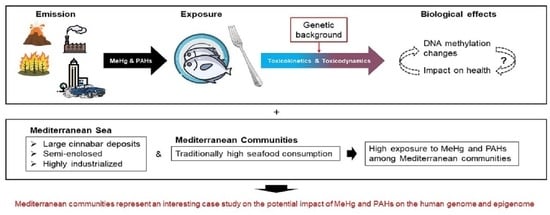
Methylmercury and Polycyclic Aromatic Hydrocarbons in Mediterranean Seafood: A Molecular Anthropological Perspective
Abstract
1. Introduction
2. Seafood Contaminants
2.1. MeHg
2.2. PAHs
3. Human Genetic Diversity
3.1. MeHg Exposure and Human Genetic Diversity
3.2. PAHs Exposure and Human Genetic Diversity
4. The Epigenetic Impact of Seafood Contaminants
4.1. The Impact of the Exposure to MeHg on DNA Methylation
4.2. The Impact of the Exposure to PAHs on DNA Methylation
5. An Anthropological Perspective
6. Mediterranean Coastal Communities as an Informative Case Study
7. Challenges
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weichselbaum, E.; Coe, S.; Buttriss, J.; Stanner, S. Fish in the Diet: A Review: Fish in the Diet. Nutr. Bull. 2013, 38, 128–177. [Google Scholar] [CrossRef]
- Thompson, L.A.; Darwish, W.S. Environmental Chemical Contaminants in Food: Review of a Global Problem. J. Toxicol. 2019, 2019, 1–14. [Google Scholar] [CrossRef]
- Marini, M.; Frapiccini, E. Persistence of Polycyclic Aromatic Hydrocarbons in Sediments in the Deeper Area of the Northern Adriatic Sea (Mediterranean Sea). Chemosphere 2013, 90, 1839–1846. [Google Scholar] [CrossRef]
- Marini, M.; Frapiccini, E. Do Lagoon Area Sediments Act as Traps for Polycyclic Aromatic Hydrocarbons? Chemosphere 2014, 111, 80–88. [Google Scholar] [CrossRef]
- Ferrante, M.; Copat, C.; Mauceri, C.; Grasso, A.; Schilirò, T.; Gilli, G. The Importance of Indicators in Monitoring Water Quality According to European Directives. Epidemiol. Prev. 2015, 5, 71–75. [Google Scholar]
- Junqué, E.; Garí, M.; Llull, R.M.; Grimalt, J.O. Drivers of the Accumulation of Mercury and Organochlorine Pollutants in Mediterranean Lean Fish and Dietary Significance. Sci. Total Environ. 2018, 634, 170–180. [Google Scholar] [CrossRef]
- Schartup, A.T.; Thackray, C.P.; Qureshi, A.; Dassuncao, C.; Gillespie, K.; Hanke, A.; Sunderland, E.M. Climate Change and Overfishing Increase Neurotoxicant in Marine Predators. Nature 2019, 572, 648–650. [Google Scholar] [CrossRef]
- Frapiccini, E.; Panfili, M.; Guicciardi, S.; Santojanni, A.; Marini, M.; Truzzi, C.; Annibaldi, A. Effects of Biological Factors and Seasonality on the Level of Polycyclic Aromatic Hydrocarbons in Red Mullet (Mullus Barbatus). Environ. Pollut. 2020, 258, 113742. [Google Scholar] [CrossRef] [PubMed]
- Castaño, A.; Cutanda, F.; Esteban, M.; Pärt, P.; Navarro, C.; Gómez, S.; Rosado, M.; López, A.; López, E.; Exley, K.; et al. Fish Consumption Patterns and Hair Mercury Levels in Children and Their Mothers in 17 EU Countries. Environ. Res. 2015, 141, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Domingo, J.L. Nutrients and Chemical Pollutants in Fish and Shellfish. Balancing Health Benefits and Risks of Regular Fish Consumption. Crit. Rev. Food Sci. Nutr. 2016, 56, 979–988. [Google Scholar] [CrossRef]
- Višnjevec, A.M.; Kocman, D.; Horvat, M. Human Mercury Exposure and Effects in Europe: Human Mercury Exposure and Effects in Europe. Environ. Toxicol. Chem. 2014, 33, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Basu, N.; Horvat, M.; Evers, D.C.; Zastenskaya, I.; Weihe, P.; Tempowski, J. A State-of-the-Science Review of Mercury Biomarkers in Human Populations Worldwide between 2000 and 2018. Environ. Health Perspect. 2018, 126, 106001. [Google Scholar] [CrossRef] [PubMed]
- Jacob, J.; Seidel, A. Biomonitoring of Polycyclic Aromatic Hydrocarbons in Human Urine. J. Chromatogr. B 2002, 778, 31–47. [Google Scholar] [CrossRef]
- Krabbenhoft, D.P.; Sunderland, E.M. Global Change and Mercury. Science 2013, 341, 1457–1458. [Google Scholar] [CrossRef]
- Selin, N.E. Global Biogeochemical Cycling of Mercury: A Review. Annu. Rev. Environ. Resour. 2009, 28, 43–63. [Google Scholar] [CrossRef]
- EEA EEA Report No 11/2018. Mercury in Europe’s Environment. A Priority for European and Global Action; Publications Office of the European Union: Luxembourg, 2018; Available online: https://www.eea.europa.eu/publications/mercury-in-europe-s-environment (accessed on 13 October 2021). [CrossRef]
- Szefer, P. Safety Assessment of Seafood with Respect to Chemical Pollutants in European Seas. Oceanol. Hydrobiol. Stud. 2013, 42, 110–118. [Google Scholar] [CrossRef]
- Rice, K.M.; Walker, E.M.; Wu, M.; Gillette, C.; Blough, E.R. Environmental Mercury and Its Toxic Effects. J. Prev. Med. Pub. Health 2014, 47, 74–83. [Google Scholar] [CrossRef]
- Zahir, F.; Rizwi, S.J.; Haq, S.K.; Khan, R.H. Low Dose Mercury Toxicity and Human Health. Environ. Toxicol. Pharmacol. 2005, 20, 351–360. [Google Scholar] [CrossRef]
- Karagas, M.R.; Choi, A.L.; Oken, E.; Horvat, M.; Schoeny, R.; Kamai, E.; Cowell, W.; Grandjean, P.; Korrick, S. Evidence on the Human Health Effects of Low-Level Methylmercury Exposure. Environ. Health Perspect. 2012, 120, 799–806. [Google Scholar] [CrossRef]
- EEA Commission Regulation (EC) No 1881/2006 of 19 December 2006 Setting Maximum Levels for Certain Contaminants in Foodstuffs (1). Off. J. Eur. Union 2006, 49, 5–24.
- WHO Evaluation of Certain Food Additives and Contaminants. Sixty-First Report of the Joint FAO/WHO Expert Committee on Food Additives; WHO: Geneva, Switzerland, 2004. [Google Scholar]
- Davidson, P.W.; van Wijngaarden, E.; Shamlaye, C.; Strain, J.J.; Myers, G.J. Putting Findings from the Seychelles Child Development Study into Perspective: The Importance of a Historical Special Issue of the Seychelles Medical and Dental Journal. NeuroToxicology 2020, 76, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Weihe, P.; Grandjean, P. Cohort Studies of Faroese Children Concerning Potential Adverse Health Effects after the Mothers’ Exposure to Marine Contaminants during Pregnancy. Acta Vet. Scand. 2012, 54, S7, 1751-0147-54-S1–S7. [Google Scholar] [CrossRef]
- EFSA Panel on Contaminants in the Food Chain (CONTAM). Scientific Opinion on the Risk for Public Health Related to the Presence of Mercury and Methylmercury in Food. EFSA J. 2012, 10, 2985. [Google Scholar] [CrossRef]
- Brambilla, G. Mercury Occurrence in Italian Seafood from the Mediterranean Sea and Possible Intake Scenarios of the Italian Coastal Population. Regul. Toxicol. Pharmacol. 2013, 9, 269–277. [Google Scholar] [CrossRef]
- Cocci, P.; Mosconi, G.; Bracchetti, L.; Nalocca, J.M.; Frapiccini, E.; Marini, M.; Caprioli, G.; Sagratini, G.; Palermo, F.A. Investigating the Potential Impact of Polycyclic Aromatic Hydrocarbons (PAHs) and Polychlorinated Biphenyls (PCBs) on Gene Biomarker Expression and Global DNA Methylation in Loggerhead Sea Turtles (Caretta Caretta) from the Adriatic Sea. Sci. Total Environ. 2018, 619–620, 49–57. [Google Scholar] [CrossRef]
- Frapiccini, E.; Annibaldi, A.; Betti, M.; Polidori, P.; Truzzi, C.; Marini, M. Polycyclic Aromatic Hydrocarbon (PAH) Accumulation in Different Common Sole (Solea Solea) Tissues from the North Adriatic Sea Peculiar Impacted Area. Mar. Pollut. Bull. 2018, 137, 61–68. [Google Scholar] [CrossRef]
- US-EPA Polycyclic Aromatic Hydrocarbons (PAHs) Fact Sheet. US EPA ARCHIVE DOCUMENT 2008. Available online: https://archive.epa.gov/epawaste/hazard/wastemin/web/pdf/pahs.pdf (accessed on 12 October 2021).
- Ferrante, M.; Zanghì, G.; Cristaldi, A.; Copat, C.; Grasso, A.; Fiore, M.; Signorelli, S.S.; Zuccarello, P.; Oliveri Conti, G. PAHs in Seafood from the Mediterranean Sea: An Exposure Risk Assessment. Food Chem. Toxicol. 2018, 115, 385–390. [Google Scholar] [CrossRef]
- Ramesh, A.; Walker, S.A.; Hood, D.B.; Guillén, M.D.; Schneider, K.; Weyand, E.H. Bioavailability and Risk Assessment of Orally Ingested Polycyclic Aromatic Hydrocarbons. Int. J. Toxicol. 2004, 23, 301–333. [Google Scholar] [CrossRef]
- Balcıoğlu, E.B. Potential Effects of Polycyclic Aromatic Hydrocarbons (PAHs) in Marine Foods on Human Health: A Critical Review. Toxin Rev. 2016, 35, 98–105. [Google Scholar] [CrossRef]
- Haritash, A.K. A Comprehensive Review of Metabolic and Genomic Aspects of PAH-Degradation. Arch. Microbiol. 2020, 26, 2033–2058. [Google Scholar]
- Castano-Vinyals, G. Biomarkers of Exposure to Polycyclic Aromatic Hydrocarbons from Environmental Air Pollution. Occup. Environ. Med. 2004, 61, e12. [Google Scholar] [CrossRef] [PubMed]
- Pruneda-Álvarez, L.G.; Pérez-Vázquez, F.J.; Ruíz-Vera, T.; Ochoa-Martínez, Á.C.; Orta-García, S.T.; Jiménez-Avalos, J.A.; Pérez-Maldonado, I.N. Urinary 1-Hydroxypyrene Concentration as an Exposure Biomarker to Polycyclic Aromatic Hydrocarbons (PAHs) in Mexican Women from Different Hot Spot Scenarios and Health Risk Assessment. Environ. Sci. Pollut. Res. 2016, 23, 6816–6825. [Google Scholar] [CrossRef]
- Iyer, S.; Wang, Y.; Xiong, W.; Tang, D.; Jedrychowski, W.; Chanock, S.; Wang, S.; Stigter, L.; Mróz, E.; Perera, F. Significant Interactions between Maternal PAH Exposure and Single Nucleotide Polymorphisms in Candidate Genes on B[ a ]P–DNA Adducts in a Cohort of Non-Smoking Polish Mothers and Newborns. Carcinogenesis 2016, 37, 1110–1115. [Google Scholar] [CrossRef]
- Gu, A.; Ji, G.; Jiang, T.; Lu, A.; You, Y.; Liu, N.; Luo, C.; Yan, W.; Zhao, P. Contributions of Aryl Hydrocarbon Receptor Genetic Variants to the Risk of Glioma and PAH-DNA Adducts. Toxicol. Sci. 2012, 128, 357–364. [Google Scholar] [CrossRef]
- Nethery, E.; Wheeler, A.J.; Fisher, M.; Sjödin, A.; Li, Z.; Romanoff, L.C.; Foster, W.; Arbuckle, T.E. Urinary Polycyclic Aromatic Hydrocarbons as a Biomarker of Exposure to PAHs in Air: A Pilot Study among Pregnant Women. J. Expo. Sci. Environ. Epidemiol. 2012, 22, 70–81. [Google Scholar] [CrossRef]
- Joneidi, Z.; Mortazavi, Y.; Memari, F.; Roointan, A.; Chahardouli, B.; Rostami, S. The Impact of Genetic Variation on Metabolism of Heavy Metals: Genetic Predisposition? Biomed. Pharmacother. 2019, 113, 108642. [Google Scholar] [CrossRef]
- Shen, H.; Tao, S.; Liu, J.; Huang, Y.; Chen, H.; Li, W.; Zhang, Y.; Chen, Y.; Su, S.; Lin, N.; et al. Global Lung Cancer Risk from PAH Exposure Highly Depends on Emission Sources and Individual Susceptibility. Sci. Rep. 2015, 4, 6561. [Google Scholar] [CrossRef] [PubMed]
- Basu, N.; Goodrich, J.M.; Head, J. Ecogenetics of Mercury: From Genetic Polymorphisms and Epigenetics to Risk Assessment and Decision-Making: Ecogenetics of Mercury. Environ. Toxicol. Chem. 2014, 33, 1248–1258. [Google Scholar] [CrossRef]
- Ruiz-Hernandez, A.; Kuo, C.-C.; Rentero-Garrido, P.; Tang, W.-Y.; Redon, J.; Ordovas, J.M.; Navas-Acien, A.; Tellez-Plaza, M. Environmental Chemicals and DNA Methylation in Adults: A Systematic Review of the Epidemiologic Evidence. Clin. Epigenetics 2015, 7, 55. [Google Scholar] [CrossRef]
- Ijomone, O.M.; Ijomone, O.K.; Iroegbu, J.D.; Ifenatuoha, C.W.; Olung, N.F.; Aschner, M. Epigenetic Influence of Environmentally Neurotoxic Metals. NeuroToxicology 2020, 81, 51–65. [Google Scholar] [CrossRef]
- Huang, Y.; Lin, S.; Wang, C.; Pi, X.; Jin, L.; Li, Z.; Wang, L.; Ren, A. Neural Tube Defects and ZIC4 Hypomethylation in Relation to Polycyclic Aromatic Hydrocarbon Exposure. Front. Cell Dev. Biol. 2020, 8, 582661. [Google Scholar] [CrossRef]
- Esposito, M.; De Roma, A.; Sansone, D.; Capozzo, D.; Iaccarino, D.; di Nocera, F.; Gallo, P. Non-Essential Toxic Element (Cd, As, Hg and Pb) Levels in Muscle, Liver and Kidney of Loggerhead Sea Turtles (Caretta Caretta) Stranded along the Southwestern Coasts of Tyrrhenian Sea. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2020, 231, 108725. [Google Scholar] [CrossRef]
- Serrano, O.; Martínez-Cortizas, A.; Mateo, M.A.; Biester, H.; Bindler, R. Millennial Scale Impact on the Marine Biogeochemical Cycle of Mercury from Early Mining on the Iberian Peninsula: Early Hg Pollution in the Iberian Region. Glob. Biogeochem. Cycles 2013, 27, 21–30. [Google Scholar] [CrossRef]
- Ancora, S.; Mariotti, G.; Ponchia, R.; Fossi, M.C.; Leonzio, C.; Bianchi, N. Trace Elements Levels in Muscle and Liver of a Rarely Investigated Large Pelagic Fish: The Mediterranean Spearfish Tetrapturus Belone (Rafinesque, 1810). Mar. Pollut. Bull. 2020, 151, 110878. [Google Scholar] [CrossRef]
- Giuliani, C.; Sazzini, M.; Bacalini, M.G.; Pirazzini, C.; Marasco, E.; Fontanesi, E.; Franceschi, C.; Luiselli, D.; Garagnani, P. Epigenetic Variability across Human Populations: A Focus on DNA Methylation Profiles of the KRTCAP3, MAD1L1 and BRSK2 Genes. Genome Biol. Evol. 2016, 8, 2760–2773. [Google Scholar] [CrossRef] [PubMed]
- Sazzini, M.; Abondio, P.; Sarno, S.; Gnecchi-Ruscone, G.A.; Ragno, M.; Giuliani, C.; De Fanti, S.; Ojeda-Granados, C.; Boattini, A.; Marquis, J.; et al. Genomic History of the Italian Population Recapitulates Key Evolutionary Dynamics of Both Continental and Southern Europeans. BMC Biol. 2020, 18, 51. [Google Scholar] [CrossRef]
- Custodio, H.M.; Broberg, K.; Wennberg, M.; Jansson, J.-H.; Vessby, B.; Hallmans, G.; Stegmayr, B.; Skerfving, S. Polymorphisms in Glutathione-Related Genes Affect Methylmercury Retention. Arch. Environ. Health Int. J. 2004, 59, 588–595. [Google Scholar] [CrossRef]
- Gundacker, C.; Komarnicki, G.; Jagiello, P.; Gencikova, A.; Dahmen, N.; Wittmann, K.; Gencik, M. Glutathione-S-Transferase Polymorphism, Metallothionein Expression, and Mercury Levels among Students in Austria. Sci. Total Environ. 2007, 385, 37–47. [Google Scholar] [CrossRef]
- Engström, K.S.; Strömberg, U.; Lundh, T.; Johansson, I.; Vessby, B.; Hallmans, G.; Skerfving, S.; Broberg, K. Genetic Variation in Glutathione-Related Genes and Body Burden of Methylmercury. Environ. Health Perspect. 2008, 116, 734–739. [Google Scholar] [CrossRef]
- Gundacker, C.; Wittmann, K.J.; Kukuckova, M.; Komarnicki, G.; Hikkel, I.; Gencik, M. Genetic Background of Lead and Mercury Metabolism in a Group of Medical Students in Austria. Environ. Res. 2009, 109, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-E.; Hong, Y.-C.; Park, H.; Ha, M.; Koo, B.S.; Chang, N.; Roh, Y.-M.; Kim, B.-N.; Kim, Y.-J.; Kim, B.-M.; et al. Interaction between GSTM1 / GSTT1 Polymorphism and Blood Mercury on Birth Weight. Environ. Health Perspect. 2010, 118, 437–443. [Google Scholar] [CrossRef]
- Goodrich, J.M.; Wang, Y.; Gillespie, B.; Werner, R.; Franzblau, A.; Basu, N. Glutathione Enzyme and Selenoprotein Polymorphisms Associate with Mercury Biomarker Levels in Michigan Dental Professionals. Toxicol. Appl. Pharmacol. 2011, 257, 301–308. [Google Scholar] [CrossRef]
- Wang, Y.; Goodrich, J.M.; Gillespie, B.; Werner, R.; Basu, N.; Franzblau, A. An Investigation of Modifying Effects of Metallothionein Single-Nucleotide Polymorphisms on the Association between Mercury Exposure and Biomarker Levels. Environ. Health Perspect. 2012, 120, 530–534. [Google Scholar] [CrossRef]
- Ng, S.; Lin, C.-C.; Hwang, Y.-H.; Hsieh, W.-S.; Liao, H.-F.; Chen, P.-C. Mercury, APOE, and Children’s Neurodevelopment. NeuroToxicology 2013, 37, 85–92. [Google Scholar] [CrossRef]
- Barcelos, G.R.M.; Grotto, D.; de Marco, K.C.; Valentini, J.; van Helvoort Lengert, A.; de Oliveira, A.Á.S.; Garcia, S.C.; Braga, G.Ú.L.; Schläwicke Engström, K.; de Syllos Cólus, I.M.; et al. Polymorphisms in Glutathione-Related Genes Modify Mercury Concentrations and Antioxidant Status in Subjects Environmentally Exposed to Methylmercury. Sci. Total Environ. 2013, 463–464, 319–325. [Google Scholar] [CrossRef]
- Julvez, J.; Smith, G.D.; Golding, J.; Ring, S.; Pourcain, B.S.; Gonzalez, J.R.; Grandjean, P. Prenatal Methylmercury Exposure and Genetic Predisposition to Cognitive Deficit at Age 8 Years. Epidemiology 2013, 24, 643–650. [Google Scholar] [CrossRef]
- De Oliveira, A.Á.S.; de Souza, M.F.; van Lengert, A.H.; de Oliveira, M.T.; de Camargo, R.B.O.G.; Braga, G.Ú.L.; de Cólus, I.M.S.; Barbosa, F.; Barcelos, G.R.M. Genetic Polymorphisms in Glutathione (GSH-) Related Genes Affect the Plasmatic Hg/Whole Blood Hg Partitioning and the Distribution between Inorganic and Methylmercury Levels in Plasma Collected from a Fish-Eating Population. BioMed Res. Int. 2014, 2014, 1–8. [Google Scholar] [CrossRef]
- Llop, S.; Pisa, F.; Tratnik, J.S.; Mazej, D.; Murcia, M.; Rebagliato, M.; Bustamante, M.; Sunyer, J.; Antonopoulou, E.; Antoniadou, I.; et al. Polymorphisms in ABC Transporter Genes and Concentrations of Mercury in Newborns – Evidence from Two Mediterranean Birth Cohorts. PLoS ONE 2014, 9, 9. [Google Scholar] [CrossRef]
- Ng, S.; Lin, C.-C.; Jeng, S.-F.; Hwang, Y.-H.; Hsieh, W.-S.; Chen, P.-C. Mercury, APOE, and Child Behavior. Chemosphere 2015, 120, 123–130. [Google Scholar] [CrossRef]
- Engström, K.; Love, T.M.; Watson, G.E.; Zareba, G.; Yeates, A.; Wahlberg, K.; Alhamdow, A.; Thurston, S.W.; Mulhern, M.; McSorley, E.M.; et al. Polymorphisms in ATP-Binding Cassette Transporters Associated with Maternal Methylmercury Disposition and Infant Neurodevelopment in Mother-Infant Pairs in the Seychelles Child Development Study. Environ. Int. 2016, 94, 224–229. [Google Scholar] [CrossRef]
- Parajuli, R.P.; Goodrich, J.M.; Chou, H.-N.; Gruninger, S.E.; Dolinoy, D.C.; Franzblau, A.; Basu, N. Genetic Polymorphisms Are Associated with Hair, Blood, and Urine Mercury Levels in the American Dental Association (ADA) Study Participants. Environ. Res. 2016, 149, 247–258. [Google Scholar] [CrossRef]
- Parajuli, R.P. Genetic Polymorphisms Are Associated with Exposure Biomarkers for Metals and Persistent Organic Pollutants among Inuit from the Inuvialuit Settlement Region, Canada. Sci. Total Environ. 2018, 10, 569–578. [Google Scholar] [CrossRef]
- Wahlberg, K.; Love, T.M.; Pineda, D.; Engström, K.; Watson, G.E.; Thurston, S.W.; Yeates, A.J.; Mulhern, M.S.; McSorley, E.M.; Strain, J.J.; et al. Maternal Polymorphisms in Glutathione-Related Genes Are Associated with Maternal Mercury Concentrations and Early Child Neurodevelopment in a Population with a Fish-Rich Diet. Environ. Int. 2018, 115, 142–149. [Google Scholar] [CrossRef]
- Lozano, M.; Murcia, M.; Soler-Blasco, R.; González, L.; Iriarte, G.; Rebagliato, M.; Lopez-Espinosa, M.-J.; Esplugues, A.; Ballester, F.; Llop, S. Exposure to Mercury among 9-Year-Old Children and Neurobehavioural Function. Environ. Int. 2021, 146, 106173. [Google Scholar] [CrossRef]
- Ji, G.; Gu, A.; Zhou, Y.; Shi, X.; Xia, Y.; Long, Y.; Song, L.; Wang, S.; Wang, X. Interactions between Exposure to Environmental Polycyclic Aromatic Hydrocarbons and DNA Repair Gene Polymorphisms on Bulky DNA Adducts in Human Sperm. PLoS ONE 2010, 5, e13145. [Google Scholar] [CrossRef]
- Etemadi, A.; Islami, F.; Phillips, D.H.; Godschalk, R.; Golozar, A.; Kamangar, F.; Malekshah, A.F.-T.; Pourshams, A.; Elahi, S.; Ghojaghi, F.; et al. Variation in PAH-Related DNA Adduct Levels among Non-Smokers: The Role of Multiple Genetic Polymorphisms and Nucleotide Excision Repair Phenotype. Int. J. Cancer 2013, 132, 2738–2747. [Google Scholar] [CrossRef]
- Clarkson, T.W.; Magos, L. The Toxicology of Mercury and Its Chemical Compounds. Crit. Rev. Toxicol. 2006, 36, 609–662. [Google Scholar] [CrossRef]
- Caito, S.W.; Jackson, B.P.; Punshon, T.; Scrimale, T.; Grier, A.; Gill, S.R.; Love, T.M.; Watson, G.E.; van Wijngaarden, E.; Rand, M.D. Editor’s Highlight: Variation in Methylmercury Metabolism and Elimination Status in Humans Following Fish Consumption. Toxicol. Sci. 2018, 161, 443–453. [Google Scholar] [CrossRef]
- Andreoli, V.; Sprovieri, F. Genetic Aspects of Susceptibility to Mercury Toxicity: An Overview. Int. J. Environ. Res. Public. Health 2017, 14, 93. [Google Scholar] [CrossRef]
- Conte, F.; Copat, C.; Longo, S.; Conti, G.O.; Grasso, A.; Arena, G.; Dimartino, A.; Brundo, M.V.; Ferrante, M. Polycyclic Aromatic Hydrocarbons in Haliotis Tuberculata (Linnaeus, 1758) (Mollusca, Gastropoda): Considerations on Food Safety and Source Investigation. Food Chem. Toxicol. 2016, 94, 57–63. [Google Scholar] [CrossRef]
- Rehman, M.Y.A.; Taqi, M.M.; Hussain, I.; Nasir, J.; Rizvi, S.H.H.; Syed, J.H. Elevated Exposure to Polycyclic Aromatic Hydrocarbons (PAHs) May Trigger Cancers in Pakistan: An Environmental, Occupational, and Genetic Perspective. Environ. Sci. Pollut. Res. 2020, 27, 42405–42423. [Google Scholar] [CrossRef]
- Feil, R.; Fraga, M.F. Epigenetics and the Environment: Emerging Patterns and Implications. Nat. Rev. Genet. 2012, 13, 97–109. [Google Scholar] [CrossRef]
- Fagny, M.; Patin, E.; MacIsaac, J.L.; Rotival, M.; Flutre, T.; Jones, M.J.; Siddle, K.J.; Quach, H.; Harmant, C.; McEwen, L.M.; et al. The Epigenomic Landscape of African Rainforest Hunter-Gatherers and Farmers. Nat. Commun. 2015, 6, 10047. [Google Scholar] [CrossRef]
- Natri, H.M.; Bobowik, K.S.; Kusuma, P.; Crenna Darusallam, C.; Jacobs, G.S.; Hudjashov, G.; Lansing, J.S.; Sudoyo, H.; Banovich, N.E.; Cox, M.P.; et al. Genome-Wide DNA Methylation and Gene Expression Patterns Reflect Genetic Ancestry and Environmental Differences across the Indonesian Archipelago. PLoS Genet. 2020, 16, e1008749. [Google Scholar] [CrossRef]
- Carja, O.; MacIsaac, J.L.; Mah, S.M.; Henn, B.M.; Kobor, M.S.; Feldman, M.W.; Fraser, H.B. Worldwide Patterns of Human Epigenetic Variation. Nat. Ecol. Evol. 2017, 1, 1577–1583. [Google Scholar] [CrossRef]
- Hodjat, M.; Rahmani, S.; Khan, F.; Niaz, K.; Navaei–Nigjeh, M.; Mohammadi Nejad, S.; Abdollahi, M. Environmental Toxicants, Incidence of Degenerative Diseases, and Therapies from the Epigenetic Point of View. Arch. Toxicol. 2017, 91, 2577–2597. [Google Scholar] [CrossRef]
- Martin, E.M.; Fry, R.C. Environmental Influences on the Epigenome: Exposure- Associated DNA Methylation in Human Populations. Annu. Rev. Public Health 2018, 39, 309–333. [Google Scholar] [CrossRef]
- Culbreth, M.; Aschner, M. Methylmercury Epigenetics. Toxics 2019, 7, 56. [Google Scholar] [CrossRef]
- Hanna, C.W.; Bloom, M.S.; Robinson, W.P.; Kim, D.; Parsons, P.J.; vom Saal, F.S.; Taylor, J.A.; Steuerwald, A.J.; Fujimoto, V.Y. DNA Methylation Changes in Whole Blood Is Associated with Exposure to the Environmental Contaminants, Mercury, Lead, Cadmium and Bisphenol A, in Women Undergoing Ovarian Stimulation for IVF. Hum. Reprod. 2012, 27, 1401–1410. [Google Scholar] [CrossRef]
- Goodrich, J.M.; Basu, N.; Franzblau, A.; Dolinoy, D.C. Mercury Biomarkers and DNA Methylation among Michigan Dental Professionals: Mercury and DNA Methylation. Environ. Mol. Mutagen. 2013, 54, 195–203. [Google Scholar] [CrossRef]
- Pavanello, S.; Bollati, V.; Pesatori, A.C.; Kapka, L.; Bolognesi, C.; Bertazzi, P.A.; Baccarelli, A. Global and Gene-Specific Promoter Methylation Changes Are Related to Anti -B[ a ]PDE-DNA Adduct Levels and Influence Micronuclei Levels in Polycyclic Aromatic Hydrocarbon-Exposed Individuals. Int. J. Cancer 2009, 125, 1692–1697. [Google Scholar] [CrossRef]
- Yang, P.; Ma, J.; Zhang, B.; Duan, H.; He, Z.; Zeng, J.; Zeng, X.; Li, D.; Wang, Q.; Xiao, Y.; et al. CpG Site–Specific Hypermethylation of P16 INK4α in Peripheral Blood Lymphocytes of PAH-Exposed Workers. Cancer Epidemiol. Biomark. Prev. 2012, 21, 182–190. [Google Scholar] [CrossRef]
- Alegría-Torres, J.A.; Barretta, F.; Batres-Esquivel, L.E.; Carrizales-Yáñez, L.; Pérez-Maldonado, I.N.; Baccarelli, A.; Bertazzi, P.A. Epigenetic Markers of Exposure to Polycyclic Aromatic Hydrocarbons in Mexican Brickmakers: A Pilot Study. Chemosphere 2013, 91, 475–480. [Google Scholar] [CrossRef]
- Bakulski, K.M.; Lee, H.; Feinberg, J.I.; Wells, E.M.; Brown, S.; Herbstman, J.B.; Witter, F.R.; Halden, R.U.; Caldwell, K.; Mortensen, M.E.; et al. Prenatal Mercury Concentration Is Associated with Changes in DNA Methylation at TCEANC2 in Newborns. Int. J. Epidemiol. 2015, 44, 1249–1262. [Google Scholar] [CrossRef]
- Cardenas, A.; Koestler, D.C.; Houseman, E.A.; Jackson, B.P.; Kile, M.L.; Karagas, M.R.; Marsit, C.J. Differential DNA Methylation in Umbilical Cord Blood of Infants Exposed to Mercury and Arsenic in Utero. Epigenetics 2015, 10, 508–515. [Google Scholar] [CrossRef]
- Maccani, J.Z.J.; Koestler, D.C.; Lester, B.; Houseman, E.A.; Armstrong, D.A.; Kelsey, K.T.; Marsit, C.J. Placental DNA Methylation Related to Both Infant Toenail Mercury and Adverse Neurobehavioral Outcomes. Environ. Health Perspect. 2015, 123, 723–729. [Google Scholar] [CrossRef]
- Cardenas, A.; Rifas-Shiman, S.L.; Agha, G.; Hivert, M.-F.; Litonjua, A.A.; DeMeo, D.L.; Lin, X.; Amarasiriwardena, C.J.; Oken, E.; Gillman, M.W.; et al. Persistent DNA Methylation Changes Associated with Prenatal Mercury Exposure and Cognitive Performance during Childhood. Sci. Rep. 2017, 7, 288. [Google Scholar] [CrossRef]
- Cediel Ulloa, A.; Gliga, A.; Love, T.M.; Pineda, D.; Mruzek, D.W.; Watson, G.E.; Davidson, P.W.; Shamlaye, C.F.; Strain, J.J.; Myers, G.J.; et al. Prenatal Methylmercury Exposure and DNA Methylation in Seven-Year-Old Children in the Seychelles Child Development Study. Environ. Int. 2021, 147, 106321. [Google Scholar] [CrossRef]
- Perera, F.; Tang, W.; Herbstman, J.; Tang, D.; Levin, L.; Miller, R.; Ho, S. Relation of DNA Methylation of 5′-CpG Island of ACSL3 to Transplacental Exposure to Airborne Polycyclic Aromatic Hydrocarbons and Childhood Asthma. PLoS ONE 2009, 4, e4488. [Google Scholar] [CrossRef]
- Herbstman, J.B.; Tang, D.; Zhu, D.; Qu, L.; Sjödin, A.; Li, Z.; Camann, D.; Perera, F.P. Prenatal Exposure to Polycyclic Aromatic Hydrocarbons, Benzo[ a ]Pyrene–DNA Adducts, and Genomic DNA Methylation in Cord Blood. Environ. Health Perspect. 2012, 120, 733–738. [Google Scholar] [CrossRef]
- Kim, Y.H.; Lee, Y.S.; Lee, D.H.; Kim, D.S. Polycyclic Aromatic Hydrocarbons Are Associated with Insulin Receptor Substrate 2 Methylation in Adipose Tissues of Korean Women. Environ. Res. 2016, 150, 47–51. [Google Scholar] [CrossRef]
- Lee, J.; Kalia, V.; Perera, F.; Herbstman, J.; Li, T.; Nie, J.; Qu, L.R.; Yu, J.; Tang, D. Prenatal Airborne Polycyclic Aromatic Hydrocarbon Exposure, LINE1 Methylation and Child Development in a Chinese Cohort. Environ. Int. 2017, 99, 315–320. [Google Scholar] [CrossRef]
- Li, J.; Zhu, X.; Yu, K.; Jiang, H.; Zhang, Y.; Wang, B.; Liu, X.; Deng, S.; Hu, J.; Deng, Q.; et al. Exposure to Polycyclic Aromatic Hydrocarbons and Accelerated DNA Methylation Aging. Environ. Health Perspect. 2018, 126, 067005. [Google Scholar] [CrossRef]
- Lin, S.; Ren, A.; Wang, L.; Santos, C.; Huang, Y.; Jin, L.; Li, Z.; Greene, N.D.E. Aberrant Methylation of Pax3 Gene and Neural Tube Defects in Association with Exposure to Polycyclic Aromatic Hydrocarbons. Clin. Epigenetics 2019, 11, 13. [Google Scholar] [CrossRef]
- Liu, K.; Jiang, J.; Lin, Y.; Liu, W.; Zhu, X.; Zhang, Y.; Jiang, H.; Yu, K.; Liu, X.; Zhou, M.; et al. Exposure to Polycyclic Aromatic Hydrocarbons, DNA Methylation and Heart Rate Variability among Non-Current Smokers. Environ. Pollut. 2021, 288, 117777. [Google Scholar] [CrossRef]
- Onishchenko, N.; Karpova, N.; Sabri, F.; Castrn, E.; Ceccatelli, S. Long-Lasting Depression-like Behavior and Epigenetic Changes of BDNF Gene Expression Induced by Perinatal Exposure to Methylmercury. J. Neurochem. 2008, 106, 1378–1387. [Google Scholar] [CrossRef]
- Grandjean, P.; Weihe, P.; White, R.F.; Debes, F.; Araki, S.; Yokoyama, K.; Murata, K.; Sørensen, N.; Dahl, R.; Jørgensen, P.J. Cognitive Deficit in 7-Year-Old Children with Prenatal Exposure to Methylmercury. Neurotoxicol. Teratol. 1997, 19, 417–428. [Google Scholar] [CrossRef]
- Axelrad, D.A.; Bellinger, D.C.; Ryan, L.M.; Woodruff, T.J. Dose–Response Relationship of Prenatal Mercury Exposure and IQ: An Integrative Analysis of Epidemiologic Data. Environ. Health Perspect. 2007, 115, 609–615. [Google Scholar] [CrossRef]
- Yokoo, E.M.; Valente, J.G.; Grattan, L.; Schmidt, S.L.; Platt, I.; Silbergeld, E.K. Low Level Methylmercury Exposure Affects Neuropsychological Function in Adults. Environ. Health 2003, 2, 8. [Google Scholar] [CrossRef]
- Akpambang, V.; Purcaro, G.; Lajide, L.; Amoo, I.; Conte, L.; Moret, S. Determination of Polycyclic Aromatic Hydrocarbons (PAHs) in Commonly Consumed Nigerian Smoked/Grilled Fish and Meat. Food Addit. Contam. 2009, 22, 1096–1103. [Google Scholar] [CrossRef]
- Forsberg, N.D.; Stone, D.; Harding, A.; Harper, B.; Harris, S.; Matzke, M.M.; Cardenas, A.; Waters, K.M.; Anderson, K.A. Effect of Native American Fish Smoking Methods on Dietary Exposure to Polycyclic Aromatic Hydrocarbons and Possible Risks to Human Health. J. Agric. Food Chem. 2012, 60, 6899–6906. [Google Scholar] [CrossRef]
- Sholts, S.B.; Smith, K.; Wallin, C.; Ahmed, T.M.; Wärmländer, S.K.T.S. Ancient Water Bottle Use and Polycyclic Aromatic Hydrocarbon (PAH) Exposure among California Indians: A Prehistoric Health Risk Assessment. Environ. Health 2017, 16, 61. [Google Scholar] [CrossRef]
- Schlebusch, C.M.; Gattepaille, L.M.; Engström, K.; Vahter, M.; Jakobsson, M.; Broberg, K. Human Adaptation to Arsenic-Rich Environments. Mol. Biol. Evol. 2015, 32, 1544–1555. [Google Scholar] [CrossRef]
- Arriaza, B.; Amarasiriwardena, D.; Cornejo, L.; Standen, V.; Byrne, S.; Bartkus, L.; Bandak, B. Exploring Chronic Arsenic Poisoning in Pre-Columbian Chilean Mummies. J. Archaeol. Sci. 2010, 37, 1274–1278. [Google Scholar] [CrossRef]
- Apata, M.; Pfeifer, S.P. Recent Population Genomic Insights into the Genetic Basis of Arsenic Tolerance in Humans: The Difficulties of Identifying Positively Selected Loci in Strongly Bottlenecked Populations. Heredity 2020, 124, 253–262. [Google Scholar] [CrossRef]
- Apata, M.; Arriaza, B.; Llop, E.; Moraga, M. Human Adaptation to Arsenic in Andean Populations of the Atacama Desert. Am. J. Phys. Anthropol. 2017, 163, 192–199. [Google Scholar] [CrossRef]
- Hubbard, T.D.; Murray, I.A.; Bisson, W.H.; Sullivan, A.P.; Sebastian, A.; Perry, G.H.; Jablonski, N.G.; Perdew, G.H. Divergent Ah Receptor Ligand Selectivity during Hominin Evolution. Mol. Biol. Evol. 2016, 33, 2648–2658. [Google Scholar] [CrossRef]
- Heyn, H.; Moran, S.; Hernando-Herraez, I.; Sayols, S.; Gomez, A.; Sandoval, J.; Monk, D.; Hata, K.; Marques-Bonet, T.; Wang, L.; et al. DNA Methylation Contributes to Natural Human Variation. Genome Res. 2013, 23, 1363–1372. [Google Scholar] [CrossRef]
- Giuliani, C.; Biggs, D.; Nguyen, T.T.; Marasco, E.; De Fanti, S.; Garagnani, P.; Le Phan, M.T.; Nguyen, V.N.; Luiselli, D.; Romeo, G. First Evidence of Association between Past Environmental Exposure to Dioxin and DNA Methylation of CYP1A1 and IGF2 Genes in Present Day Vietnamese Population. Environ. Pollut. 2018, 242, 976–985. [Google Scholar] [CrossRef]
- Droghini, E.; Annibaldi, A.; Prezioso, E.; Tramontana, M.; Frapiccini, E.; De Marco, R.; Illuminati, S.; Truzzi, C.; Spagnoli, F. Mercury Content in Central and Southern Adriatic Sea Sediments in Relation to Seafloor Geochemistry and Sedimentology. Molecules 2019, 24, 4467. [Google Scholar] [CrossRef]
- Koenig, S.; Solé, M.; Fernández-Gómez, C.; Díez, S. New Insights into Mercury Bioaccumulation in Deep-Sea Organisms from the NW Mediterranean and Their Human Health Implications. Sci. Total Environ. 2013, 442, 329–335. [Google Scholar] [CrossRef]
- Cammilleri, G.; Vazzana, M.; Arizza, V.; Giunta, F.; Vella, A.; Lo Dico, G.; Giaccone, V.; Giofrè, S.V.; Giangrosso, G.; Cicero, N.; et al. Mercury in Fish Products: What’s the Best for Consumers between Bluefin Tuna and Yellowfin Tuna? Nat. Prod. Res. 2018, 32, 457–462. [Google Scholar] [CrossRef]
- Cinnirella, S.; Bruno, D.E.; Pirrone, N.; Horvat, M.; Živković, I.; Evers, D.C.; Johnson, S.; Sunderland, E.M. Mercury Concentrations in Biota in the Mediterranean Sea, a Compilation of 40 Years of Surveys. Sci. Data 2019, 6, 205. [Google Scholar] [CrossRef]
- Gallo, P.; De Carlo, E.; Marigliano, L.; Maglio, P.; Amato, A.; Improta, A.; Caruso, C.; De Roma, A. Food Safety Assessment of Heavy Metals in Uncommon and Abyssal Fish and Cephalopod from the Tyrrhenian Sea. J. Consum. Prot. Food Saf. 2018, 13, 399–402. [Google Scholar] [CrossRef]
- Annibaldi, A.; Truzzi, C.; Carnevali, O.; Pignalosa, P.; Api, M.; Scarponi, G.; Illuminati, S. Determination of Hg in Farmed and Wild Atlantic Bluefin Tuna (Thunnus Thynnus L.) Muscle. Molecules 2019, 24, 1273. [Google Scholar] [CrossRef]
- Copat, C.; Conti, G.O.; Fallico, R.; Sciacca, S.; Ferrante, M. Heavy Metals in Fish from the Mediterranean Sea. In The Mediterranean Diet; Elsevier: Amsterdam, The Netherlands, 2015; pp. 547–562. ISBN 978-0-12-407849-9. [Google Scholar]
- Perugini, M.; Visciano, P.; Manera, M.; Abete, M.C.; Gavinelli, S.; Amorena, M. Contamination of Different Portions of Raw and Boiled Specimens of Norway Lobster by Mercury and Selenium. Environ. Sci. Pollut. Res. 2013, 20, 8255–8262. [Google Scholar] [CrossRef]
- Perugini, M.; Visciano, P.; Manera, M.; Zaccaroni, A.; Olivieri, V.; Amorena, M. Heavy Metal (As, Cd, Hg, Pb, Cu, Zn, Se) Concentrations in Muscle and Bone of Four Commercial Fish Caught in the Central Adriatic Sea, Italy. Environ. Monit. Assess. 2014, 186, 2205–2213. [Google Scholar] [CrossRef]
- Perugini, M.; Zezza, D.; Tulini, S.M.R.; Abete, M.C.; Monaco, G.; Conte, A.; Olivieri, V.; Amorena, M. Effect of Cooking on Total Mercury Content in Norway Lobster and European Hake and Public Health Impact. Mar. Pollut. Bull. 2016, 109, 521–525. [Google Scholar] [CrossRef]
- Storelli, M.M.; Barone, G. Toxic Metals (Hg, Pb, and Cd) in Commercially Important Demersal Fish from Mediterranean Sea: Contamination Levels and Dietary Exposure Assessment: Toxic Metals in Fish. J. Food Sci. 2013, 78, T362–T366. [Google Scholar] [CrossRef]
- Di Lena, G.; Casini, I.; Caproni, R.; Orban, E. Total Mercury Levels in Crustacean Species from Italian Fishery. Food Addit. Contam. Part B 2018, 11, 175–182. [Google Scholar] [CrossRef]
- Bajt, O.; Ramšak, A.; Milun, V.; Andral, B.; Romanelli, G.; Scarpato, A.; Mitrić, M.; Kupusović, T.; Kljajić, Z.; Angelidis, M.; et al. Assessing Chemical Contamination in the Coastal Waters of the Adriatic Sea Using Active Mussel Biomonitoring with Mytilus Galloprovincialis. Mar. Pollut. Bull. 2019, 141, 283–298. [Google Scholar] [CrossRef]
- Di Lena, G.; Casini, I.; Caproni, R.; Fusari, A.; Orban, E. Total Mercury Levels in Commercial Fish Species from Italian Fishery and Aquaculture. Food Addit. Contam. Part B 2017, 10, 118–127. [Google Scholar] [CrossRef]
- Galgani, F.; Martínez-Gómez, C.; Giovanardi, F.; Romanelli, G.; Caixach, J.; Cento, A.; Scarpato, A.; BenBrahim, S.; Messaoudi, S.; Deudero, S.; et al. Assessment of Polycyclic Aromatic Hydrocarbon Concentrations in Mussels (Mytilus Galloprovincialis) from the Western Basin of the Mediterranean Sea. Environ. Monit. Assess. 2011, 172, 301–317. [Google Scholar] [CrossRef]
- Arienzo, M.; Toscanesi, M.; Trifuoggi, M.; Ferrara, L.; Stanislao, C.; Donadio, C.; Grazia, V.; Gionata, D.V.; Carella, F. Contaminants Bioaccumulation and Pathological Assessment in Mytilus Galloprovincialis in Coastal Waters Facing the Brownfield Site of Bagnoli, Italy. Mar. Pollut. Bull. 2019, 140, 341–352. [Google Scholar] [CrossRef]
- Conti, G.O.; Copat, C.; Ledda, C.; Fiore, M.; Fallico, R.; Sciacca, S.; Ferrante, M. Evaluation of Heavy Metals and Polycyclic Aromatic Hydrocarbons (PAHs) in Mullus Barbatus from Sicily Channel and Risk-Based Consumption Limits. Bull. Environ. Contam. Toxicol. 2012, 88, 946–950. [Google Scholar] [CrossRef]
- US-EPA Guidance for Assessing Chemical Contamination Data for Use in Fish Advisories, Vol. II. Risk Assessment and Fish Consumption Limits EPA/823-B94-004 2000. Available online: https://www.epa.gov/sites/default/files/2015-06/documents/volume2.pdf (accessed on 12 October 2021).
- Copat, C.; Vinceti, M.; D’Agati, M.G.; Arena, G.; Mauceri, V.; Grasso, A.; Fallico, R.; Sciacca, S.; Ferrante, M. Mercury and Selenium Intake by Seafood from the Ionian Sea: A Risk Evaluation. Ecotoxicol. Environ. Saf. 2014, 100, 87–92. [Google Scholar] [CrossRef]
- Barone, G.; Storelli, A.; Garofalo, R.; Busco, V.P.; Quaglia, N.C.; Centrone, G.; Storelli, M.M. Assessment of Mercury and Cadmium via Seafood Consumption in Italy: Estimated Dietary Intake (EWI) and Target Hazard Quotient (THQ). Food Addit. Contam. Part A 2015, 32, 1277–1286. [Google Scholar] [CrossRef]
- Díez, S.; Montuori, P.; Pagano, A.; Sarnacchiaro, P.; Bayona, J.M.; Triassi, M. Hair Mercury Levels in an Urban Population from Southern Italy: Fish Consumption as a Determinant of Exposure. Environ. Int. 2008, 6, 162–167. [Google Scholar] [CrossRef]
- Den Hond, E.; Govarts, E.; Willems, H.; Smolders, R.; Casteleyn, L.; Kolossa-Gehring, M.; Schwedler, G.; Seiwert, M.; Fiddicke, U.; Castaño, A.; et al. First Steps toward Harmonized Human Biomonitoring in Europe: Demonstration Project to Perform Human Biomonitoring on a European Scale. Environ. Health Perspect. 2015, 123, 255–263. [Google Scholar] [CrossRef]
- EUMOFA The EU Fish Market. European Market Observatory for Fisheries and Aquaculture Products. European Commission, Directorate-General for Maritime Affairs and Fisheries, Director-General, 2018. Available online: http://agricultura.gencat.cat/web/.content/de_departament/de02_estadistiques_observatoris/27_butlletins/02_butlletins_nd/documents_nd/fitxers_estatics_nd/2018/0218_2018_Pesca_Productes-pesquers-mercats-UE-2018-exportacions.pdf (accessed on 13 October 2021). [CrossRef]
- Sekovanic, A.; Piasek, M.; Orct, T.; Grgec, A.S.; Saric, M.M.; Stasenko, S.; Jurasovic, J. Mercury Exposure Assessment in Mother–Infant Pairs from Continental and Coastal Croatia. Biomolecules 2020, 27, 821. [Google Scholar] [CrossRef]
- Elhamri, H.; Idrissi, L.; Coquery, M.; Azemard, S.; Abidi, A.E.; Benlemlih, M.; Saghi, M.; Cubadda, F. Hair Mercury Levels in Relation to Fish Consumption in a Community of the Moroccan Mediterranean Coast. Food Addit. Contam. 2007, 24, 1236–1246. [Google Scholar] [CrossRef][Green Version]
- Giangrosso, G.; Cammilleri, G.; Macaluso, A.; Vella, A.; D’Orazio, N.; Graci, S.; Lo Dico, G.M.; Galvano, F.; Giangrosso, M.; Ferrantelli, V. Hair Mercury Levels Detection in Fishermen from Sicily (Italy) by ICP-MS Method after Microwave-Assisted Digestion. Bioinorg. Chem. Appl. 2016, 2016, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Barbone, F.; Valent, F.; Pisa, F.; Daris, F.; Fajon, V.; Logar, M.; Horvat, M. Prenatal low-level methyl mercury exposure and child development in an Italian coastal area. Seychelles Med Dent. J. 2004, 7, 149–154. [Google Scholar] [CrossRef]
- Giuliani, C.; Bacalini, M.G.; Sazzini, M.; Pirazzini, C.; Franceschi, C.; Garagnani, P.; Luiselli, D. The Epigenetic Side of Human Adaptation: Hypotheses, Evidences and Theories. Ann. Hum. Biol. 2015, 42, 1–9. [Google Scholar] [CrossRef]
- Lam, L.L.; Emberly, E.; Fraser, H.B.; Neumann, S.M.; Chen, E.; Miller, G.E.; Kobor, M.S. Factors Underlying Variable DNA Methylation in a Human Community Cohort. Proc. Natl. Acad. Sci. USA 2012, 109, 17253–17260. [Google Scholar] [CrossRef]
- Bosch, A.C.; O’Neill, B.; Sigge, G.O.; Kerwath, S.E.; Hoffman, L.C. Heavy Metals in Marine Fish Meat and Consumer Health: A Review: Heavy Metals in Marine Fish Meat. J. Sci. Food Agric. 2016, 96, 32–48. [Google Scholar] [CrossRef]
- Maggi, C.; Berducci, M.T.; Di Lorenzo, B.; Dattolo, M.; Cozzolino, A.; Mariotti, S.; Fabrizi, V.; Spaziani, R.; Virno Lamberti, C. Temporal Evolution of the Environmental Quality of the Vallona Lagoon (Northern Mediterranean, Adriatic Sea). Mar. Pollut. Bull. 2017, 125, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Balali-Mood, M.; Naseri, K.; Tahergorabi, Z.; Khazdair, M.R.; Sadeghi, M. Toxic Mechanisms of Five Heavy Metals: Mercury, Lead, Chromium, Cadmium, and Arsenic. Front. Pharmacol. 2021, 12, 643972. [Google Scholar] [CrossRef]
- Alegría-Torres, J.A.; Carrizales-Yánez, L.; Díaz-Barriga, F.; Rosso-Camacho, F.; Motta, V.; Tarantini, L.; Bollati, V. DNA Methylation Changes in Mexican Children Exposed to Arsenic from Two Historic Mining Areas in San Luis Potosí: DNA Methylation Changes. Environ. Mol. Mutagen. 2016, 57, 717–723. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.K.; Paul, S.; Adak, S.; Giri, A.K. Reduced LINE-1 Methylation Is Associated with Arsenic-Induced Genotoxic Stress in Children. BioMetals 2016, 29, 731–741. [Google Scholar] [CrossRef]
- Lambrou, A.; Baccarelli, A.; Wright, R.O.; Weisskopf, M.; Bollati, V.; Amarasiriwardena, C.; Vokonas, P.; Schwartz, J. Arsenic Exposure and DNA Methylation among Elderly Men. Epidemiology 2012, 23, 668–676. [Google Scholar] [CrossRef]
- Hossain, M.B.; Vahter, M.; Concha, G.; Broberg, K. Low-Level Environmental Cadmium Exposure Is Associated with DNA Hypomethylation in Argentinean Women. Environ. Health Perspect. 2012, 120, 879–884. [Google Scholar] [CrossRef]
- Li, C.; Yang, X.; Xu, M.; Zhang, J.; Sun, N. Epigenetic Marker (LINE-1 Promoter) Methylation Level Was Associated with Occupational Lead Exposure. Clin. Toxicol. 2013, 51, 225–229. [Google Scholar] [CrossRef]
- Non, A.L.; Thayer, Z.M. Epigenetics for Anthropologists: An Introduction to Methods: Epigenetic Methods for Anthropologists. Am. J. Hum. Biol. 2015, 27, 295–303. [Google Scholar] [CrossRef]
- Samson, C.A.; Whitford, W.; Snell, R.G.; Jacobsen, J.C.; Lehnert, K. Contaminating DNA in Human Saliva Alters the Detection of Variants from Whole Genome Sequencing. Sci. Rep. 2020, 10, 19255. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-C.; Wang, Q.; Chung, W.K.; Andrulis, I.L.; Daly, M.B.; John, E.M.; Keegan, T.H.; Knight, J.; Bradbury, A.R.; Kappil, M.A.; et al. Correlation of DNA Methylation Levels in Blood and Saliva DNA in Young Girls of the LEGACY Girls Study. Epigenetics 2014, 9, 929–933. [Google Scholar] [CrossRef]
- Langie, S.A.S.; Szarc vel Szic, K.; Declerck, K.; Traen, S.; Koppen, G.; Van Camp, G.; Schoeters, G.; Vanden Berghe, W.; De Boever, P. Whole-Genome Saliva and Blood DNA Methylation Profiling in Individuals with a Respiratory Allergy. PLoS ONE 2016, 11, e0151109. [Google Scholar] [CrossRef]
- Murata, Y.; Fujii, A.; Kanata, S.; Fujikawa, S.; Ikegame, T.; Nakachi, Y.; Zhao, Z.; Jinde, S.; Kasai, K.; Bundo, M.; et al. Evaluation of the Usefulness of Saliva for DNA Methylation Analysis in Cohort Studies. Neuropsychopharmacol. Rep. 2019, 39, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Braun, P.R.; Han, S.; Hing, B.; Nagahama, Y.; Gaul, L.N.; Heinzman, J.T.; Grossbach, A.J.; Close, L.; Dlouhy, B.J.; Howard, M.A.; et al. Genome-Wide DNA Methylation Comparison between Live Human Brain and Peripheral Tissues within Individuals. Transl. Psychiatry 2019, 9, 47. [Google Scholar] [CrossRef]
- Van Dongen, J.; Ehli, E.A.; Jansen, R.; van Beijsterveldt, C.E.M.; Willemsen, G.; Hottenga, J.J.; Kallsen, N.A.; Peyton, S.A.; Breeze, C.E.; Kluft, C.; et al. Genome-Wide Analysis of DNA Methylation in Buccal Cells: A Study of Monozygotic Twins and MQTLs. Epigenetics Chromatin 2018, 11, 54. [Google Scholar] [CrossRef] [PubMed]
- McGregor, K.; Bernatsky, S.; Colmegna, I.; Hudson, M.; Pastinen, T.; Labbe, A.; Greenwood, C.M.T. An Evaluation of Methods Correcting for Cell-Type Heterogeneity in DNA Methylation Studies. Genome Biol. 2016, 17, 84. [Google Scholar] [CrossRef]
- Narváez, D.M.; Groot, H.; Diaz, S.M.; Palma, R.M.; Muñoz, N.; Cros, M.-P.; Hernández-Vargas, H. Oxidative Stress and Repetitive Element Methylation Changes in Artisanal Gold Miners Occupationally Exposed to Mercury. Heliyon 2017, 3, e00400. [Google Scholar] [CrossRef] [PubMed]
- Olsson, A.C.; Fevotte, J.; Fletcher, T.; Cassidy, A.; ’t Mannetje, A.; Zaridze, D.; Szeszenia-Dabrowska, N.; Rudnai, P.; Lissowska, J.; Fabianova, E.; et al. Occupational Exposure to Polycyclic Aromatic Hydrocarbons and Lung Cancer Risk: A Multicenter Study in Europe. Occup. Environ. Med. 2010, 67, 98–103. [Google Scholar] [CrossRef]
- Patel, A.B.; Shaikh, S.; Jain, K.R.; Desai, C.; Madamwar, D. Polycyclic Aromatic Hydrocarbons: Sources, Toxicity, and Remediation Approaches. Front. Microbiol. 2020, 11, 562813. [Google Scholar] [CrossRef]
- Liu, Y.; Li, X.; Zhang, B.; Fu, Y.; Yang, A.; Zhang, H.; Zhang, H.; Niu, Y.; Nie, J.; Yang, J. CYP1A1 Methylation Mediates the Effect of Smoking and Occupational Polycyclic Aromatic Hydrocarbons Co-Exposure on Oxidative DNA Damage among Chinese Coke-Oven Workers. Environ. Health 2019, 18, 69. [Google Scholar] [CrossRef]
- Khansakorn, N.; Wongwit, W.; Tharnpoophasiam, P.; Hengprasith, B.; Suwannathon, L.; Chanprasertyothin, S.; Sura, T.; Kaojarern, S.; Sritara, P.; Sirivarasai, J. Genetic Variations of Glutathione S-Transferase Influence on Blood Cadmium Concentration. J. Toxicol. 2012, 2012, 1–6. [Google Scholar] [CrossRef]
- Chen, H.-I.; Chiu, Y.-W.; Hsu, Y.K.; Li, W.-F.; Chen, Y.-C.; Chuang, H.-Y. The Association of Metallothionein-4 Gene Polymorphism and Renal Function in Long-Term Lead-Exposed Workers. Biol. Trace Elem. Res. 2010, 137, 55–62. [Google Scholar] [CrossRef]
- Nariya, A.; Pathan, A.; Shah, N.; Patel, A.; Chettiar, S.; Vyas, J.; Shaikh, I.; Jhala, D. Association of Delta-Aminolevulinic Acid Dehydratase Polymorphism with Blood Lead and Hemoglobin Level in Lead Exposed Workers. Annu. Res. Rev. Biol. 2017, 17, 1–7. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, J.; Sun, P. Effects of Delta-Aminolevulinic Acid Dehydratase Polymorphisms on Susceptibility to Lead in Han Subjects from Southwestern China. Int. J. Environ. Res. Public. Health 2012, 9, 2326–2338. [Google Scholar] [CrossRef] [PubMed]
- Miniero, R.; Beccaloni, E.; Carere, M.; Ubaldi, A.; Mancini, L.; Marchegiani, S.; Cicero, M.R.; Scenati, R.; Lucchetti, D.; Ziemacki, G.; et al. Mercury (Hg) and Methyl Mercury (MeHg) Concentrations in Fish from the Coastal Lagoon of Orbetello, Central Italy. Mar. Pollut. Bull. 2013, 76, 365–369. [Google Scholar] [CrossRef]
- Storelli, M.M.; Giacominelli-Stuffler, R.; Storelli, A.; D’Addabbo, R.; Palermo, C.; Marcotrigiano, G.O. Survey of Total Mercury and Methylmercury Levels in Edible Fish from the Adriatic Sea. Food Addit. Contam. 2003, 20, 1114–1119. [Google Scholar] [CrossRef]
- Saavedra, Y.; González, A.; Fernández, P.; Blanco, J. The Effect of Size on Trace Metal Levels in Raft Cultivated Mussels (Mytilus Galloprovincialis). Sci. Total Environ. 2004, 318, 115–124. [Google Scholar] [CrossRef]
- Puel, D.; Zsürger, N.; Breittmayer, J.P.H. Statistical Assessment of a Sampling Pattern for Evaluation of Changes in Mercury and Zinc Concentrations InPatella Coerulea. Bull. Environ. Contam. Toxicol. 1987, 38, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Vives, I.; Grimalt, J.; Fernandez, P.; Rosseland, B. Polycyclic Aromatic Hydrocarbons in Fish from Remote and High Mountain Lakes in Europe and Greenland. Sci. Total Environ. 2004, 324, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Perugini, M.; Visciano, P.; Giammarino, A.; Manera, M.; Di Nardo, W.; Amorena, M. Polycyclic Aromatic Hydrocarbons in Marine Organisms from the Adriatic Sea, Italy. Chemosphere 2007, 66, 1904–1910. [Google Scholar] [CrossRef] [PubMed]
- Storelli, M.M.; Barone, G.; Perrone, V.G.; Storelli, A. Risk Characterization for Polycyclic Aromatic Hydrocarbons and Toxic Metals Associated with Fish Consumption. J. Food Compos. Anal. 2013, 31, 115–119. [Google Scholar] [CrossRef]
- Rahmanpour, S.; Farzaneh Ghorghani, N.; Lotfi Ashtiyani, S.M. Polycyclic Aromatic Hydrocarbon (PAH) in Four Fish Species from Different Trophic Levels in the Persian Gulf. Environ. Monit. Assess. 2014, 186, 7047–7053. [Google Scholar] [CrossRef] [PubMed]
- Vuorinen, P.J.; Keinänen, M.; Vuontisjärvi, H.; Baršienė, J.; Broeg, K.; Förlin, L.; Gercken, J.; Kopecka, J.; Köhler, A.; Parkkonen, J.; et al. Use of Biliary PAH Metabolites as a Biomarker of Pollution in Fish from the Baltic Sea. Mar. Pollut. Bull. 2006, 53, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Piccardo, M.T.; Coradeghini, R.; Valerio, F. Polycyclic Aromatic Hydrocarbon Pollution in Native and Caged Mussels. Mar. Pollut. Bull. 2001, 42, 951–956. [Google Scholar] [CrossRef]
- Perugini, M.; Visciano, P.; Manera, M.; Turno, G.; Lucisano, A.; Amorena, M. Polycyclic Aromatic Hydrocarbons in Marine Organisms from the Gulf of Naples, Tyrrhenian Sea. J. Agric. Food Chem. 2007, 55, 2049–2054. [Google Scholar] [CrossRef]
- Guerranti, C.; Grazioli, E.; Focardi, S.; Renzi, M.; Perra, G. Levels of Chemicals in Two Fish Species from Four Italian Fishing Areas. Mar. Pollut. Bull. 2016, 111, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Bajc, Z.; Kirbiš, A. Trace Element Concentrations in Mussels (Mytilus Galloprovincialis) from the Gulf of Trieste, Slovenia. J. Food Prot. 2019, 82, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Noisel, N.; Bouchard, M.; Carrier, G.; Plante, M. Comparison of a Toxicokinetic and a Questionnaire-Based Approach to Assess Methylmercury Intake in Exposed Individuals. J. Expo. Sci. Environ. Epidemiol. 2011, 21, 328–335. [Google Scholar] [CrossRef]
- Branco, V.; Caito, S.; Farina, M.; Teixeira da Rocha, J.; Aschner, M.; Carvalho, C. Biomarkers of Mercury Toxicity: Past, Present, and Future Trends. J. Toxicol. Environ. Health Part B 2017, 20, 119–154. [Google Scholar] [CrossRef] [PubMed]
- Kusanagi, E.; Takamura, H.; Chen, S.-J.; Adachi, M.; Hoshi, N. Children’s Hair Mercury Concentrations and Seafood Consumption in Five Regions of Japan. Arch. Environ. Contam. Toxicol. 2018, 74, 259–272. [Google Scholar] [CrossRef]
- Suzuki, K.; Yoshinaga, J. Inhalation and Dietary Exposure to Polycyclic Aromatic Hydrocarbons and Urinary 1-Hydroxypyrene in Non-Smoking University Students. Int. Arch. Occup. Environ. Health 2007, 81, 115–121. [Google Scholar] [CrossRef]
- Domingo, J.L.; Nadal, M. Human Dietary Exposure to Polycyclic Aromatic Hydrocarbons: A Review of the Scientific Literature. Food Chem. Toxicol. 2015, 86, 144–153. [Google Scholar] [CrossRef]
- Doddamani, A.; Ballala, A.B.K.; Madhyastha, S.P.; Kamath, A.; Kulkarni, M.M. A Cross-Sectional Study to Identify the Determinants of Non-Communicable Diseases among Fishermen in Southern India. BMC Public Health 2021, 21, 414. [Google Scholar] [CrossRef]
- Zytoon, M.A. Occupational Injuries and Health Problems in the Egyptian Mediterranean Fisheries. Saf. Sci. 2012, 50, 113–122. [Google Scholar] [CrossRef]
- Doza, S.; Bovbjerg, V.E.; Vaughan, A.; Nahorniak, J.S.; Case, S.; Kincl, L.D. Health-Related Exposures and Conditions among US Fishermen. J. Agromedicine 2021, 1–8. [Google Scholar] [CrossRef]
- Wu, Y.; Li, S.; Hu, K.; Yang, J. Evidence of the Moderating Role of Hair Cortisol and Hair Cortisone in the Relationship between Work Stress and Depression Symptoms among Chinese Fishermen. J. Affect. Disord. 2021, 294, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Skuladottir, G.V.; Nilsson, E.K.; Mwinyi, J.; Schiöth, H.B. One-Night Sleep Deprivation Induces Changes in the DNA Methylation and Serum Activity Indices of Stearoyl-CoA Desaturase in Young Healthy Men. Lipids Health Dis. 2016, 15, 137. [Google Scholar] [CrossRef]
- Matosin, N.; Cruceanu, C.; Binder, E.B. Preclinical and Clinical Evidence of DNA Methylation Changes in Response to Trauma and Chronic Stress. Chronic Stress 2017, 1, 247054701771076. [Google Scholar] [CrossRef] [PubMed]
- Mahna, D.; Puri, S.; Sharma, S. DNA Methylation Signatures: Biomarkers of Drug and Alcohol Abuse. Mutat. Res. Mutat. Res. 2018, 777, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Christiansen, C.; Castillo-Fernandez, J.E.; Domingo-Relloso, A.; Zhao, W.; El-Sayed Moustafa, J.S.; Tsai, P.-C.; Maddock, J.; Haack, K.; Cole, S.A.; Kardia, S.L.R.; et al. Novel DNA Methylation Signatures of Tobacco Smoking with Trans-Ethnic Effects. Clin. Epigenetics 2021, 13, 36. [Google Scholar] [CrossRef]


| Study | Genes | Biomarker | Sample |
|---|---|---|---|
| [50] | GCLC; GSTP1 | Erythrocyte Hg | Swedish cases of acute myocardial infarction/stroke and controls |
| [51] | GSTM1; GSTT1 | Hair Hg | Students in Austria |
| [52] | GCLM; GSTP1 | Erythrocyte Hg | Fish-eating Swedish individuals |
| [53] | GSTP1; MT4; GSTM1; GCLC; GSTT1 | Blood and hair Hg | Students in Austria |
| [54] | GSTM1; GSTT1 | Maternal and cord blood Hg | Korean mothers and their infants |
| [55] | GSTT1; GSS; GSTP1; SEPP1 | Hair and urinary Hg | Michigan dental professionals |
| [56] | MT1M; MT2A; MT1A | Hair and urinary Hg | Michigan dental professionals |
| [57] | APOE | Cord blood Hg | Children in Taiwan |
| [58] | GCLM; GSTM1 | Blood and hair Hg | Amazonian population in Brazil cronically exposed to MeHg from fish |
| [59] | TF | Umbilical cord Hg | Children from Bristol |
| [60] | GCLC; GCLM; GSTM1 | Plasmatic Hg and MeHg; whole blood Hg | Fish-eating communities of Brazilian Amazon |
| [61] | ABCB1; ABCC1; ABCC2 | Cord blood Hg | Pregnant women from Greece, Italy and Spain |
| [62] | APOE | Cord blood Hg | Children in Taiwan |
| [63] | ABCB1; ABCC1; ABCC2 | Hair Hg | Seychellois mother–child pairs with a diet rich in fish of mixed African, European and East Asian origin |
| [64] | GLRX2; GSTA4; GSTM3; GSTO1; SELS; MT1M; (see Table S1 for the whole list) | Blood and urinary Hg | American dental professionals |
| [65] | CBS; TXNRD2; SEPHS2; CYP1A2; CBS; MTRR; (see Table S1 for the whole list) | Blood Hg | Inuit from Canada |
| [66] | GCLC; GCLM; GSTP1 | Maternal blood and hair Hg and cord blood Hg | Seychellois mother–child pairs with a diet rich in fish of mixed African, European and East Asian origin |
| [67] | BDNF; GSTP1 | Hair Hg | Children in Valentia |
| Study | Genes | Biomarker | Sample |
|---|---|---|---|
| [68] | XRCC1 | Sperm PAH–DNA adducts | Infertile adult men from Nanjing, China |
| [69] | MPO; NAT2; ERCC5 | Blood PAH–DNA adducts | Non-smoking healthy women from eastern Golestan Province, Iran |
| [36] | CYP1A1; GSTT2; CYP1B | Cord blood B(a)P–DNA adducts | Mother–infant pairs from Krakow, Poland |
| Study | Genes | Biomarker | Tissue (DNA) | Sample |
|---|---|---|---|---|
| [82] | GSTM1 | Whole blood Hg | Whole blood | Women from San Francisco |
| [83] | SEPP1 | Hair Hg | Buccal mucosa | Michigan dental professionals |
| [87] | TCEANC2; ANGTP2; PRPF18; FOXD2 | Cord whole blood Hg and MeHg | Cord blood | Newborns from Baltimore, USA |
| [88] | PARM1; PFKFB3; LGMN; CCDC68; LRBA; FBXO31; (see Table S2 for the whole list) | Maternal toenail Hg | Cord blood | Mother–infant pairs from USA |
| [89] | EMID2 | Infant toenail Hg | Placenta | Rhode Island infants |
| [90] | PON1 | Maternal red blood cell Hg | Cord blood and children buffy coat | Mother–children pairs from Massachusetts, USA |
| [91] | GRIN2B; NR3C1 | Maternal hair Hg | Children saliva | Children from Europe and US populations |
| Study | Genes | Biomarker | Tissue (DNA) | Sample |
|---|---|---|---|---|
| [92] | ACSL3 | PAM | Umbilical cord white blood cell | Nonsmoking Dominican and African American mother-infants pairs |
| [93] | Global DNA methylation | Pyr and B(a)P in PAM | Umbilical cord blood leukocytes | Nonsmoking women from NYC |
| [94] | IRS2 | Nap, Ace, Fl, Phe and Ant in VAT | VAT | Nonsmoking women from Korea with myoma |
| [95] | LINE1 | B(a)P-DNA adducts | Buffy coat | Nonsmoking pregnant women from Tongliang County, China |
| [96] | 239 quality-controlled autosome CpGs | Urinary ΣOH-PAHs, 9-OH-Phe and 1-OH-Pyr | Whole blood | Nonsmoking healthy Chinese individuals |
| [97] | PAX3 | ΣH_PAHs in maternal serum | Fetal neural tissue | Mother–fetus pairs |
| [44] | ZIC4 | ΣH_PAHs in fetal liver tissue | Fetal neural tissue | NTD fetuses |
| [98] | PLEC1 | Urinary ΣOH-PAHs | Nonsmoking healthy Chinese individuals |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Giovanni, A.; Giuliani, C.; Marini, M.; Luiselli, D. Methylmercury and Polycyclic Aromatic Hydrocarbons in Mediterranean Seafood: A Molecular Anthropological Perspective. Appl. Sci. 2021, 11, 11179. https://doi.org/10.3390/app112311179
De Giovanni A, Giuliani C, Marini M, Luiselli D. Methylmercury and Polycyclic Aromatic Hydrocarbons in Mediterranean Seafood: A Molecular Anthropological Perspective. Applied Sciences. 2021; 11(23):11179. https://doi.org/10.3390/app112311179
Chicago/Turabian StyleDe Giovanni, Andrea, Cristina Giuliani, Mauro Marini, and Donata Luiselli. 2021. "Methylmercury and Polycyclic Aromatic Hydrocarbons in Mediterranean Seafood: A Molecular Anthropological Perspective" Applied Sciences 11, no. 23: 11179. https://doi.org/10.3390/app112311179
APA StyleDe Giovanni, A., Giuliani, C., Marini, M., & Luiselli, D. (2021). Methylmercury and Polycyclic Aromatic Hydrocarbons in Mediterranean Seafood: A Molecular Anthropological Perspective. Applied Sciences, 11(23), 11179. https://doi.org/10.3390/app112311179