Visual-Saliency-Based Abnormality Detection for MRI Brain Images—Alzheimer’s Disease Analysis
Abstract
:1. Introduction
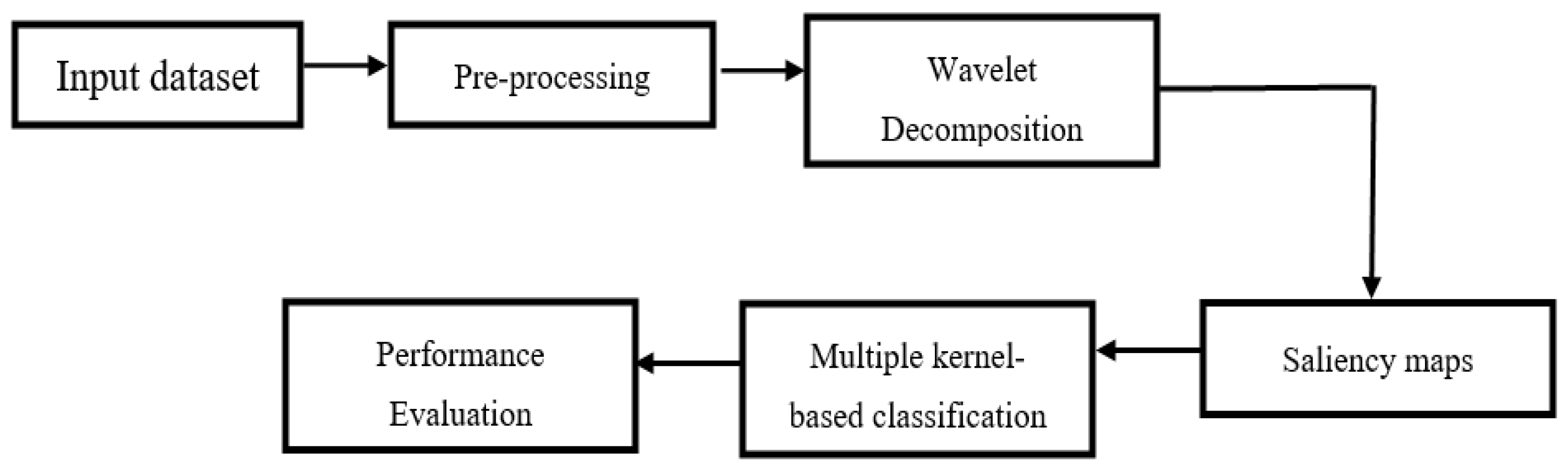
2. Proposed Methodology
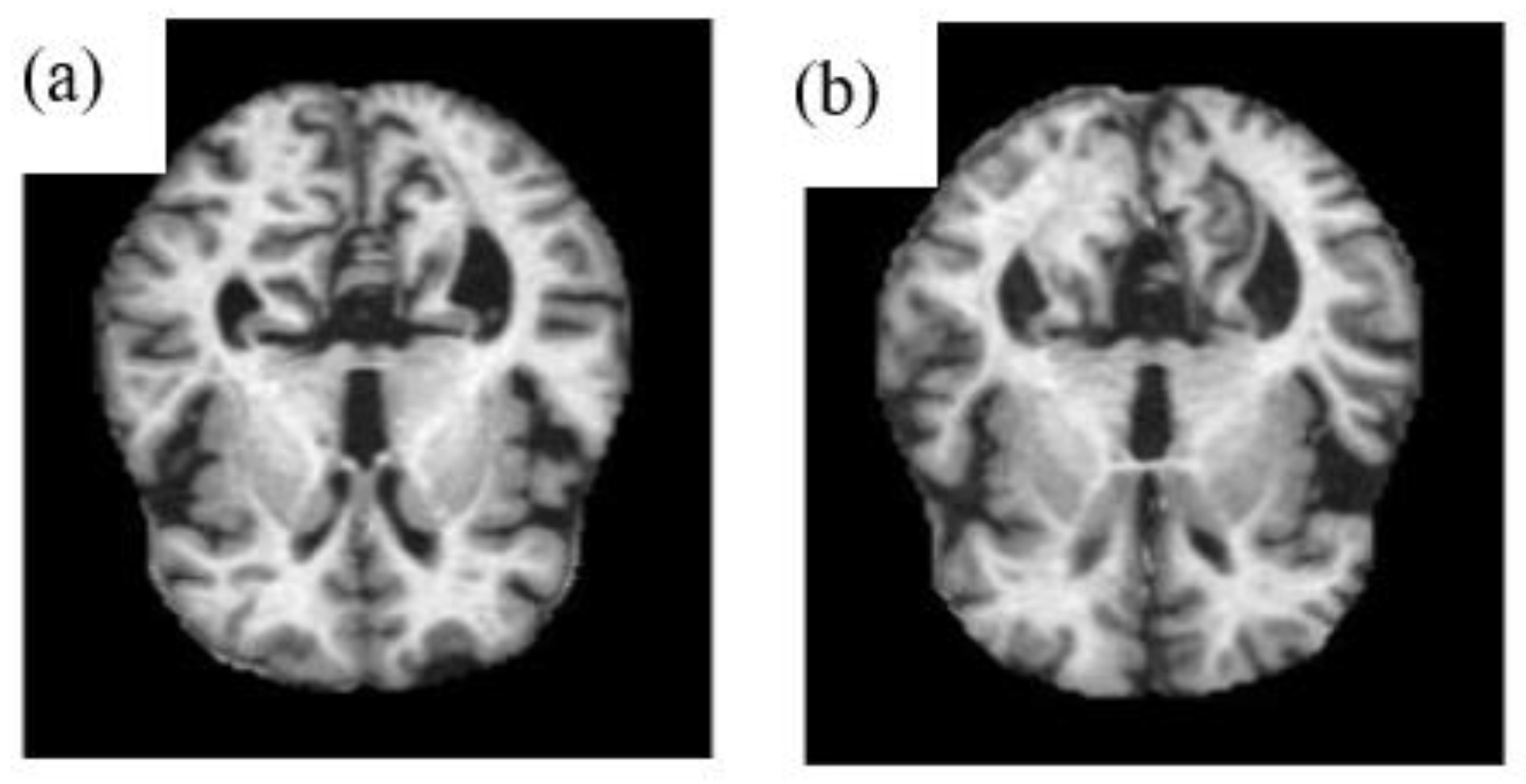
2.1. Pre-Processing
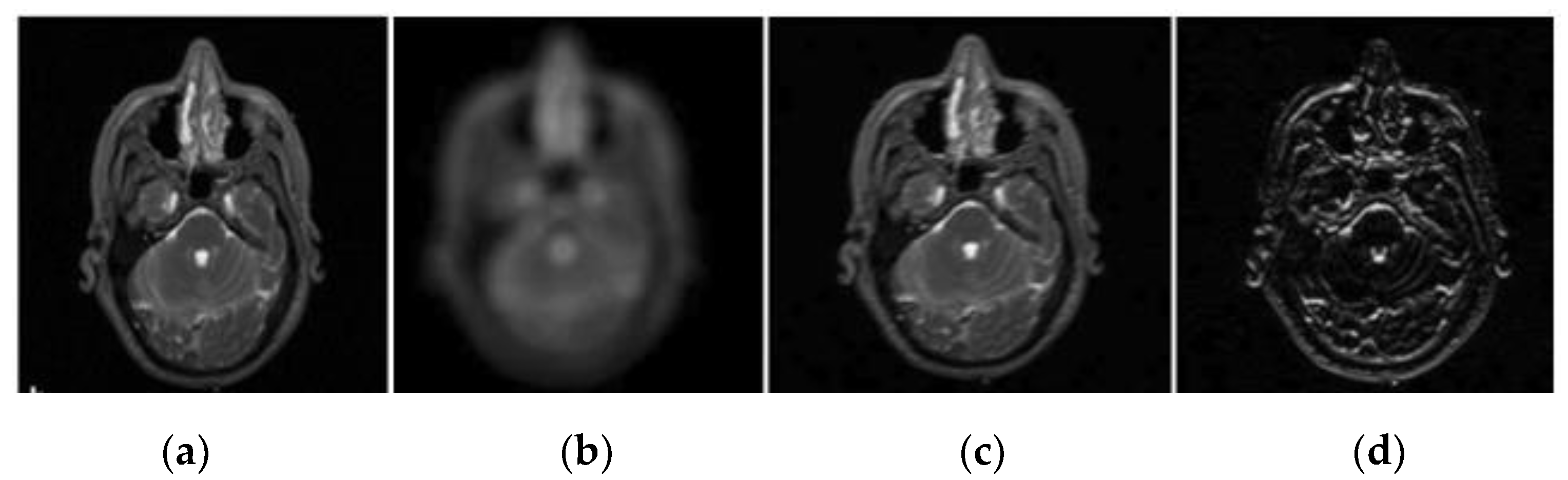
2.2. Wavelet Decomposition
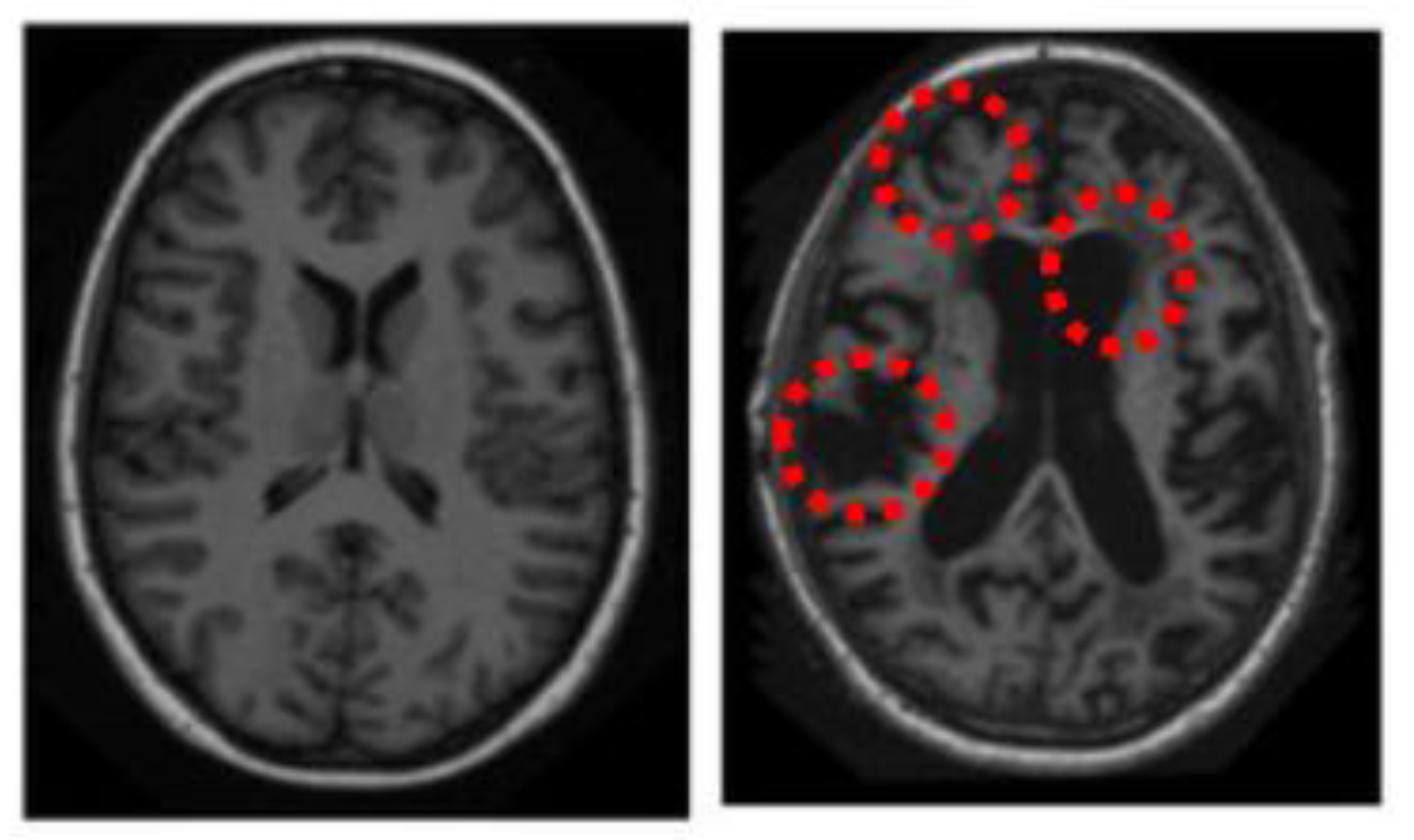
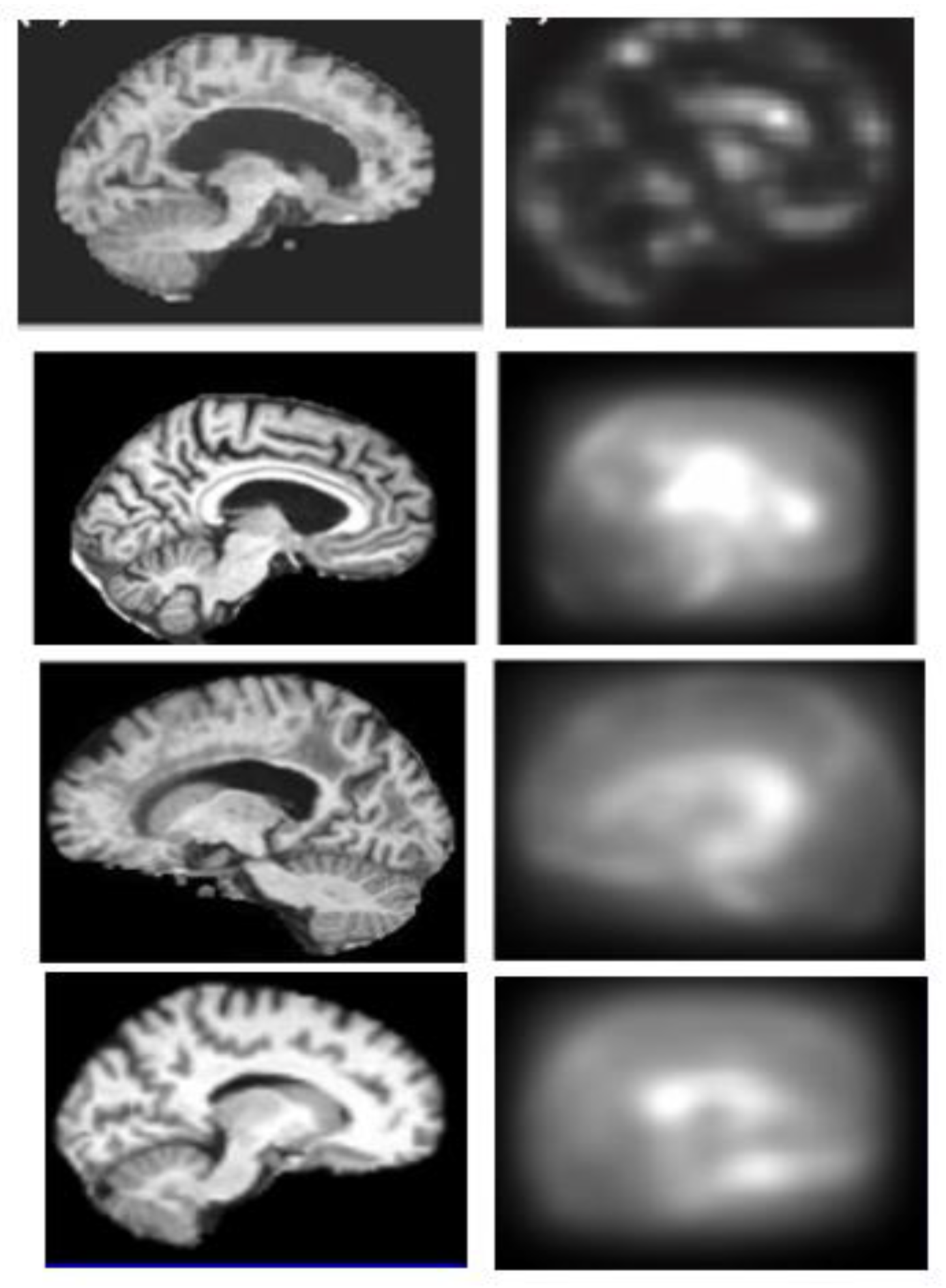
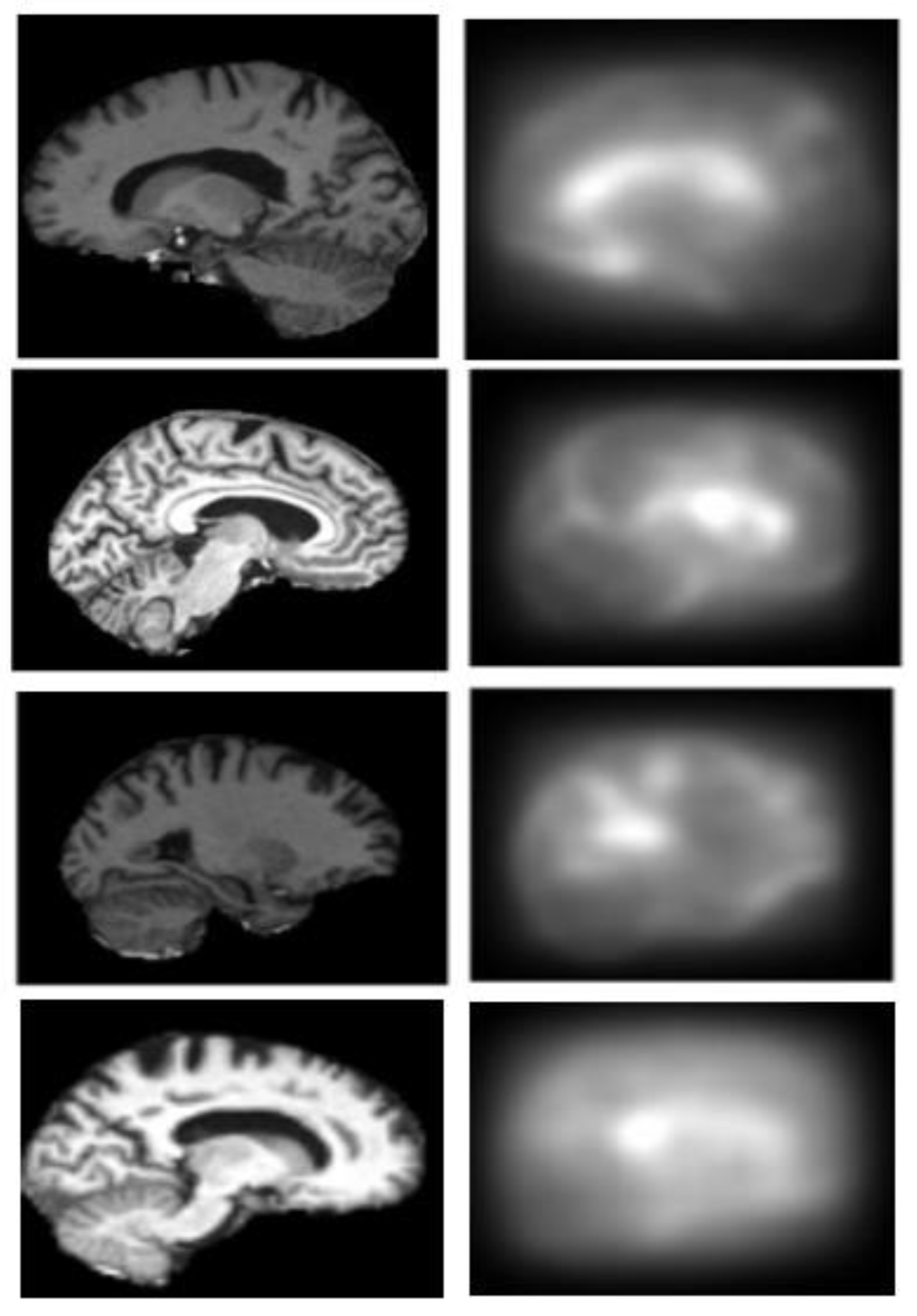
2.3. Generation of Saliency Maps
- Input: 3D MRI brain volume with
- is the Alzheimer’s disease pattern, is the normal pattern.
- Step 1: Find the bottom-up and top-down saliency map;
- Step 2: Compute saliency map;
- Step 3: Classification of AD and non-AD interpretation.
2.3.1. Top-Down Saliency Maps (
- Build probability map;
- Regularize into a fixed range (0….1);
- If probability (g)> = 0.5 then
- else
- ;
- .
2.3.2. Bottom-Up Saliency Maps (
Elliptical Local Binary Pattern
2.3.3. Final Saliency Map
2.4. Multiple-Kernel Learning (MKL)
- Step 1:
- Initialize the range of kernels for MKL and SEMKL;
- Step 2:
- Compute the basic kernel matrixes using Equation (10);
- Step 3:
- Solve the projective direction according to Equation (11);
- Step 4:
- Using the projective direction ‘w’, combine the basic kernels;
- Step 5:
- Utilizing the combined kernel, the classification problem is approached via SVM.
3. Results
3.1. Dataset
- Division 1:
- Age 60–80, CDR=1, 7/6, 86 images, 20 D, 66 N;
- Division 2:
- Age 60–96, CDR=1, 7/6, 126 images, 28 D, 98 N;
- Division 3:
- Age 60–80, CDR= 0.5, 21/30, 136 images, 70 D, 66 N;
- Division 4:
- Age 90–96, CDR= {2,1,0.5}, 198 images, 100 D, 98 N.
3.2. Training and Testing
3.3. Quantitative Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Payan, A.; Montana, G. Predicting Alzheimer’s Disease: A Neuroimaging Study with 3D Convolutional Neural Networks. arXiv 2015, arXiv:1502.02506v1. [Google Scholar]
- Rossor, M.N.; Fox, N.; Mummery, C.J.; Schott, J.; Warren, J. The Diagnosis of Young-Onset Dementia. Lancet Neurol. 2010, 9, 793–806. [Google Scholar] [CrossRef] [Green Version]
- Sandanalakshmi, R.; Sardius, V. Selected Saliency Based Analysis for the Diagnosis of Alzheimer’s Disease Using Structural Magnetic Resonance Image. J. Med. Imaging Health Inform. 2016, 6, 177–184. [Google Scholar] [CrossRef]
- Marcus, C.; Mena, E.; Subramaniam, R.M. Brain PET in the Diagnosis of Alzheimer’s Disease. Clin. Nucl. Med. 2014, 39, e413–e426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombardi, G.; Crescioli, G.; Cavedo, E.; Lucenteforte, E.; Casazza, G.; Bellatorre, A.G.; Lista, C.; Costantino, G.; Frisoni, G.; Virgili, G.; et al. Structural Magnetic Resonance Imaging for the Early Diagnosis of Dementia due to Alzheimer’s Disease in People with Mild Cognitive Impairment. Cochrane Database Syst. Rev. 2020, 3, CD009628. [Google Scholar] [CrossRef] [PubMed]
- Hojjati, S.H.; Ebrahimzadeh, A.; Babajani-Feremi, A. Identification of the Early stage of Alzheimer’s Disease Using Structural MRI and Resting-State fMRI. Front. Neurol. 2019, 10, 904. [Google Scholar] [CrossRef] [Green Version]
- Beheshti, I.; Demirel, H.; Farokhian, F.; Yang, C.; Matsuda, H. Structural MRI-based Detection of Alzheimer’s Disease Using Feature Ranking and Classification Error. Comput. Methods Programs Biomed. 2016, 137, 177–193. [Google Scholar] [CrossRef]
- Arevalo-Rodriguez, I.; Smailagic, N.; Figuls, M.R.I.; Ciapponi, A.; Sanchez-Perez, E.; Giannakou, A.; Pedraza, O.L.; Bonfill Cosp, X.; Cullum, S. Mini-Mental State Examination (MMSE) for the Detection of Alzheimer’s Disease and other Dementias in People with Mild Cognitive Impairment (MCI). Cochrane Database Syst. Rev. 2015, 2015, CD010783. [Google Scholar] [CrossRef]
- Calhoun, V.D.; Adali, T. Time-Varying Brain Connectivity in fMRI Data: Whole-brain Data-Driven Approaches for Capturing and Characterizing Dynamic States. IEEE Signal Process. Mag. 2016, 33, 52–66. [Google Scholar] [CrossRef]
- Guo, H.; Grajauskas, L.; Habash, B.; D’Arcy, R.C.; Song, X. Functional MRI Technologies in the Study of Medication Treatment Effect on Alzheimer’s Disease. Aging Med. 2018, 1, 75–95. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Suk, H.I.; Shen, D. A Novel Matrix-Similarity Based Loss Function for Joint Regression and Classification in AD Diagnosis. NeuroImage 2014, 100, 91–105. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Wu, X.; Chen, K.; Yao, L. Multi-Modality Sparse Representation-Based Classification for Alzheimer’s Disease and Mild Cognitive Impairment. Comput. Methods Programs Biomed. 2015, 122, 182–190. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, D.; Shen, D. Ensemble Sparse Classification of Alzheimer’s Disease. NeuroImage 2012, 60, 1106–1116. [Google Scholar] [CrossRef] [Green Version]
- Rathore, S.; Habes, M.; Iftikhar, M.A.; Shacklett, A.; Davatzikos, C. A Review on Neuroimaging-Based Classification Studies and Associated Feature Extraction Methods for Alzheimer’s Disease and Its Prodromal Stages. NeuroImage 2017, 155, 530–548. [Google Scholar] [CrossRef]
- Chyzhyk, D.; Graña, M. Optimal Hyperbox Shrinking in Dendritic Computing Applied to Alzheimer’s Disease Detection in MRI. In Proceedings of the Advances in Intelligent and Soft Computing, Salamanca, Spain, 6–8 April 2011; Volume 87, pp. 543–550. [Google Scholar]
- Chyzhyk, D.; Graña, M.; Savio, A.; Maiora, J. Hybrid Dendritic Computing with Kernel-LICA Applied to Alzheimer’s Disease Detection in MRI. Neurocomputing 2012, 75, 72–77. [Google Scholar] [CrossRef]
- Khajehnejad, M.; Saatlou, F.H.; Mohammadzade, H. Alzheimer’s Disease Early Diagnosis Using Manifold-Based Semi-Supervised Learning. Brain Sci. 2017, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, D.L.; Garcia-Arteaga, J.D.; Cárdenas-Robledo, S.; Romero, E. Characterization of Brain Anatomical Patterns by Comparing Region Intensity Distributions: Applications to the Description of Alzheimer’s Disease. Brain Behav. 2018, 8, e00942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basaia, S.; Agosta, F.; Wagner, L.; Canu, E.; Magnani, G.; Santangelo, R.; Filippi, M. Automated Classification of Alzheimer’s Disease and Mild Cognitive Impairment Using a Single MRI and Deep Neural Networks. NeuroImage Clin. 2019, 21, 101645. [Google Scholar] [CrossRef] [PubMed]
- Rueda, A.; Gonzalez, F.A.; Romero, E. Extracting Salient Brain Patterns for Imaging-Based Classification of Neurodegenerative Diseases. IEEE Trans. Med. Imaging 2014, 33, 1262–1274. [Google Scholar] [CrossRef] [PubMed]
- Andrushia, A.D.; Thangarjan, R. Saliency-Based Image Compression Using Walsh–Hadamard Transform (WHT). In Biologically Rationalized Computing Techniques for Image Processing Applications; Lecture Notes in Computational Vision and Biomechanics; Hemanth, J., Balas, V.E., Eds.; Springer: Berlin/Heidelberg, Germany, 2018; Volume 25, pp. 21–42. [Google Scholar]
- Andrushia, D.; Thangarajan, R. Visual Attention-Based Leukocyte Image Segmentation Using Extreme Learning Machine. Int. J. Adv. Intell. Paradig. 2015, 7, 172. [Google Scholar] [CrossRef]
- Andrushia, A.D.; Thangarajan, R. An Efficient Visual Saliency Detection Model Based on Ripplet Transform. Sadhana 2017, 42, 671–685. [Google Scholar] [CrossRef] [Green Version]
- Wei, D.; Zhuang, K.; Ai, L.; Chen, Q.; Yang, W.; Liu, W.; Wang, K.; Sun, J.; Qiu, J. Structural and Functional Brain Scans from the Cross-Sectional Southwest University Adult Lifespan Dataset. Sci. Data 2018, 5, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Ayadi, W.; Elhamzi, W.; Charfi, I.; Atri, M. A Hybrid Feature Extraction Approach for Brain MRI Classification Based on Bag-of-Words. Biomed. Signal Process. Control 2019, 48, 144–152. [Google Scholar] [CrossRef]
- Blennow, K.; Hampel, H.; Weiner, M.W.; Zetterberg, H. Cerebrospinal Fluid and Plasma Biomarkers in Alzheimer Disease. Nat. Rev. Neurol. 2010, 6, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Braak, E. Evolution of Neuronal Changes in the Course of Alzheimer’s Disease. J. Neural Transmission. Suppl. 1998, 53, 127–140. [Google Scholar] [CrossRef]
- Ben Ahmed, O.; Larabi, M.C.; Paccalin, M.; Fernandez-Maloigne, C. Saliency Guided Computer-aided Diagnosis for Neurodegenerative Dementia. In Proceedings of the 10th International Joint Conference on Biomedical Engineering Systems and Technologies, Porto, Portugal, 21–23 February 2017; pp. 140–147. [Google Scholar]
- George, M.; Zwiggelaar, R. Comparative Study on Local Binary Patterns for Mammographic Density and Risk Scoring. J. Imaging 2019, 5, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koutras, P.; Panagiotaropoulou, G.; Tsiami, A.; Maragos, P. Audio-Visual Temporal Saliency Modeling Validated by fMRI Data. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Salt Lake City, UT, USA, 18–22 June 2018; pp. 2081–208110. [Google Scholar]
- Gevaert, C.M.; Persello, C.; Vosselman, G. Optimizing Multiple Kernel Learning for the Classification of UAV Data. Remote Sens. 2016, 8, 1025. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Jin, R.; Yang, H.; King, I.; Lyu, M.R. Simple and Efficient Multiple Kernel Learning by Group Lasso. In Proceedings of the 27th International Conference on Machine Learning (ICML-10), Haifa, Israel, 21–24 June 2010; pp. 1175–1182. [Google Scholar]
- Wilson, C.M.; Li, K.; Yu, X.; Kuan, P.F.; Wang, X. Multiple-Kernel Learning for Genomic Data Mining and Prediction. BMC Bioinform. 2019, 20, 426. [Google Scholar] [CrossRef] [Green Version]
- Ben-Ahmed, O.; Lecellier, F.; Paccalin, M.; Fernandez-Maloigne, C. Multi-View Visual Saliency-Based MRI Classification for Alzheimer’s Disease Diagnosis. In Proceedings of the 2017 Seventh International Conference on Image Processing Theory, Tools and Applications (IPTA), Montreal, QC, Canada, 28 November–1 December 2017; pp. 1–6. [Google Scholar]
- Rakotomamonjy, A.; Bach, F.R.; Canu, S.; Grandvalet, Y. SimpleMkl. J. Mach. Learn. Res. 2008, 9, 2491–2521. [Google Scholar]
- Marcus, D.S.; Wang, T.H.; Parker, J.; Csernansky, J.G.; Morris, J.C.; Buckner, R.L. Open Access Series of Imaging Studies (OASIS): Cross-sectional MRI Data in Young, Middle Aged, Nondemented, and Demented Older Adults. J. Cogn. Neurosci. 2007, 19, 1498–1507. [Google Scholar] [CrossRef] [Green Version]
- OASIS Brains—Open Access Series of Imaging Studies. Available online: https://www.oasis-brains.org (accessed on 15 July 2019).
- Toews, M.; Wells, W.; Collins, D.L.; Arbel, T. Feature-based morphometry: Discovering group-related anatomical patterns. NeuroImage 2010, 49, 2318–2327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrea, R.; Fabio, A.G.; Eduardo, R. Saliency-Based Characterization of Group Differences for Magnetic Resonance Disease Classification. Dyna 2013, 80, 21–28. [Google Scholar]
- Yang, W.; Xia, H.; Xia, B.; Lui, L.M.; Huang, X. ICA-Based Feature Extraction and Automatic Classification of AD-Related MRI Data. In Proceedings of the 2010 Sixth International Conference on Natural Computation, Yantai, China, 10–12 August 2010; Volume 3, pp. 1261–1265. [Google Scholar]
- Jha, D.; Alam, S.; Pyun, J.Y.; Lee, K.H.; Kwon, G.R. Alzheimer’s Disease Detection Using Extreme Learning Machine, Complex Dual Tree Wavelet Principal Coefficients and Linear Discriminant Analysis. J. Med Imaging Health Inform. 2018, 8, 881–890. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, S.; Sui, Y.; Yang, M.; Bin Liu, B.; Cheng, H.; Sun, J.; Jia, W.; Phillips, P.; Gorriz, J.M. Multivariate Approach for Alzheimer’s Disease Detection Using Stationary Wavelet Entropy and Predator-Prey Particle Swarm Optimization. J. Alzheimer’s Dis. 2018, 65, 855–869. [Google Scholar] [CrossRef]
- Feng, J.; Zhang, S.W.; Chen, L. Alzheimer’s Disease Neuroimaging Initiative (ADNI). Identification of Alzheimer’s Disease Based on Wavelet Transformation Energy Feature of the Structural MRI Image and NN Classifier. Artif. Intell. Med. 2020, 108, 101940. [Google Scholar] [CrossRef]
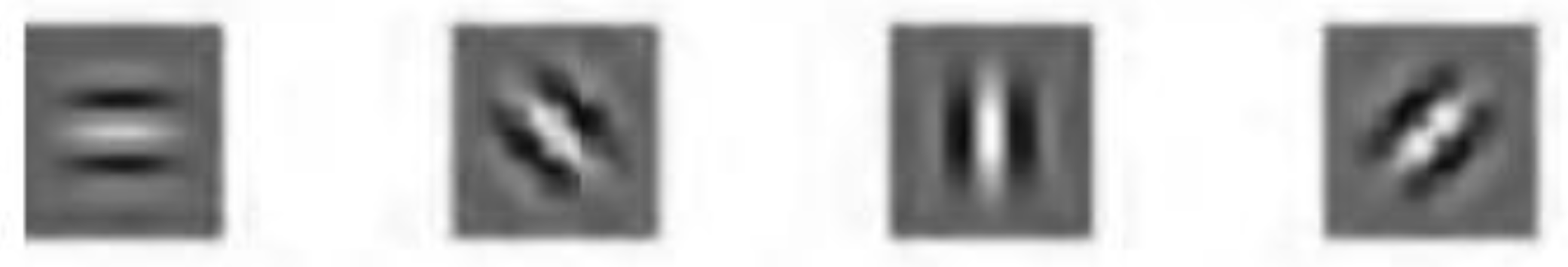
| Condition | Numbers | Gender | Socioeconomic Status | Age | MMSE | CDR | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Range | Mean | Range | Mean | 0 | 0.5 | 1 | 2 | ||||
| AD | 100 | M/F | 2.94 | 66–96 | 78.08 | 15–30 | 24 | 0 | 31 | 17 | 1 |
| Normal | 100 | M/F | 2.88 | 65–94 | 77.77 | 26–30 | 28.96 | 49 | 0 | 0 | 0 |
| Accuracy (A) | Sensitivity (S) | Specificity (SP) | F-Measure (Fm) | |
|---|---|---|---|---|
| Division 1 | 91.18 | 88.21 | 89.34 | 90.91 |
| Division 2 | 88.26 | 86.15 | 87.33 | 87.81 |
| Division 3 | 87.54 | 85.78 | 86.13 | 86.91 |
| Division 4 | 84.23 | 82.91 | 83.21 | 84.12 |
| Approach | Accuracy (A) | Sensitivity (S) | Specificity (SP) | F-Measure (Fm) |
|---|---|---|---|---|
| Toews et al. [38] | 71.45 | 67.54 | 72.65 | 73.56 |
| Yang et al. [40] | 67.15 | 62.65 | 73.11 | 69.13 |
| Chyzhyk et al. [16] | 69 | 81 | 56 | 70.12 |
| Chyzhyk et al. [17] | 74.25 | 96 | 52.5 | 74.89 |
| Andrea R et al. [39] | 67.68 | 72 | 63.27 | 68.01 |
| Feng J et al. [43] | 86.4 | 82.11 | 89.91 | -- |
| Jha et al. [41] | 78.48 | 75.35 | 79.98 | -- |
| Zhang et al. [42] | 72.86 | 69.55 | 75.49 | -- |
| Proposed | 89.12 | 86.71 | 87.31 | 88.93 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrushia, A.D.; Sagayam, K.M.; Dang, H.; Pomplun, M.; Quach, L. Visual-Saliency-Based Abnormality Detection for MRI Brain Images—Alzheimer’s Disease Analysis. Appl. Sci. 2021, 11, 9199. https://doi.org/10.3390/app11199199
Andrushia AD, Sagayam KM, Dang H, Pomplun M, Quach L. Visual-Saliency-Based Abnormality Detection for MRI Brain Images—Alzheimer’s Disease Analysis. Applied Sciences. 2021; 11(19):9199. https://doi.org/10.3390/app11199199
Chicago/Turabian StyleAndrushia, A. Diana, K. Martin Sagayam, Hien Dang, Marc Pomplun, and Lien Quach. 2021. "Visual-Saliency-Based Abnormality Detection for MRI Brain Images—Alzheimer’s Disease Analysis" Applied Sciences 11, no. 19: 9199. https://doi.org/10.3390/app11199199
APA StyleAndrushia, A. D., Sagayam, K. M., Dang, H., Pomplun, M., & Quach, L. (2021). Visual-Saliency-Based Abnormality Detection for MRI Brain Images—Alzheimer’s Disease Analysis. Applied Sciences, 11(19), 9199. https://doi.org/10.3390/app11199199







