant differences are indicated by different letters (p < 0.05). * indicates undetected expression level.
3.3. Subcellular Localization of SlPG49
Onion epidermal cells are a good material for the transient expression of exogenous genes because of their clear structure, lack of chloroplasts, and convenient materials. A transient expression assay in onion epidermal cells with CaMV35S::SlPG49-GFP plasmids was conducted using particle bombardment to examine the subcellular localization of SlPG49. The results showed that green fluorescence signals were widely distributed in onion epidermal cells transformed with SlPG49-GFP (Figure 3A–C), and the fusion protein signals of SlPG49-GFP could be detected in the cell membrane, cell wall, and extracellular space in the plasma wall-isolated onion epidermal cells (Figure 3D,F). These results indicate that SlPG49 is a secreted protein, and localizes in the cell membrane and the cell wall.
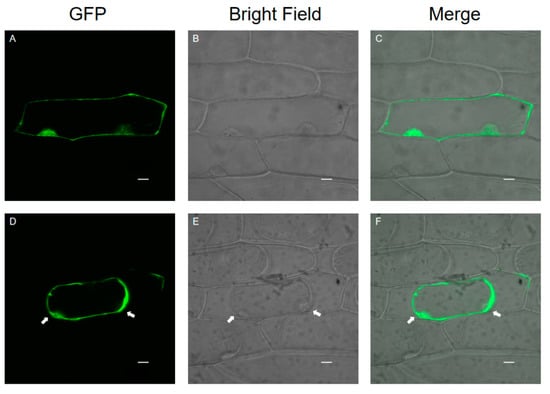
Figure 3.
Subcellular localization of SlPG49-GFP fusion protein in living onion epidermal cells: (A–C) localization of SlPG49-GFP; (D–F) localization of SlPG49-GFP in plasmolyzed onion epidermal cells; cells were plasmolyzed by treatment with 1 mol·L−1 mannitol for 5 min. White arrowheads show the membranes separated from the cell wall. Bars = 20 µm.
3.4. Enzymatic Activity of SlPG49 In Vitro
SlPG49 was heterologously expressed, and its activity was tested to characterize whether it encoded an active PG. SlPG49 was predicted to contain a signal peptide (Figure 4A). A truncated CDS, including sequences encoding GH28 domain, was cloned into the pMAL-c2x vector (Figure 4B) to improve protein solubility, using pMAL-MBP as a negative control. The predicted sizes of SlPG49 and MBP tag protein were 49.03 kDa and 43 kDa, respectively. The results of SDS-PAGE gels and Western blot illustrated that the sizes of the tag protein band and fusion protein band were consistent with the prediction (Figure 4C,D), indicating that SlPG49 protein was successfully isolated and purified. The purified proteins were incubated with polygalacturonic acid to analyze its hydrolytic activity. The results demonstrated that SlPG49 showed high enzymatic activity (Figure 4E), indicating that SlPG49 encodes a bona fide PG.
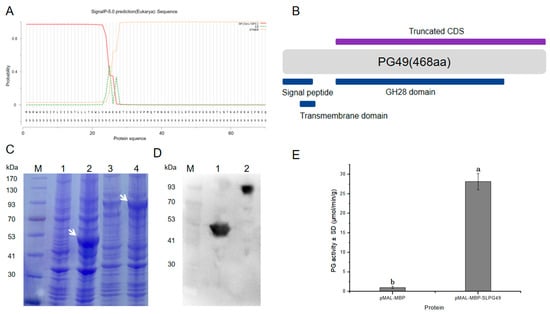
Figure 4.
SlPG49 displays PG activity in vitro: (A,B) signal peptide analysis and schematic protein structure of SlPG49; (C) SDS-PAGE analysis; M, protein maker; 1, not induced pMAL-MBP; 2, induced pMAL-MBP; 3, not induced pMAL-MBP-SlPG49; 4, induced pMAL-MBP-SlPG49. White arrowheads show the bands of pMAL-MBP-SlPG49 and pMAL-MBP; (D) western blot analysis; 1: purified pMAL-MBP; 2: purified pMAL-MBP-SlPG49; (E) purified pMAL-MBP-SlPG49 shows PG activity in vitro. Data are presented as mean ± SE with at least three biological replicates. Significant differences are indicated by different letters (p < 0.05).
4. Discussion
According to the latest classification system, PGs can be divided into seven clades (A–G), and most members of clade E can be detected at a high overall expression in all tissues [45]. In the present work, phylogenetic analysis showed that SlPG49 was located in clade E and closed to ATPG5 (Figure 1D). Interestingly, the expression of ATPG5 was significantly increased when an Arabidopsis endo-PG (AtPGLR) was knocked out, indicating that ATPG5 has the function of replenishing AtPGLR, which may also be an endo-PG [29]. Therefore, in view of the close evolutionary relationship between SlPG49 and ATPG5, we speculated that SlPG49 was probably an endo-PG. As mentioned earlier, PG has four conserved domains (Ⅰ–Ⅳ). Generally, genes encoding the proteins of these four domains are defined as PG genes. Notably, domain Ⅲ is relatively poorly conserved [46]. The results of multiple sequence alignments show that the conservation of domain III is indeed lower than that of domains Ⅰ, Ⅱ, and Ⅳ (Figure 1D). Although they are different from the typical domain Ⅲ, the nine PGs from clade E (including SlPG49) are highly conserved in domain Ⅲ. It has been reported that the positively charged domain Ⅳ may participate in an ionic interaction with the carboxyl terminal of the substrate [47]. Here, although R (Arginine) in the domain Ⅳ of SlPG49 is replaced by H (Histidine), it is still positively charged. Therefore, we speculated that the structure and function of SlPG49 may not have been changed.
A previous study showed that the enzymatic activity of PG increased during the ripening period of the tomato fruit, suggesting that these enzymes may be critical for initiating the ripening process [21]. In our previous research, SlPG49 was predicted to have a high expression in red ripening fruits [18]. In this work, qPCR analyses confirmed that SlPG49 is extremely highly expressed in fruits at the red ripening stage (Figure 2), demonstrating that SlPG49 may be involved in tomato fruit ripening and softening.
Pectin is synthesized in the Golgi apparatus and secreted into the cell wall in the state of methyl esterification and acetylation, and can be modified and degraded in the cell wall [26]. PGs are synthesized in the endoplasmic reticulum and secreted into the cell wall after maturation in the trans-Golgi network [48,49]. In accordance with their function, it is predicted that PGs exhibit extracellular localization. Previous studies have demonstrated that a PG protein in Brassica campestris, BcMF23, was a secreted protein and exhibited extracellular localization by subcellular localization of onion epidermis transient expression [50]. In transgenic Arabidopsis, ATPGX3 and AtPG45 were located in the cell wall [51,52]. The present transient expression result in the onion epidermis confirmed that SlPG49 was a secreted protein and may play a great role in the cell wall or extracellular matrix (Figure 3), similar to PGs in A. thaliana. To date, only nine PG genes have been identified in A. thaliana, with five that have been proven to show PG activity in vitro [51,53,54,55]. However, there is no report of in vitro activity of PGs in tomatoes. In this work, SlPG49 was successfully induced and purified by prokaryotic expression, and it was proved that SlPG49 has the activity of degrading polygalacturonic acid in vitro (Figure 4E), demonstrating that SlPG49 is another bonafide PG. Studies have shown that the increase in PG activity promotes the degradation of pectin in the cell walls of strawberries, apples, bananas, and other fruits, and accelerates fruit ripening and softening [37,38,39]. This study shows that the expression of SlPG49 increases significantly during the softening stage, and it has the activity of degrading one of the main components of pectin (polygalacturonic acid), indicating that it may play an important role in tomato fruit softening.
In summary, a new PG gene, SlPG49, has been identified in S. lycopersicum. The sequence analysis suggested that SlPG49 possesses the basic characteristics of PG family members. The results of expression pattern, subcellular localization, and enzymatic activity analysis indicated that SlPG49 may function in the degradation of cell wall polygalacturonic acid during fruit softening. Further studies will focus on elucidating the precise biological function and regulation of SlPG49 in fruit ripening.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/app112311450/s1, Table S1: Primers used in this study; Table S2: Sequences used in this study.
Author Contributions
Conceptualization, Y.Y. and Y.Z.; methodology, W.L. and L.X.; validation, Y.Y. and Y.Z.; formal analysis, Y.Y.; investigation, R.X.; data curation, W.L.; writing—original draft preparation, W.L. and L.X.; writing—review and editing, W.L. and L.X.; visualization, Y.S.; supervision, Y.Z.; project administration, Z.Z. and Y.Y.; funding acquisition, Z.Z. and Y.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by Natural Science Foundation of Zhejiang Province [No. LQ16C150005], and the National Natural Science Foundation of China [No. 31501769].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Forlani, S.; Masiero, S.; Mizzotti, C. Fruit ripening: The role of hormones, cell wall modifications, and their relationship with pathogens. J. Exp. Bot. 2019, 70, 2993–3006. [Google Scholar] [CrossRef]
- Zhang, J.; Feng, S.; Jing, Y.; Wang, C.; Lu, T.; Wang, H.; Yu, C. The Formation of Fruit Quality in Cucumis sativus L. Front. Plant Sci. 2021, 12, 729448. [Google Scholar] [CrossRef]
- Giovannoni, J.; Nguyen, C.; Ampofo, B.; Zhong, S.; Fei, Z. The epigenome and transcriptional dynamics of fruit ripening. Annu. Rev. Plant Biol. 2017, 68, 61–84. [Google Scholar] [CrossRef]
- Klee, H.J.; Giovannoni, J.J. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 2011, 45, 41–59. [Google Scholar] [CrossRef]
- Jarvis, M.C.; Briggs, S.P.H.; Knox, J.P. Intercellular adhesion and cell separation in plants. Plant Cell Environ. 2003, 26, 977–989. [Google Scholar] [CrossRef]
- Brummell, D.A. Cell wall disassembly in ripening fruit. Funct. Plant Biol. 2006, 33, 103–119. [Google Scholar] [CrossRef]
- Senechal, F.; Wattier, C.; Rusterucci, C.; Pelloux, J. Homogalacturonan-modifying enzymes: Structure, expression, and roles in plants. J. Exp. Bot. 2014, 65, 5125–5160. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Yeats, T.H.; Uluisik, S.; Rose, J.K.; Seymour, G.B. Fruit Softening: Revisiting the Role of Pectin. Trends Plant Sci. 2018, 23, 302–310. [Google Scholar] [CrossRef]
- Tucker, G.; Yin, X.R.; Zhang, A.D.; Wang, M.M.; Zhu, Q.G.; Liu, X.F.; Xie, X.L.; Chen, K.S.; Grierson, D. Ethylene and fruit softening. Food Qual. Saf. 2017, 1, 253–267. [Google Scholar]
- Villarreal, N.M.; Hernan, G.R.; Gustavo, A.M.; Civello, P.M. Polygalacturonase activity and Expression of related genes during ripening of strawberry cultivars with contrasting fruit firmness. Postharvest Biol. Technol. 2008, 47, 141–150. [Google Scholar] [CrossRef]
- Chen, H.; Cao, S.; Fang, X.; Mu, H.; Yang, H.; Wang, X.; Xu, Q.; Gao, H. Changes in fruit firmness, cell wall composition, and cell wall degrading enzymes in postharvest blueberries duringstorage. Sci. Hortic. 2015, 188, 44–48. [Google Scholar] [CrossRef]
- Ng, J.K.; Schröder, R.; Brummell, D.A.; Sutherland, P.W.; Hallett, I.C.; Smith, B.G.; Melton, L.D.; Johnston, J.W. Lower cell wall pectin solubilisation and galactose loss during early fruit development in apple (Malus x domestica) cultivar ‘Scifresh’ are associated with slower softening rate. J. Plant Physiol. 2015, 176, 129–137. [Google Scholar] [CrossRef]
- Konarska, A. The structure of the fruit peel in two varieties of Malus domestica Borkh. (Rosaceae) before and after storage. Protoplasma 2013, 250, 701–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Yan, J.; Wang, S.; Xu, M.; Huang, K.; Chen, G.; Ding, Y. Genome-wide identification of the pectate lyase-like (PLL) gene family and functional analysis of two PLL genes in rice. Mol. Genet. Genom. 2018, 293, 1317–1331. [Google Scholar] [CrossRef] [PubMed]
- Silva-Sanzana, C.; Celiz-Balboa, J.; Garzo, E.; Marcus, S.E.; Parra-Rojas, J.P.; Rojas, B.; Olmedo, P.; Rubilar, M.A.; Rios, I.; Chorbadjian, R.A.; et al. Pectin methylesterases modulate plant homogalacturonan Status in Defenses against the Aphid Myzus persicae. Plant Cell 2019, 31, 1913–1929. [Google Scholar] [CrossRef] [Green Version]
- Gou, J.Y.; Miller, L.M.; Hou, G.; Yu, X.H.; Chen, X.Y.; Liu, C.J. Acetylesterase-Mediated Deacetylation of Pectin Impairs Cell Elongation, Pollen Germination, and Plant Reproduction. Plant Cell 2012, 24, 50–65. [Google Scholar] [CrossRef] [Green Version]
- Orfila, C.; Degan, F.D.; Jørgensen, B.; Scheller, H.V.; Ray, P.M.; Ulvskov, P. Expression of mung bean pectin acetyl esterase in potato tubers: Effect on acetylation of cell wall polymers and tuber mechanical properties. Planta 2012, 236, 185–196. [Google Scholar] [CrossRef]
- Ke, X.; Wang, H.; Li, Y.; Zhu, B.; Zang, Y.; He, Y.; Cao, J.; Zhu, Z.; Yu, Y. Genome-Wide identification and analysis of polygalacturonase genes in Solanum lycopersicum. Int. J. Mol. Sci. 2018, 19, 2290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, A.; Miller, J.G.; Murray, L.; Sadler, I.H.; Fry, S.C. The pectic disaccharides lepidimoic acid and β-d-xylopyranosyl-(1→3)-d-galacturonic acid occur in cress-seed exudate but lack allelochemical activity. Ann. Bot. 2016, 117, 607–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Li, G.; Zhao, X. Comparative analysis of pectate lyase in relation to softening in strawberry fruits. Can. J. Plant Sci. 2016, 96, 604–612. [Google Scholar] [CrossRef] [Green Version]
- Prasanna, V.; Prabha, T.N.; Tharanathan, R.N. Fruit Ripening Phenomena-An Overview. Crit. Rev. Food Sci. 2007, 47, 1–19. [Google Scholar] [CrossRef]
- Markovic, O.; Janecek, S. Pectin degrading glycoside hydrolases of family 28: Sequence-structural features, specificities and evolution. Protein Eng. 2001, 14, 615–631. [Google Scholar] [CrossRef]
- Torki, M.; Mandaron, P.; Mache, R.; Falconet, D. Characterization of a ubiquitous expressed gene family encoding polygalacturonase in Arabidopsis thaliana. Gene 2000, 242, 427–436. [Google Scholar] [CrossRef]
- Park, K.C.; Kwon, S.J.; Kim, N.S. Intron loss mediated structural dynamics and functional differentiation of the polygalacturonase gene family in land plants. Genes Genom. 2010, 32, 570–577. [Google Scholar] [CrossRef]
- Yang, Y.; Yu, Y.; Liang, Y.; Anderson, C.T.; Cao, J. A profusion of molecular scissors for pectins: Classification, expression, and functions of plant polygalacturonases. Front. Plant Sci. 2018, 9, 1208. [Google Scholar] [CrossRef] [Green Version]
- Asl, L.K.; Dhondt, S.; Boudolf, V.; Beemster, G.T.S.; Beeckman, T.; Inzé, D.; Govaerts, W.; De Veylder, L. Model-based analysis of Arabidopsis leaf epidermal cells reveals distinct division and expansion patterns for pavement and guard cell. Plant Physiol. 2011, 156, 2172–2183. [Google Scholar]
- Andriankaja, M.; Dhondt, S.; DeBodt, S.; Vanhaeren, H.; Coppens, F.; De Milde, L.; Mühlenbock, P.; Skirycz, A.; Gonzalez, N.; Beemster, G.T.; et al. Exit from proliferation during leaf development in Arabidopsis thaliana: A not-so-gradual process. Dev. Cell 2012, 22, 64–78. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Lv, M.; Liang, Y.; Xiong, X.; Cao, J. Molecular cloning and characterization of a novel polygalacturonase gene, BcMF24, involved in pollen development of Brassica campestris ssp. chinensis. Plant Mol. Biol. Rep. 2014, 32, 476–486. [Google Scholar] [CrossRef]
- Hocq, L.; Guinand, S.; Habrylo, O.; Voxeur, A.; Tabi, W.; Safran, J.; Fournet, F.; Domon, J.M.; Mollet, J.C.; Pilard, S.; et al. The exogenous application of AtPGLR, an endo-polygalacturonase, triggers pollen tube burst and repair. Plant J. 2020, 103, 617–633. [Google Scholar] [CrossRef]
- Wakasa, Y.; Kudo, H.; Ishikawa, R.; Akada, S.; Senda, M.; Niizeki, M.; Harada, T. Low expression of an endopolygalacturonase gene in apple fruit with long-term storage potential. Postharvest Biol. Technol. 2006, 39, 193–198. [Google Scholar] [CrossRef]
- Hadfield, K.A.; Bennett, A.B. Polygalacturonases: Many genes in search of a function. Plant Physiol. 1998, 117, 337–343. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Qiu, L.; Huang, L.; Yu, X.; Lu, G.; Cao, J. Isolation and Characterization of an Anther-Specific Polygalacturonase Gene, BcMF16, in Brassica campestris ssp. chinensis. Plant Mol. Biol. Rep. 2012, 30, 330–338. [Google Scholar] [CrossRef]
- Sander, L.; Child, R.; Ulvskov, P.; Albrechtsen, M.; Borkhardt, B. Analysis of a dehiscence zone endo-polygalacturonase in oilseed rape (Brassica napus) and Arabidopsis thaliana: Evidence for roles in cell separation in dehiscence and abscission zones, and in stylar tissues during pollen tube growth. Plant Mol. Biol. 2001, 46, 469–479. [Google Scholar] [CrossRef]
- Liu, Y.Z.; Dong, T.; Lei, Y.; Deng, X.X.; Gu, Q.Q. Isolation of a polygalacturonase gene from Citrus sinensis fruit and its expression relative to fruit mastication trait, fruit development, and calcium or boron treatments. Plant Mol. Biol. Rep. 2011, 29, 51–59. [Google Scholar] [CrossRef]
- Li, J.; Yuan, R. NAA and ethylene regulate expression of genes related to ethylene biosynthesis, perception, and cell wall degradation during fruit abscission and ripening in ‘delicious’ apples. J. Plant Growth Regul. 2008, 27, 283–295. [Google Scholar] [CrossRef]
- Redondo-Nevado, J.; Moyano, E.; Medina-Escobar, N.; Caballero, J.L.; Muñoz-Blanco, J. A fruit-specific and developmentally regulated endopolygalacturonase gene from strawberry (Fragaria × ananassa cv. Chandler). J. Exp. Bot. 2001, 52, 1941–1945. [Google Scholar] [CrossRef] [Green Version]
- Posé, S.; Kirby, A.R.; Paniagua, C.; Waldron, K.W.; Morris, V.J.; Quesada, M.A.; Mercado, J.A. The nanostructural characterization of strawberry pectins in pectate lyase or polygalacturonase silenced fruits elucidates their role in softening. Carbohydr. Polym. 2015, 132, 134–145. [Google Scholar] [CrossRef]
- Atkinson, R.G.; Sutherland, P.W.; Johnston, S.L.; Gunaseelan, K.; Hallett, I.C.; Mitra, D.; Brummell, D.A.; Schröder, R.; Johnston, J.W.; Schaffer, R.J. Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus × domestica) fruit. BMC Plant Biol. 2012, 12, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asif, M.H.; Nath, P. Expression of multiple forms of polygalacturonase gene during ripening in banana fruit. Plant Physiol. Biochem. 2005, 43, 177–184. [Google Scholar] [CrossRef]
- Wang, Z.Y.; MacRae, E.A.; Wright, M.A.; Bolitho, K.M.; Ross, G.S.; Atkinson, R.G. Polygalacturonase gene expression in kiwifruit: Relationship to fruit softening and ethylene production. Plant Mol. Biol. 2000, 42, 317–328. [Google Scholar] [CrossRef]
- Sekine, D.; Munemura, I.; Gao, M.; Mitsuhashi, W.; Toyomasu, T.; Murayama, H. Cloning of cDNAs encoding cell-wall hydrolases from pear (Pyrus communis) fruit and their involvement in fruit softening and development of melting texture. Physiol. Plant. 2006, 126, 163–174. [Google Scholar] [CrossRef]
- Smith, C.; Watson, C.; Ray, J.; Bird, C.; Morris, P.; Schuch, W.; Grierson, D. Antisense RNA inhibition of polygalacturonase gene expression in transgenic tomatoes. Nature 1988, 334, 724–726. [Google Scholar] [CrossRef]
- Geng, R.; Ke, X.; Wang, C.; He, Y.; Wang, H.; Zhu, Z. Genome-wide identification and expression analysis of transcription factors in Solanum lycopersicum. Agri Gene 2017, 6, 14–23. [Google Scholar] [CrossRef]
- Kong, F.; Wang, J.; Cheng, L.; Liu, S.; Wu, J.; Peng, Z.; Lu, G. Genome-wide analysis of the mitogen-activated protein kinase gene family in Solanum lycopersicum. Gene 2012, 499, 108–120. [Google Scholar] [CrossRef]
- Liang, Y.; Yu, Y.; Cui, J.; Lyu, M.; Xu, L.; Cao, J. A comparative analysis of the evolution, expression, and cis-regulatory element of polygalacturonase genes in grasses and dicots. Funct. Integr. Genom. 2016, 16, 641–656. [Google Scholar] [CrossRef]
- Park, K.C.; Kwon, S.J.; Kim, P.H.; Bureau, T.; Kim, N.S. Gene structure dynamics and divergence of the polygalacturonase gene family of plants and fungus. Genome 2008, 51, 30–40. [Google Scholar] [CrossRef]
- Bussink, H.J.D.; Buxton, F.P.; Visse, J. Expression and sequence comparison of the Aspergillus-niger and Aspergillus-tubigensis genes encoding polygalacturonase-II. Curr. Genet. 1991, 19, 467–474. [Google Scholar] [CrossRef]
- Caroli, M.D.; Lenucci, M.S.; Sansebastiano, G.P.D.; Dalessandro, G.; Lorenzo, G.D.; Piro, G. Protein trafficking to the cell wall occurs through mechanisms distinguishable from default sorting in tobacco. Plant J. 2011, 65, 295–308. [Google Scholar] [CrossRef]
- Caroli, M.D.; Lenucci, M.S.; Sansebastiano, G.P.D.; Dalessandro, G.; Lorenzo, G.D.; Piro, G. Dynamic protein trafficking to the cell wall. Plant Signal. Behav. 2011, 6, 1012–1015. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Huang, L.; Yu, X.; Xiong, X.; Yue, X.; Liu, T.; Liang, Y.; Lv, M.; Cao, J. Characterization of BcMF23a and BcMF23b, two putative pectin methylesterase genes related to pollen development in Brassica campestris ssp. chinensis. Mol. Biol. Rep. 2017, 44, 139–148. [Google Scholar] [CrossRef]
- Yang, Y.; Anderson, C.T.; Cao, J. Polygalacturonase45 cleaves pectin and links cell proliferation and morphogenesis to leaf curvature in Arabidopsis thaliana. Plant J. 2021, 106, 1493–1508. [Google Scholar] [CrossRef]
- Rui, Y.; Xiao, C.; Yi, H.; Kandemir, B.; Wang, J.Z.; Puri, V.M.; Anderson, C.T. POLYGALACTURONASE INVOLVED IN EXPANSION3 functions in seedling development, rosette growth, and stomatal dynamics in Arabidopsis thaliana. Plant Cell 2017, 29, 2413–2432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babu, Y.; Musielak, T.; Henschen, A.; Bayer, M. Suspensor length determines developmental progression of the embryo in Arabidopsis. Plant Physiol. 2013, 162, 1448–1458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rhee, S.Y.; Osborne, E.; Poindexter, P.D.; Somerville, C.R. Microspore separation in the quartet 3 mutants of Arabidopsis is impaired by a defect in a developmentally regulated polygalacturonase required for pollen mother cell wall degradation. Plant Physiol. 2003, 133, 1170–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, M.; Kay, P.; Wilson, S.; Swain, S.M. Arabidopsis dehiscence zone polygalacturonase1 (ADPG1), ADPG2, and QUARTET2 are polygalacturonases required for cell separation during reproductive development in Arabidopsis. Plant Cell 2009, 21, 216–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).