Evaluation of the Adhesive Potential of Bacteria Isolated from Meat-Related Sources
Abstract
1. Introduction
2. Material and Methods
2.1. Bacterial Strains and Inoculation
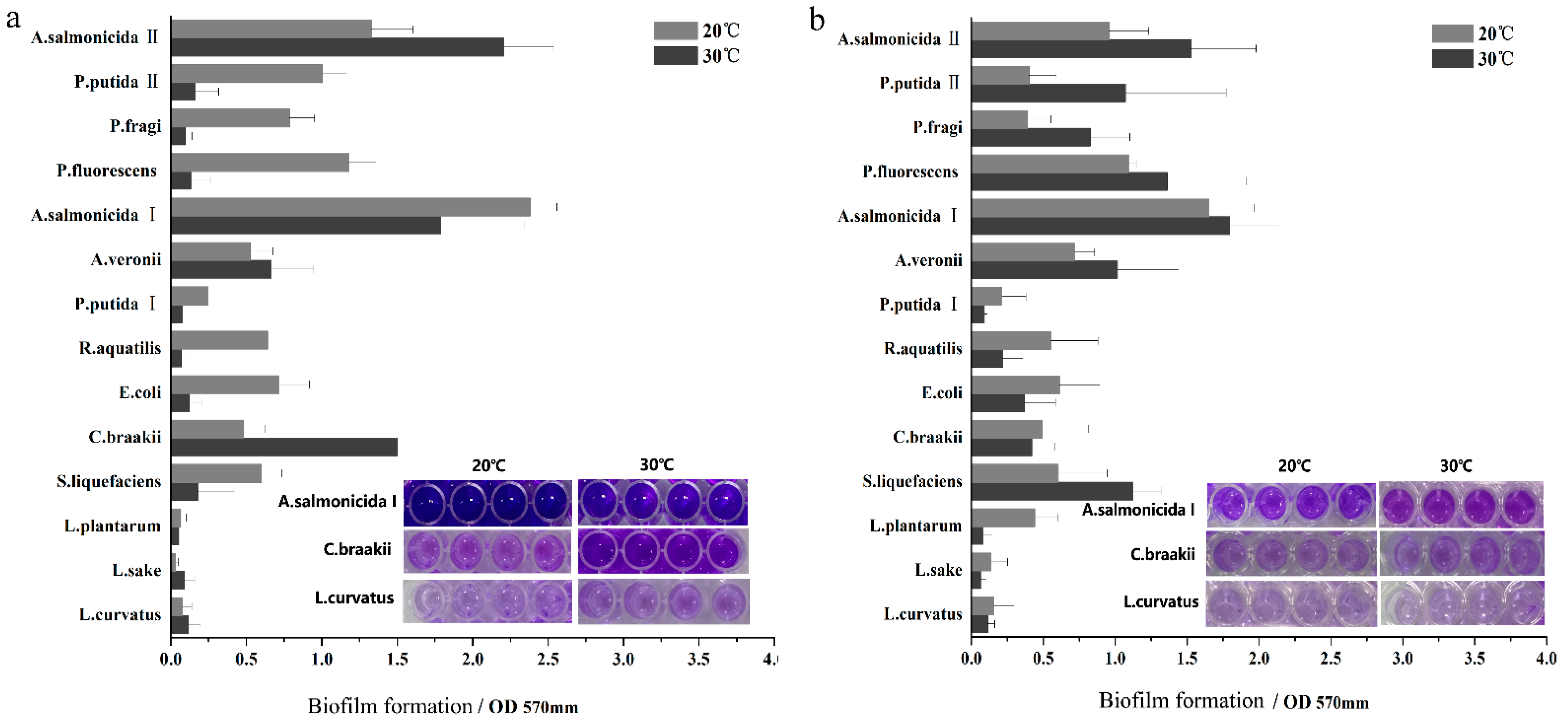
2.2. Biofilm Assay
2.3. Motility Assay
2.4. Cell Surface Hydrophobicity and Electron-Donor/Acceptor Properties
2.5. Statistical Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
| LAB | Lactic acid bacteria |
| TSA | Tryptone soy agar |
| TSB | Tryptone soy broth |
| BSP | Buffered saline peptone water |
| CFU | Colony-forming units |
| QS | Quorum sensing |
References
- Garrett, T.R.; Bhakoo, M.; Zhang, Z. Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. 2008, 18, 1049–1056. [Google Scholar] [CrossRef]
- Petrova, O.E.; Sauer, K. Sticky situations: Key components that control bacterial surface attachment. J. Bacteriol. 2012, 194, 2413–2425. [Google Scholar] [CrossRef]
- Tuson, H.H.; Weibel, D.B. Bacteria-surface Interactions. Soft Matter 2013, 9, 4368. [Google Scholar] [CrossRef] [PubMed]
- Houry, A.; Briandet, R.; Aymerich, S.; Gohar, M. Involnement of motility and flagella in Bacillus cereus biofilm formation. Microbiology 2010, 156, 1009–1018. [Google Scholar] [CrossRef]
- Yang, Y.; Zhou, M.; Hou, H.; Zhu, J.; Yao, F.; Zhang, X.; Zhu, X.; Hardwidge, P.R.; Zhu, G. Quorum-sensing gene luxS regulates flagella expression and Shiga-like toxin production in F18ab Escherichia coli. Can. J. Microbiol. 2014, 60, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Chaves, R.D.; Silva, A.R.; Sant’Ana, A.S.; Campana, F.B.; Massaguer, P.R. Gas-producing and spoilage potential of Enterobacteriaceae and lactic acid bacteria isolated from chilled vacuum-packaged beef. Int. J. Food Sci. Technol. 2012, 47, 1750–1756. [Google Scholar] [CrossRef]
- Beazhidalgo, R.; Agüeria, D.; Latifeugenín, F.; Yeannes, M.I.; Figueras, M.J. Molecular characterization of Shewanella and Aeromonas isolates associated with spoilage of Common carp (Cyprinus carpio). FEMS Microbiol. Lett. 2015, 362, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Hernández, I. Bacteriophages against Serratia as fish spoilage control Technology. Front. Microbiol. 2017, 8, 449. [Google Scholar] [CrossRef]
- Raposo, A.; Pérez, E.; de Faria, C.T.; Ferrus, M.A.; Carrascosa, A. Food spoilage by Pseudomonas spp.-an overview. Food Borne Pathog. Antibiot. Resist. 2017, 3, 41–58. [Google Scholar]
- Wang, Y.; Yi, L.; Wang, Y.; Wang, Y.; Cai, Y.; Zhao, W.; Ding, C. Isolation, phylogenetic group, drug resistance, biofilm formation, and adherence genes of Escherichia coli from poultry in central China. Poult. Sci. 2016, 95, 252. [Google Scholar] [CrossRef]
- Caixeta, D.S.; Scarpa, T.H.; Brugnera, D.F.; Freire, D.O.; Alves, E.; de Abreu, L.R.; Piccoli, R.H. Chemical sanitizers to control biofilms formed by two Pseudomonas species on stainless steel surface. Ciência E Tecnol. Aliment. 2012, 32, 142–150. [Google Scholar] [CrossRef][Green Version]
- Wang, H.; Ye, K.; Zhang, Q.; Dong, Y.; Xu, X.-L.; Zhou, G.-H. Biofilm formation of meat-borne Salmonella enterica and inhibition by the cell-free supernatant from Pseudomonas aeruginosa. Food Control 2013, 32, 650–658. [Google Scholar] [CrossRef]
- Wang, H.; Ding, S.; Dong, Y.; Ye, K.; Xu, X.; Zhou, G. Biofilm formation of Salmonella serotypes in simulated meat processing environments and its relationship to cell characteristics. J. Food Prot. 2013, 76, 1784–1789. [Google Scholar] [CrossRef]
- Darilmaz, D.O.; Beyatli, Y.; Yuksekdag, Z.N. Aggregation and hydrophobicity properties of 6 dairy propionibacteria strains isolated from homemade Turkish cheeses. J. Food Sci. 2012, 77, M20–M24. [Google Scholar] [CrossRef]
- Dias, C.; Borges, A.; Saavedra, M.J.; Simões, M. Biofilm formation and multidrug-resistant Aeromonas spp. from wild animals. J. Glob. Antimicrob. Resist. 2018, 12, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Vélez, M.P.; De Keersmaecker, S.C.; Vanderleyden, J. Adherence factors of Lactobacillus in the human gastrointestinal tract. FEMS Microbiol. Lett. 2007, 276, 140–148. [Google Scholar] [CrossRef]
- Houdt, R.V.; Michiels, C.W. Biofilm formation and the food industry, a focus on the bacterial outer surface. J. Appl. Microbiol. 2010, 109, 1117–1131. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Uppuluri, P.; Chaturvedi, A.K.; Srinivasan, A.; Banerjee, M.; Ramasubramaniam, A.K.; Köhler, J.R.; Kadosh, D.; Lopez-Ribot, J.L. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010, 6, e1000828. [Google Scholar] [CrossRef] [PubMed]
- Abee, T.; Kovács, Á.T.; Kuipers, O.P.; van der Veen, S. Biofilm formation and dispersal in Gram-positive bacteria. Curr. Opin. Biotechnol. 2011, 22, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Kearns, D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010, 8, 634–644. [Google Scholar] [CrossRef]
- Mattingly, A.E.; Weaver, A.A.; Dimkovikj, A.; Shrout, J.D. Assessing travel conditions: Environmental and host influences on bacterial surface motility. J. Bacteriol. 2018, 200, e00014-18. [Google Scholar] [CrossRef]
- Watnick, P.I.; Lauriano, C.M.; Klose, K.E.; Croal, L.; Kolter, R. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 2001, 39, 223–235. [Google Scholar] [CrossRef]
- Guo, K.; Freguia, S.; Dennis, P.G.; Chen, X.; Donose, B.C.; Keller, J.; Gooding, J.J.; Rabaey, K. Effects of surface charge and hydrophobicity on anodic biofilm formation, community composition, and current generation in bioelectrochemical systems. Environ. Sci. Technol. 2013, 47, 7563–7570. [Google Scholar] [CrossRef]
- Pezzoni, M.; Pizarro, R.A.; Costa, C.S. Exposure to low doses of UVA increases biofilm formation in Pseudomonas aeruginosa. Biofouling 2018, 34, 673–684. [Google Scholar] [CrossRef]
- Wang, G.Y.; Li, M.; Ma, F.; Wang, H.-H.; Xu, X.-L.; Zhou, G.-H. Physicochemical properties of Pseudomonas fragi isolates response to modified atmosphere packaging. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Giaouris, E.; Chapot-Chartier, M.P.; Briandet, R. Surface physicochemical analysis of natural Lactococcus lactis strains reveals the existence of hydrophobic and low charged strains with altered adhesive properties. Int. J. Food Microbiol. 2009, 131, 2–9. [Google Scholar] [CrossRef]
- Hayashi, H.; Tsuneda, S.; Hirata, A.; Sasaki, H. Soft particle analysis of bacterial cells and its interpretation of cell adhesion behaviors in terms of DLVO theory. Colloids Surfaces B 2001, 22, 149–157. [Google Scholar] [CrossRef]
- Briandet, R.; Herry, J.; Bellon-Fontaine, M. Determination of the van der Waals, electron donor and electron acceptor surface tension components of static Gram-positive microbial biofilms. Colloids Surfaces B 2001, 21, 299–310. [Google Scholar] [CrossRef]
- Bellon-Fontaine, M.; Rault, J.; Oss, C. Microbial adhesion to solvents: A novel method to determine the electron-donor/electron-acceptor or Lewis acid-base properties of microbial cells. Colloids Surfaces B Biointerfaces 1996, 7, 47–53. [Google Scholar] [CrossRef]
- Grasland, B.; Mitalane, J.; Briandet, R.; Quemener, E.; Meylheuc, T.; Linossier, I.; Vallee-Rehel, K.; Haras, D. Bacterial biofilm in seawater: Cell surface properties of early-attached marine bacteria. Biofouling 2003, 19, 307–313. [Google Scholar] [CrossRef] [PubMed]

| Strain Number | Bacteria Isolates | Source | Affinity/% | ||
|---|---|---|---|---|---|
| Xylene | Ethyl Acetate | Chloroform | |||
| NCM1577 | L. curvatus | Cooked ham | 4.42 ± 4.64 a | 6.90 ± 2.61 bc | 3.88 ± 1.43 a |
| NCM1578 | L. sake | Cooked ham | 19.28 ± 2.92 b | 4.09 ± 1.52 abc | 4.08 ± 2.63 a |
| NCM1579 | L. plantarum | Cooked ham | 16.02 ± 3.16 b | 0.75 ± 0.35 a | 15.64 ± 2.97 b |
| NCM1580 | S. liquefaciens | Cooked ham | 75.64 ± 6.16 f | 3.39 ± 1.55 ab | 63.96 ± 2.19 cd |
| NCM1594 | C. braakii | Chilled chicken meat | 73.55 ± 6.05 f | 3.09 ± 2.60 ab | 15.45 ± 1.54 b |
| NCM1582 | E. Coli | Chilled chicken meat | 29.85 ± 6.35 c | 6.44 ± 1.07 bc | 22.77 ± 3.30 b |
| NCM1583 | R. aquatilis | Cooked ham | 56.52 ± 13.87 e | 17.40 ± 2.71 d | 59.45 ± 12.52 c |
| NCM1586 | P. putida I | Chilled chicken meat | 72.37 ± 8.05 f | 31.47 ± 7.73 f | 70.04 ± 9.51 d |
| NCM1587 | A. veronii | Chilled chicken meat | 71.73 ± 5.16 f | 8.21 ± 3.31 c | 79.41 ± 4.67e |
| NCM1589 | A. salmonicida I | Chilled chicken meat | 54.31 ± 6.08 e | 26.11 ± 1.91 e | 78.63 ± 7.14 e |
| NCM1590 | P. fluorescens | Chicken conveyor belt surface | 40.72 ± 2.69 d | 38.15 ± 1.92 g | 82.05 ± 8.62 ef |
| NCM1591 | P. fragi | Chilled chicken meat | 90.13 ± 6.77 g | 61.77 ± 3.20 i | 92.90 ± 1.64 g |
| NCM1592 | P. putida II | Chicken conveyor belt surface | 70.96 ± 17.14 f | 57.38 ± 1.57 h | 92.36 ± 3.41 g |
| NCM1596 | A. salmonicida II | Chicken conveyor belt surface | 81.14 ± 12.00 fg | 64.96 ± 3.78 i | 89.24 ± 5.83 fg |
| Index | Chloroform | Ethyl Acetate | Swarming | Swimming | 24 h 20 °C Adhesion | 24 h 30 °C Adhesion | 72 h 20 °C Adhesion | 72 h 30 °C Adhesion |
|---|---|---|---|---|---|---|---|---|
| Xylene | 0.73 ** | 0.54 ** | 0.20 | 0.09 | 0.33 ** | 0.37 ** | 0.48 ** | 0.25 |
| Chloroform | 0.77 ** | −0.11 | −0.33 ** | 0.21 | 0.63 ** | 0.68 ** | 0.46 ** | |
| Ethyl acetate | −0.07 | −0.28 * | 0.23 | 0.50 ** | 0.50 ** | 0.27 * | ||
| Swarming | 0.81 ** | 0.23 | 0.14 | 0.02 | 0.14 | |||
| Swimming | 0.08 | −0.05 | −0.17 | −0.00 | ||||
| 24 h 20 °C adhesion | 0.58 ** | 0.50 ** | 0.47 ** | |||||
| 24 h 30 °C adhesion | 0.80 ** | 0.63 ** | ||||||
| 72 h 20 °C adhesion | 0.61 ** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ning, Z.; Xue, B.; Wang, H. Evaluation of the Adhesive Potential of Bacteria Isolated from Meat-Related Sources. Appl. Sci. 2021, 11, 10652. https://doi.org/10.3390/app112210652
Ning Z, Xue B, Wang H. Evaluation of the Adhesive Potential of Bacteria Isolated from Meat-Related Sources. Applied Sciences. 2021; 11(22):10652. https://doi.org/10.3390/app112210652
Chicago/Turabian StyleNing, Zhenzhen, Bei Xue, and Huhu Wang. 2021. "Evaluation of the Adhesive Potential of Bacteria Isolated from Meat-Related Sources" Applied Sciences 11, no. 22: 10652. https://doi.org/10.3390/app112210652
APA StyleNing, Z., Xue, B., & Wang, H. (2021). Evaluation of the Adhesive Potential of Bacteria Isolated from Meat-Related Sources. Applied Sciences, 11(22), 10652. https://doi.org/10.3390/app112210652





