Soft Computing Paradigms to Find the Numerical Solutions of a Nonlinear Influenza Disease Model
Abstract
:1. Introduction
- The proposed ANN-GA-ASM provides effective solutions of the nonlinear influenza system.
- Consistent, stable and reliable outcomes from the nonlinear influenza system validate the value of the proposed ANN-GA-ASM.
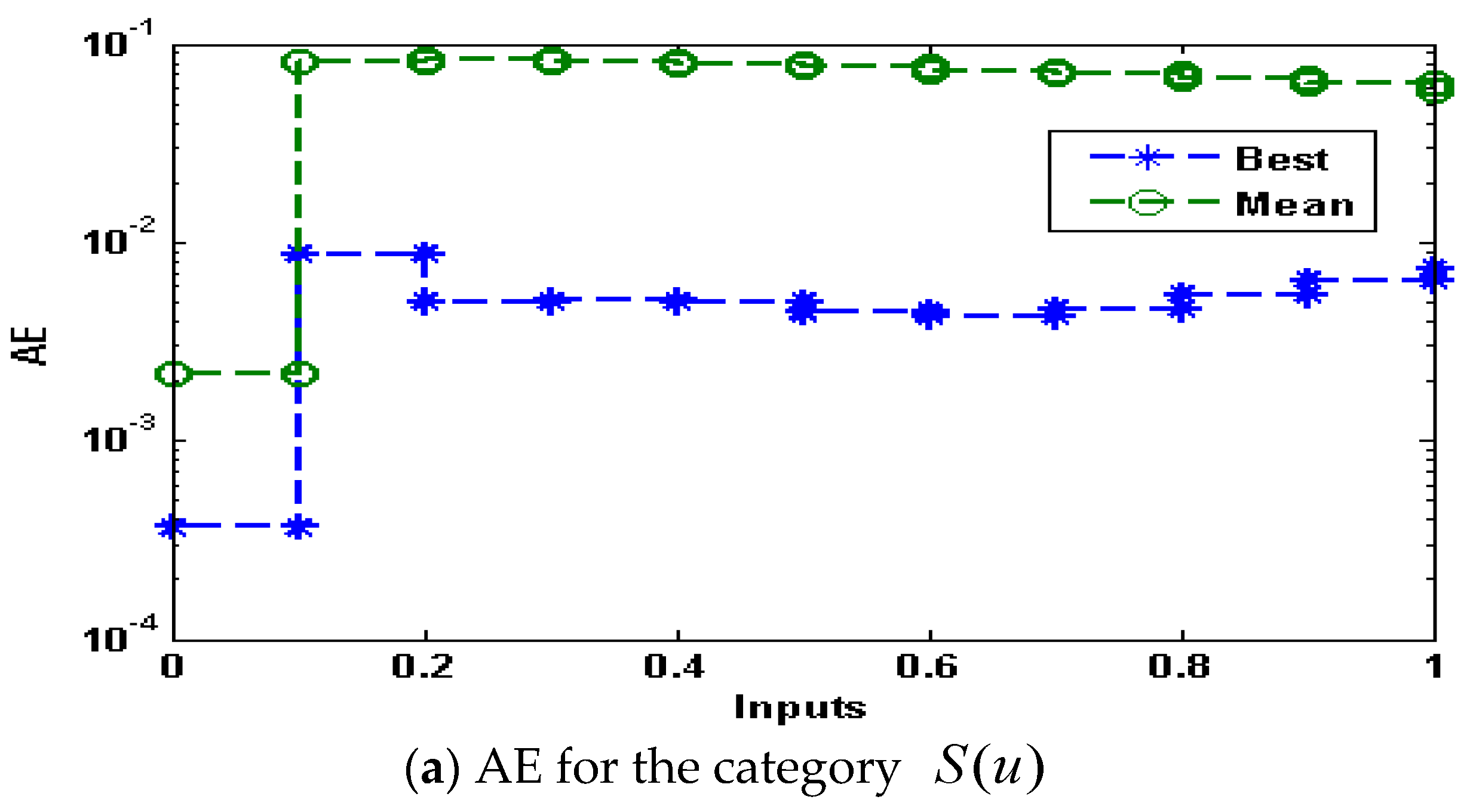
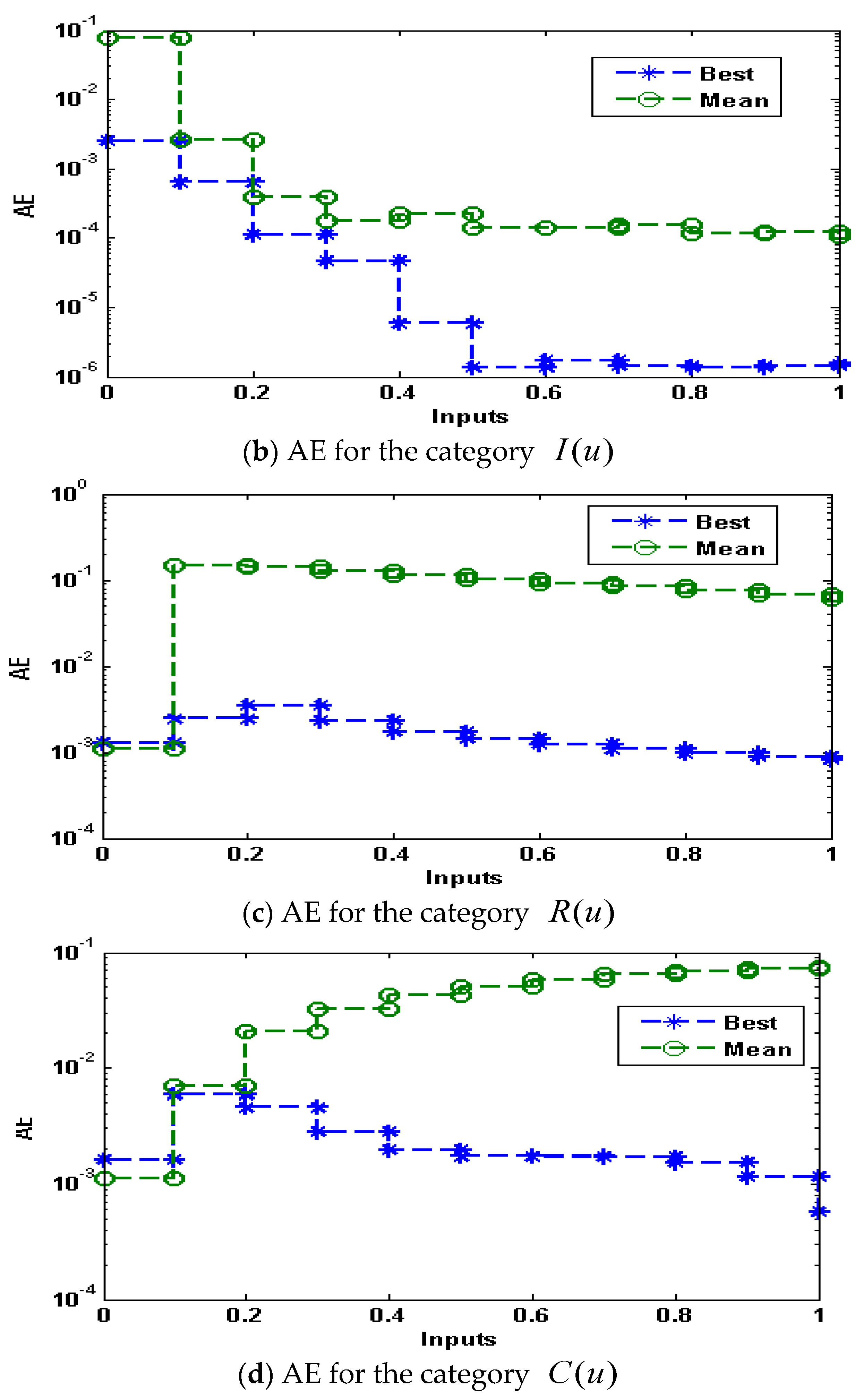
- The absolute error (AE) values are in the good agreements, indicating the reliability of the proposed ANN-GA-ASM.
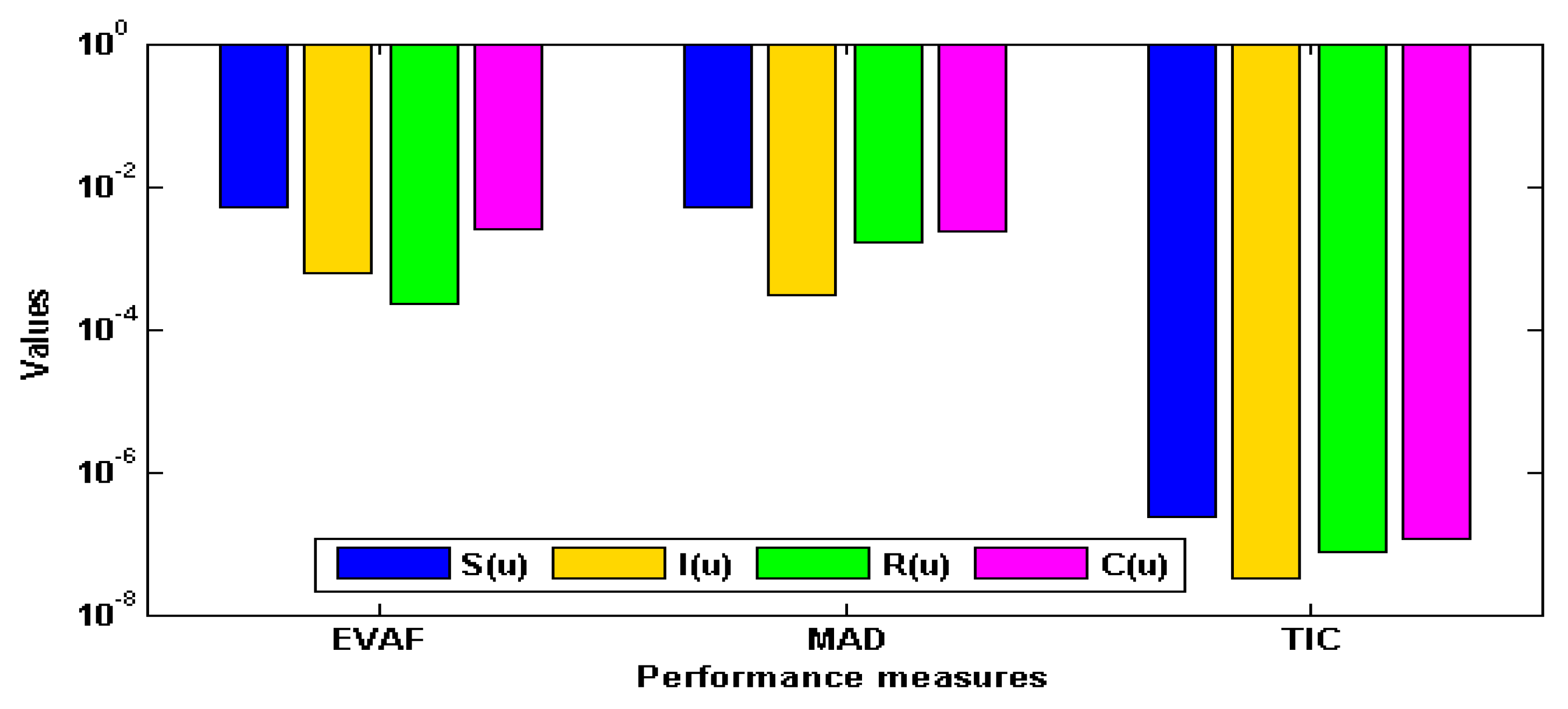
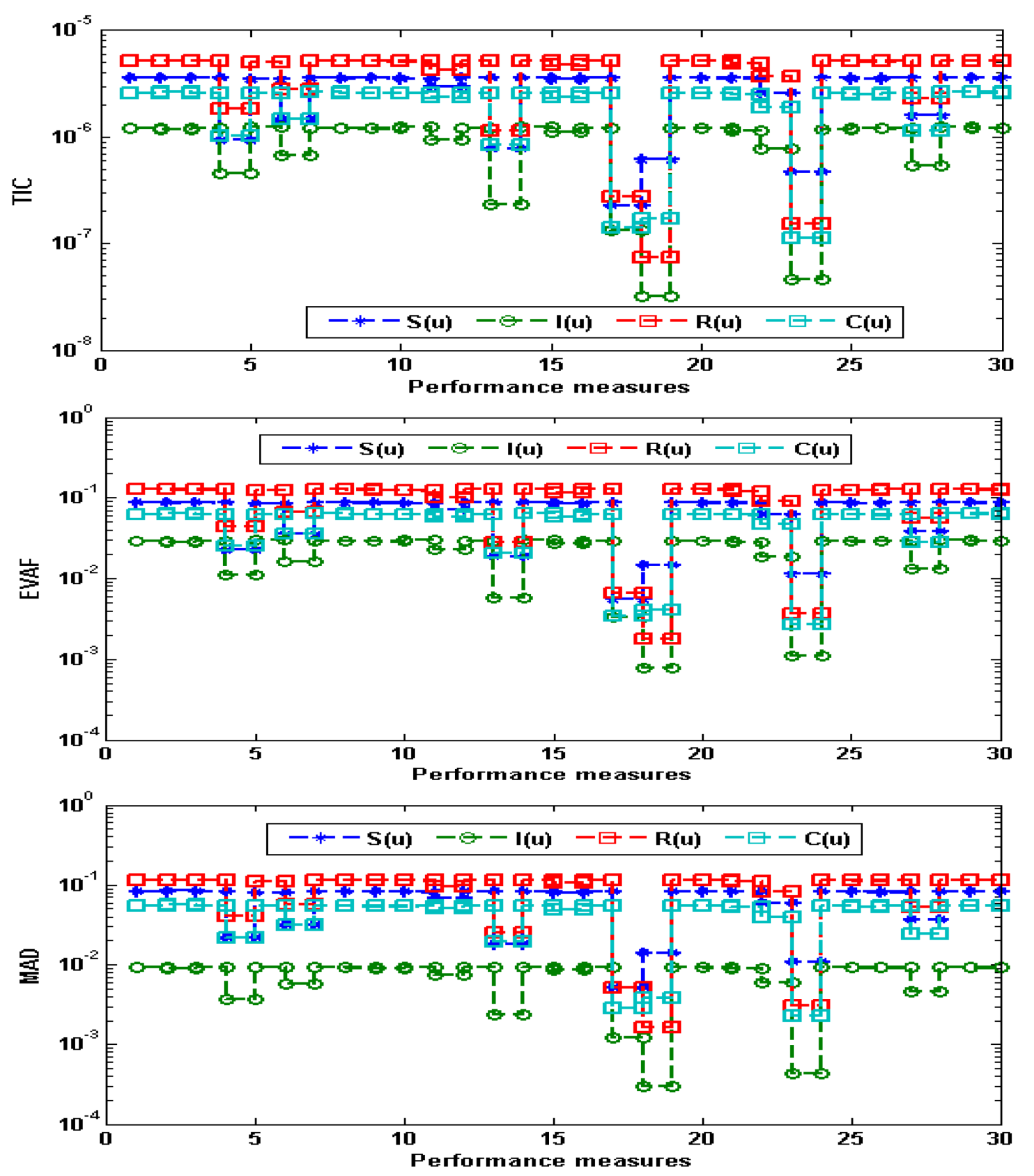
- The performance is certified through ANN-GA-ASM using different statistical observations to solve the nonlinear influenza system for thirty independent trials.
- The designed ANN-GA-ASM is effortlessly implemented to solve the nonlinear influenza system with smooth operations, inclusive and easy to understand.
2. Methodology
2.1. ANN Structure
2.2. Optimization Performances: GA-ASM
2.3. Performance Measures
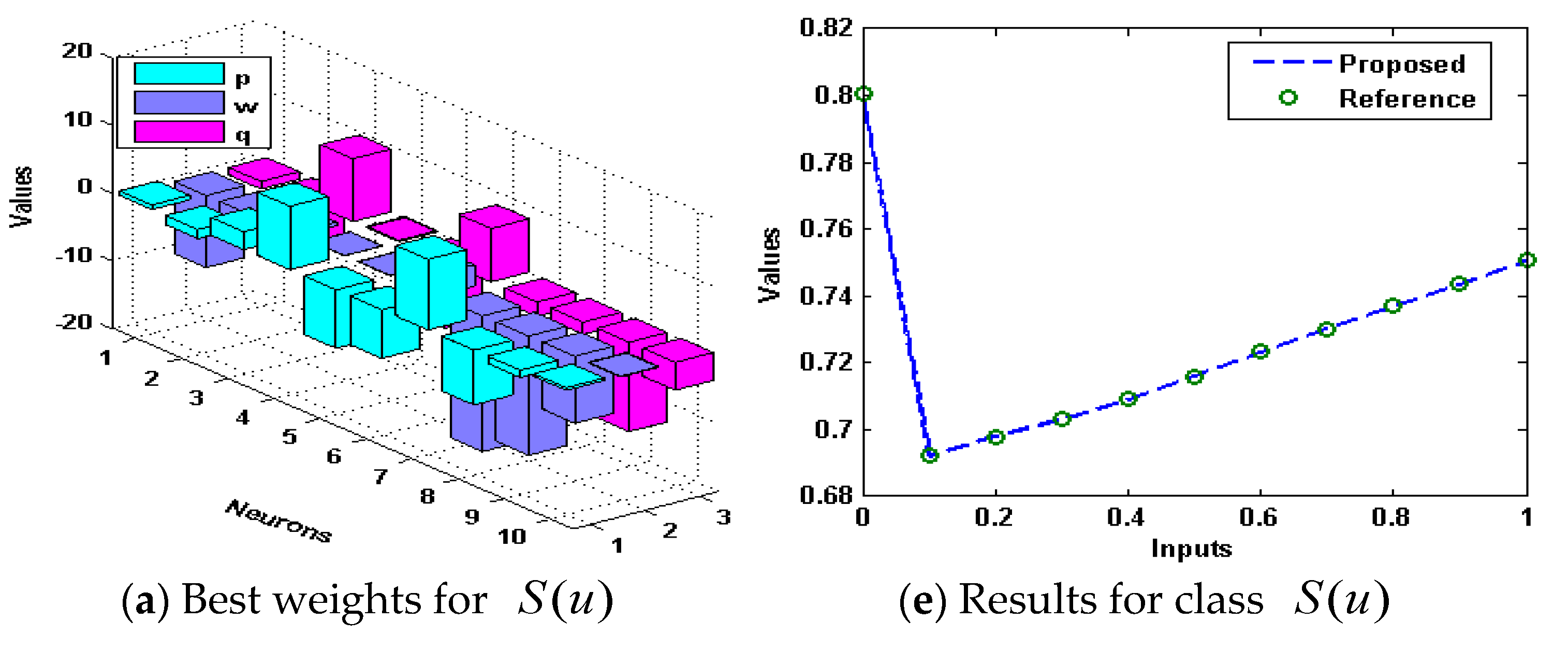
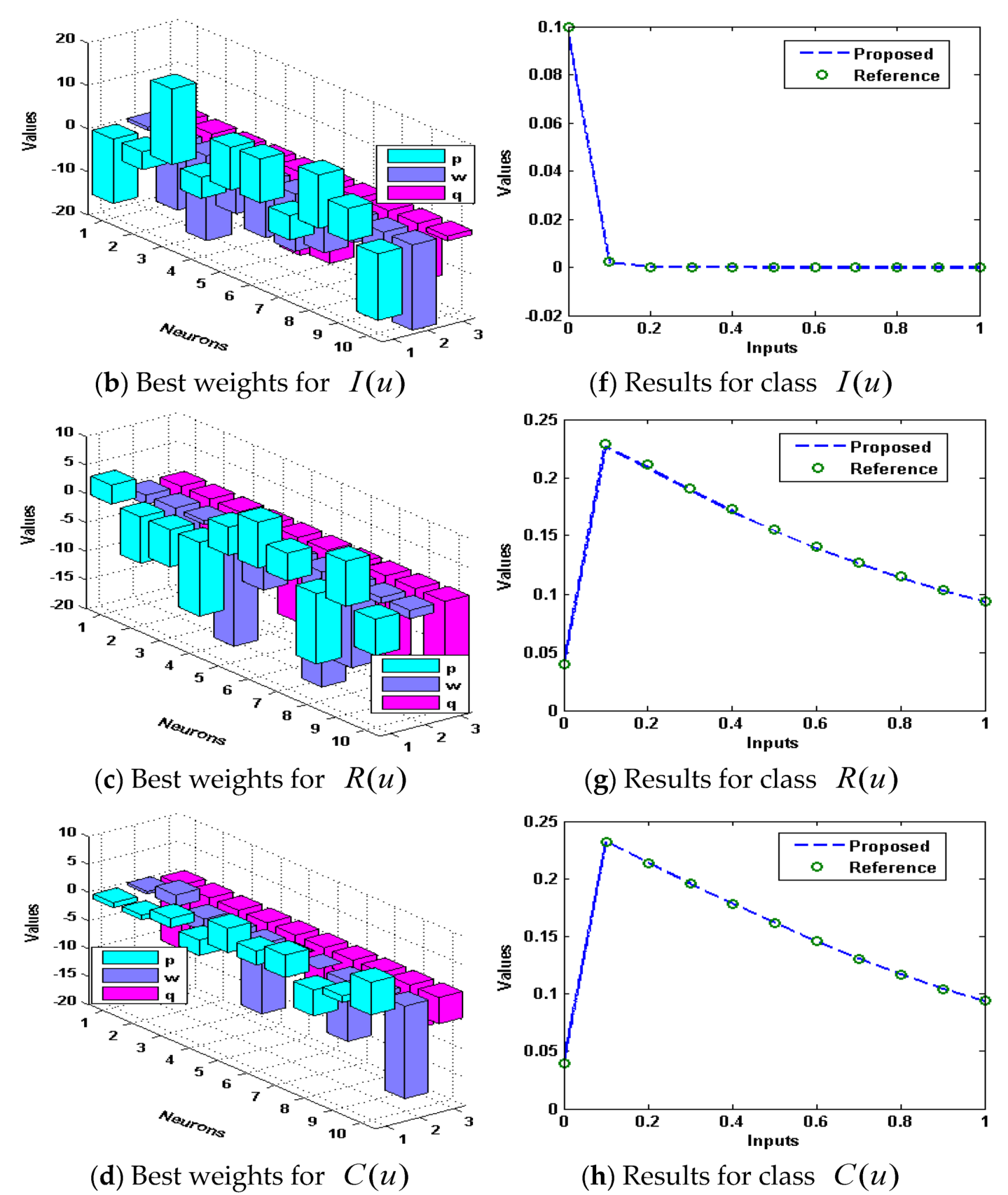
3. Simulations and Results
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Nomenclature
| ANN | Artificial Neural Networks |
| SIQR | Susceptible–infectious–quarantine–recovered |
| (SITR) | Susceptible–infectious–treatment–recovered |
| ANN-GA-ASM- | ANN optimized with GA-ASM |
| TIC | ‘Theil’s inequality coefficient |
| S.I.R | Semi interquartile |
| Min | Minimum |
| Med | Median |
| G.MAD | Mean of MAD |
| GA | Genetic Algorithms |
| ASM | Active-set Algorithm |
| GA-ASM | Hybrid of GA and ASM |
| MAD | Mean Absolute deviation |
| VAF | Variance account for |
| EVAF | Error in VAF |
| Max | Maximum |
| G.TIC | Mean of TIC |
| G.EVAF | Mean of EVAF |
Appendix A
Appendix B
Appendix C
References
- World Health Organization (WHO). Influenza Overview. Available online: http://www.who.int/mediacentre/factsheets/fs211/en/ (accessed on 16 May 2021).
- Astuti, F.; Suryanto, A.; Darti, I. Multi-step differential transform method for solving the influenza virus model with disease resistance. IOP Conf. Ser. Mater. Sci. Eng. 2019, 546, 052013. [Google Scholar] [CrossRef]
- Erdem, M.; Safan, M.; Castillo-Chavez, C. Mathematical analysis of an SIQR influenza model with imperfect quarantine. Bull. Math. Biol. 2017, 79, 1612–1636. [Google Scholar] [CrossRef] [PubMed]
- Alzahrani, E.O.; Khan, M.A. Comparison of numerical techniques for the solution of a fractional epidemic model. Eur. Phys. J. Plus 2020, 135, 110. [Google Scholar] [CrossRef]
- Sun, L.; DePuy, G.W.; Evans, G.W. Multi-objective optimization models for patient allocation during a pandemic influenza outbreak. Comput. Oper. Res. 2014, 51, 350–359. [Google Scholar] [CrossRef]
- Ghanbari, B.; Gómez-Aguilar, J.F. Analysis of two avian influenza epidemic models involving fractal-fractional derivatives with power and Mittag-Leffler memories. Chaos Interdiscip. J. Nonlinear Sci. 2019, 29, 123113. [Google Scholar] [CrossRef] [PubMed]
- Arenas, A.J.; González-Parra, G.; Chen-Charpentier, B.M. Construction of nonstandard finite difference schemes for the SI and SIR epidemic models of fractional order. Math. Comput. Simul. 2016, 121, 48–63. [Google Scholar] [CrossRef]
- Tchuenche, J.M.; Dube, N.; Bhunu, C.P.; Smith, R.J.; Bauch, C.T. The impact of media coverage on the transmission dynamics of human influenza. BMC Public Health 2011, 11, S5. [Google Scholar] [CrossRef] [Green Version]
- Schulze-Horsel, J.; Schulze, M.; Agalaridis, G.; Genzel, Y.; Reichl, U. Infection dynamics and virus-induced apoptosis in cell culture-based influenza vaccine production—Flow cytometry and mathematical modeling. Vaccine 2009, 27, 2712–2722. [Google Scholar] [CrossRef]
- Hovav, S.; Tsadikovich, D. A network flow model for inventory management and distribution of influenza vaccines through a healthcare supply chain. Oper. Res. Health Care 2015, 5, 49–62. [Google Scholar] [CrossRef]
- Patel, R.; Longini, I.M.; Halloran, M.E. Finding optimal vaccination strategies for pandemic influenza using genetic algorithms. J. Theor. Biol. 2005, 234, 201–212. [Google Scholar] [CrossRef]
- Kanyiri, C.W.; Luboobi, L.; Kimathi, M. Application of optimal control to influenza pneumonia coinfection with antiviral resistance. Comput. Math. Methods Med. 2020, 2020, 5984095. [Google Scholar] [CrossRef]
- Jódar, L.; Villanueva, R.J.; Arenas, A.J.; González, G.C. Nonstandard numerical methods for a mathematical model for influenza disease. Math. Comput. Simul. 2008, 79, 622–633. [Google Scholar] [CrossRef]
- Casagrandi, R.; Bolzoni, L.; Levin, S.A.; Andreasen, V. The SIRC model and influenza A. Math. Biosci. 2006, 200, 152–169. [Google Scholar] [CrossRef]
- Zachreson, C.; Fair, K.M.; Harding, N.; Prokopenko, M. Interfering with influenza: Nonlinear coupling of reactive and static mitigation strategies. J. R. Soc. Interface 2020, 17, 20190728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, X.; Yu, Y.; Meng, F.; Xu, Y. Modelling the dynamics of avian influenza with nonlinear recovery rate and psychological effect. J. Appl. Anal. Comput. 2020, 10, 1170–1192. [Google Scholar] [CrossRef]
- Chong, K.C.; Liang, J.; Jia, K.M.; Kobayashi, N.; Wang, M.H.; Wei, L.; Lau, S.Y.F.; Sumi, A. Latitudes mediate the association between influenza activity and meteorological factors: A nationwide modelling analysis in 45 Japanese prefectures from 2000 to 2018. Sci. Total Environ. 2020, 703, 134727. [Google Scholar] [CrossRef] [PubMed]
- Handel, A.; Liao, L.E.; Beauchemin, C.A. Progress and trends in mathematical modelling of influenza A virus infections. Curr. Opin. Syst. Biol. 2018, 12, 30–36. [Google Scholar] [CrossRef]
- McCarthy, Z.; Athar, S.; Alavinejad, M.; Chow, C.; Moyles, I.; Nah, K.; Kong, J.D.; Agrawal, N.; Jaber, A.; Keane, L.; et al. Quantifying the annual incidence and underestimation of seasonal influenza: A modelling approach. Theor. Biol. Med Model. 2020, 17, 11. [Google Scholar] [CrossRef] [PubMed]
- Sabir, Z.; Raja, M.A.Z.; Baleanu, D. Fractional mayer neuro-swarm heuristic solver for multi-fractional order doubly singular model based on lane–emden equation. Fractals 2021, 29, 2040033. [Google Scholar]
- Umar, M.; Sabir, Z.; Raja, M.A.Z.; Shoaib, M.; Gupta, M.; Sánchez, Y.G. A stochastic intelligent computing with neuro-evolution heuristics for nonlinear SITR system of novel COVID-19 dynamics. Symmetry 2020, 12, 1628. [Google Scholar] [CrossRef]
- Tao, Z.; Huiling, L.; Wenwen, W.; Xia, Y. GA-SVM based feature selection and parameter optimization in hospitalization expense modeling. Appl. Soft Comput. 2019, 75, 323–332. [Google Scholar] [CrossRef]
- Sayed, S.; Nassef, M.; Badr, A.; Farag, I. A Nested Genetic Algorithm for feature selection in high-dimensional cancer Microarray datasets. Expert Syst. Appl. 2019, 121, 233–243. [Google Scholar] [CrossRef]
- Hemanth, D.J.; Anitha, J. Modified Genetic Algorithm approaches for classification of abnormal Magnetic Resonance Brain tumour images. Appl. Soft Comput. 2019, 75, 21–28. [Google Scholar] [CrossRef]
- Armaghani, D.J.; Hasanipanah, M.; Mahdiyar, A.; Majid, M.Z.A.; Amnieh, H.B.; Tahir, M.M.D. Airblast prediction through a hybrid genetic algorithm-ANN model. Neural Comput. Appl. 2018, 29, 619–629. [Google Scholar] [CrossRef]
- Jiang, Y.; Wu, P.; Zeng, J.; Zhang, Y.; Zhang, Y.; Wang, S. Multi-parameter and multi-objective optimisation of articulated monorail vehicle system dynamics using genetic algorithm. Veh. Syst. Dyn. 2019, 58, 74–91. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, B.; Wang, S.; Liu, F.; Wang, Y.; Shu, X. A dynamic ant-colony genetic algorithm for cloud service composition optimization. Int. J. Adv. Manuf. Technol. 2019, 102, 355–368. [Google Scholar] [CrossRef]
- Hassoon, M.; Kouhi, M.S.; Zomorodi-Moghadam, M.; Abdar, M. Rule optimization of boosted C5.0 classification using genetic algorithm for liver disease prediction. In Proceedings of the 2017 International Conference on Computer and Applications (ICCA), Doha, Qatar, 6–7 September 2017; pp. 299–305. [Google Scholar]
- Quirynen, R.; Knyazev, A.; Di Cairano, S. Block structured preconditioning within an active-set method for real-time optimal control. In Proceedings of the 2018 European Control Conference (ECC), Limassol, Cyprus, 12–15 June 2018; pp. 1154–1159. [Google Scholar]
- Gao, Y.; Song, H.; Wang, X.; Zhang, K. Primal-dual active set method for pricing American better-of option on two assets. Commun. Nonlinear Sci. Numer. Simul. 2020, 80, 104976. [Google Scholar] [CrossRef]
- Deuerlein, J.W.; Piller, O.; Elhay, S.; Simpson, A.R. Content-based active-set method for the pressure-dependent model of water distribution systems. J. Water Resour. Plan. Manag. 2019, 145, 04018082. [Google Scholar] [CrossRef] [Green Version]
- Swathika, O.V.G.; Das, A.; Gupta, Y.; Mukhopadhyay, S.; Hemamalini, S.; Satapathy, S.C.; Bhateja, V.; Udgata, S.K.; Pattnaik, P.K. Optimization of overcurrent relays in microgrid using interior point method and active set method. In Proceedings of the Advances in Intelligent Systems and Computing; Springer Science and Business Media LLC: Singapore, 2017; Volume 516, pp. 89–97. [Google Scholar]
- Klaučo, M.; Kalúz, M.; Kvasnica, M. Machine learning-based warm starting of active set methods in embedded model predictive control. Eng. Appl. Artif. Intell. 2019, 77, 1–8. [Google Scholar] [CrossRef]
- Abide, S.; Barboteu, M.; Cherkaoui, S.; Danan, D.; Dumont, S. Inexact primal–dual active set method for solving elastodynamic frictional contact problems. Comput. Math. Appl. 2021, 82, 36–59. [Google Scholar] [CrossRef]
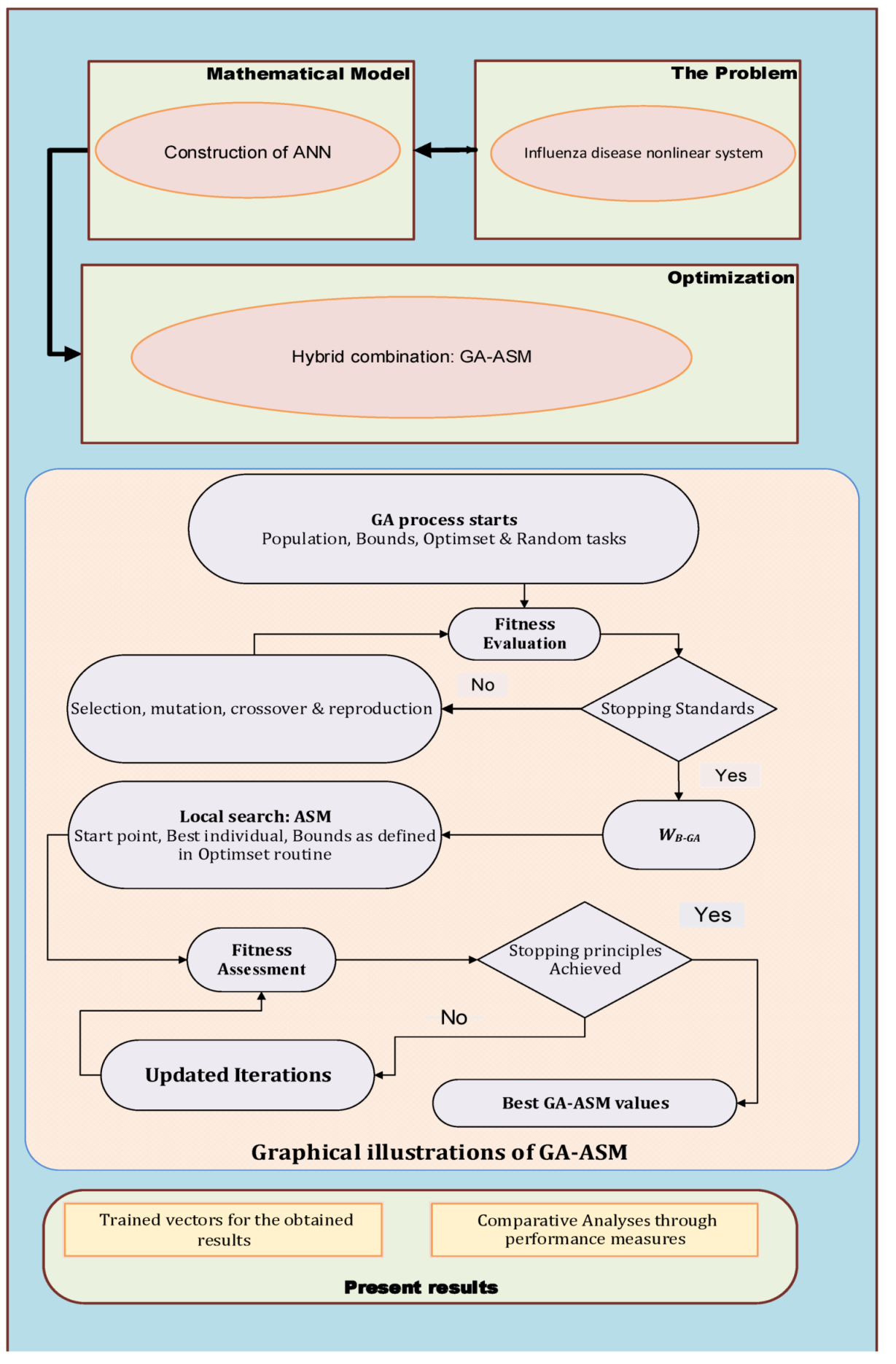
| GA process starts |
| Inputs: Select the chromosomes with the same numbers as decision variable of |
| ANN models for solving influenza disease system: |
| W = [a, w, b] |
| Population: The chromosomes set is given as: |
| , for , , |
| and |
| Output: Global vectors are WB-GA |
| Initialization: For variables selection, modify the weight vector. |
| Fitness calculation: Determine the fitness E of each member of the population |
| (P) using Equations (4)–(9) |
| • Stopping standards: Terminate if [E = 10−21], [TolCon = TolFun = 10−21]. |
| [StallLimit = 140], [PopSize = 210] and [Generations = 120] are achieved. |
| Move to storage |
| Ranking: Rank precise weight vector in the designated population based on |
| appropriateness on fitness E, i.e., better the fitness more be the rank of the |
| individual. |
| Storage: Save WB-GA, iterations, FIT, function counts and time. |
| End of GA |
| ASM |
| Inputs: WB-GA. |
| Output: WGA-ASM is the best GA-ASM weight vectors. |
| Initialize: WB-GA, iterations, assignments and other values. |
| Termination: Stop if [E = 10−20], [Iterations = 1200], [TolX = TolCon = 10−20] |
| [TolFun = 10−21] and [MaxFunEvals = 279,000] meets. |
| Evaluation of FIT: Calculate W and E using networks 4–9. |
| Adjustments: Adjust ‘fmincon’ for ASM, calculate E using 4–9 systems. |
| Accumulate: Transmute WGA-ASM, function counts, time iterations and FIT |
| ASM End |
| Min | Max | Med | Mean | S.I.R | SD | |
|---|---|---|---|---|---|---|
| 0 | 5.6403 × 10−5 | 1.0752 × 10−2 | 6.4347 × 10−4 | 2.1832 × 10−3 | 9.2647 × 10−4 | 3.1586 × 10−3 |
| 0.1 | 8.6933 × 10−3 | 1.0204 × 10−1 | 9.9764 × 10−2 | 8.0571 × 10−2 | 1.7817 × 10−2 | 3.2865 × 10−2 |
| 0.2 | 5.0136 × 10−3 | 1.0330 × 10−1 | 1.0171 × 10−2 | 8.2758 × 10−2 | 1.5101 × 10−2 | 3.3698 × 10−2 |
| 0.3 | 5.0721 × 10−3 | 1.0168 × 10−1 | 1.0031 × 10−2 | 8.1157 × 10−2 | 1.3434 × 10−2 | 3.3551 × 10−2 |
| 0.4 | 4.9908 × 10−3 | 1.0007 × 10−1 | 9.6958 × 10−2 | 7.9023 × 10−2 | 1.2838 × 10−2 | 3.2084 × 10−2 |
| 0.5 | 4.4433 × 10−3 | 9.7345 × 10−2 | 9.3189 × 10−2 | 7.6406 × 10−2 | 1.2362 × 10−2 | 3.0706 × 10−2 |
| 0.6 | 4.2461 × 10−3 | 9.3883 × 10−2 | 9.0095 × 10−2 | 7.3296 × 10−2 | 1.2040 × 10−2 | 2.9743 × 10−2 |
| 0.7 | 4.6397 × 10−3 | 8.9911 × 10−2 | 8.6530 × 10−2 | 6.9938 × 10−2 | 1.1763 × 10−2 | 2.8846 × 10−2 |
| 0.8 | 5.4428 × 10−3 | 8.5574 × 10−2 | 8.2248 × 10−2 | 6.6416 × 10−2 | 1.1654 × 10−2 | 2.7814 × 10−2 |
| 0.9 | 6.3978 × 10−3 | 8.0976 × 10−2 | 7.8024 × 10−2 | 6.2713 × 10−2 | 1.1469 × 10−2 | 2.6591 × 10−2 |
| 1 | 7.3410 × 10−3 | 7.6194 × 10−2 | 7.3517 × 10−2 | 5.8825 × 10−2 | 1.1127 × 10−2 | 2.5110 × 10−2 |
| Min | Max | Med | Mean | S.I.R | SD | |
|---|---|---|---|---|---|---|
| 0 | 2.5065 × 10−3 | 9.9741 × 10−2 | 9.6765 × 10−2 | 2.1832 × 10−3 | 1.8123 × 10−2 | 3.2534 × 10−2 |
| 0.1 | 2.0358 × 10−4 | 3.8280 × 10−3 | 2.6376 × 10−3 | 8.0571 × 10−2 | 2.9036 × 10−4 | 7.8410 × 10−4 |
| 0.2 | 1.0562 × 10−4 | 1.5922 × 10−3 | 2.8609 × 10−4 | 8.2758 × 10−2 | 9.2734 × 10−5 | 3.2756 × 10−4 |
| 0.3 | 7.9627 × 10−6 | 7.5276 × 10−4 | 1.4348 × 10−4 | 8.1157 × 10−2 | 1.0830 × 10−4 | 1.7174 × 10−4 |
| 0.4 | 5.8824 × 10−6 | 1.1768 × 10−3 | 1.4911 × 10−4 | 7.9023 × 10−2 | 6.2778 × 10−5 | 2.5473 × 10−4 |
| 0.5 | 5.2022 × 10−7 | 8.4523 × 10−4 | 8.5497 × 10−5 | 7.6406 × 10−2 | 5.2659 × 10−5 | 1.7651 × 10−4 |
| 0.6 | 3.9676 × 10−7 | 9.6438 × 10−4 | 6.6933 × 10−5 | 7.3296 × 10−2 | 5.8891 × 10−5 | 1.9772 × 10−4 |
| 0.7 | 1.4420 × 10−6 | 1.0347 × 10−3 | 8.1183 × 10−5 | 6.9938 × 10−2 | 5.7567 × 10−5 | 2.2108 × 10−4 |
| 0.8 | 1.3782 × 10−6 | 1.0473 × 10−3 | 6.5095 × 10−5 | 6.6416 × 10−2 | 5.3914 × 10−5 | 1.9876 × 10−4 |
| 0.9 | 1.0788 × 10−6 | 9.9345 × 10−4 | 6.9847 × 10−5 | 6.2713 × 10−2 | 4.8355 × 10−5 | 1.8417 × 10−4 |
| 1 | 3.7397 × 10−7 | 8.6629 × 10−4 | 6.3295 × 10−5 | 5.8825 × 10−2 | 3.6254 × 10−5 | 1.5961 × 10−4 |
| Min | Max | Med | Mean | S.I.R | SD | |
|---|---|---|---|---|---|---|
| 0 | 6.2366 × 10−6 | 1.4851 × 10−2 | 1.8505 × 10−4 | 2.1832 × 10−3 | 3.8502 × 10−4 | 2.9318 × 10−3 |
| 0.1 | 2.4607 × 10−3 | 1.9038 × 10−1 | 1.8634 × 10−1 | 8.0571 × 10−2 | 3.2715 × 10−2 | 6.2894 × 10−2 |
| 0.2 | 2.3532 × 10−3 | 1.7746 × 10−1 | 1.7489 × 10−1 | 8.2758 × 10−2 | 2.4669 × 10−2 | 5.9498 × 10−2 |
| 0.3 | 2.3392 × 10−3 | 1.6039 × 10−1 | 1.5817 × 10−1 | 8.1157 × 10−2 | 2.0756 × 10−2 | 5.4314 × 10−2 |
| 0.4 | 1.7447 × 10−3 | 1.4608 × 10−1 | 1.4106 × 10−1 | 7.9023 × 10−2 | 1.8914 × 10−2 | 4.8288 × 10−2 |
| 0.5 | 1.4329 × 10−3 | 1.3259 × 10−1 | 1.2623 × 10−1 | 7.6406 × 10−2 | 1.7328 × 10−2 | 4.3191 × 10−2 |
| 0.6 | 6.3834 × 10−4 | 1.1983 × 10−1 | 1.1406 × 10−1 | 7.3296 × 10−2 | 1.5985 × 10−2 | 3.9221 × 10−2 |
| 0.7 | 1.0809 × 10−3 | 1.0806 × 10−1 | 1.0395 × 10−1 | 6.9938 × 10−2 | 1.4990 × 10−2 | 3.5981 × 10−2 |
| 0.8 | 9.6811 × 10−4 | 9.7331 × 10−2 | 9.4041 × 10−2 | 6.6416 × 10−2 | 1.3740 × 10−2 | 3.3132 × 10−2 |
| 0.9 | 6.8779 × 10−4 | 8.7620 × 10−2 | 8.5302 × 10−2 | 6.2713 × 10−2 | 1.2596 × 10−2 | 3.0469 × 10−2 |
| 1 | 3.8749 × 10−4 | 7.8853 × 10−2 | 7.7195 × 10−2 | 5.8825 × 10−2 | 1.0930 × 10−2 | 2.7874 × 10−2 |
| Min | Max | Med | Mean | S.I.R | SD | |
|---|---|---|---|---|---|---|
| 0 | 1.1372 × 10−5 | 6.9224 × 10−3 | 2.1165 × 10−4 | 1.1227 × 10−3 | 6.5694 × 10−4 | 1.8441 × 10−3 |
| 0.1 | 1.5570 × 10−4 | 1.5199 × 10−2 | 7.0904 × 10−3 | 7.0882 × 10−3 | 3.7494 × 10−4 | 2.5091 × 10−3 |
| 0.2 | 2.3633 × 10−3 | 2.9209 × 10−2 | 2.4420 × 10−2 | 2.0540 × 10−2 | 4.4120 × 10−3 | 7.6724 × 10−3 |
| 0.3 | 2.5936 × 10−4 | 4.1491 × 10−2 | 3.9420 × 10−2 | 3.2301 × 10−2 | 7.0413 × 10−3 | 1.3088 × 10−2 |
| 0.4 | 1.8337 × 10−4 | 5.4041 × 10−2 | 5.1929 × 10−2 | 4.2548 × 10−2 | 8.7280 × 10−3 | 1.7298 × 10−2 |
| 0.5 | 1.6757 × 10−3 | 6.4379 × 10−2 | 6.2491 × 10−2 | 5.1205 × 10−2 | 9.0134 × 10−3 | 2.0582 × 10−2 |
| 0.6 | 1.7138 × 10−3 | 7.3162 × 10−2 | 7.1056 × 10−2 | 5.8351 × 10−2 | 9.3617 × 10−3 | 2.3249 × 10−2 |
| 0.7 | 1.6948 × 10−3 | 8.0311 × 10−2 | 7.8098 × 10−2 | 6.4106 × 10−2 | 9.7669 × 10−3 | 2.5506 × 10−2 |
| 0.8 | 1.5227 × 10−3 | 8.6049 × 10−2 | 8.3577 × 10−2 | 6.8593 × 10−2 | 1.0268 × 10−2 | 2.7492 × 10−4 |
| 0.9 | 1.1457 × 10−3 | 9.0577 × 10−2 | 8.7617 × 10−2 | 7.1938 × 10−2 | 1.0876 × 10−2 | 2.9302 × 10−2 |
| 1 | 5.6992 × 10−4 | 9.4077 × 10−2 | 9.1159 × 10−2 | 7.4264 × 10−2 | 1.1513 × 10−2 | 3.1009 × 10−2 |
| Index | (G-MAD) | (G-TIC) | (G-EVAF) | |||
|---|---|---|---|---|---|---|
| Min | S.I.R | Min | S.I.R | Min | S.I.R | |
| 8.2308 × 10−2 | 1.1452 × 10−2 | 3.5709 × 10−6 | 5.0771 × 10−7 | 9.5011 × 10−1 | 2.7588 × 10−1 | |
| 9.1646 × 10−3 | 1.6069 × 10−3 | 1.2032 × 10−6 | 2.2481 × 10−7 | 9.3349 × 10−1 | 2.9071 × 10−1 | |
| 1.1467 × 10−2 | 1.6332 × 10−2 | 5.1525 × 10−6 | 7.5221 × 10−8 | 8.7737 × 10−1 | 2.0805 × 10−2 | |
| 5.4491 × 10−2 | 7.7396 × 10−3 | 2.5843 × 10−7 | 3.4476 × 10−7 | 1.0349 × 10−2 | 2.1164 × 10−1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sabir, Z.; Ag Ibrahim, A.A.; Raja, M.A.Z.; Nisar, K.; Umar, M.; Rodrigues, J.J.P.C.; Mahmoud, S.R. Soft Computing Paradigms to Find the Numerical Solutions of a Nonlinear Influenza Disease Model. Appl. Sci. 2021, 11, 8549. https://doi.org/10.3390/app11188549
Sabir Z, Ag Ibrahim AA, Raja MAZ, Nisar K, Umar M, Rodrigues JJPC, Mahmoud SR. Soft Computing Paradigms to Find the Numerical Solutions of a Nonlinear Influenza Disease Model. Applied Sciences. 2021; 11(18):8549. https://doi.org/10.3390/app11188549
Chicago/Turabian StyleSabir, Zulqurnain, Ag Asri Ag Ibrahim, Muhammad Asif Zahoor Raja, Kashif Nisar, Muhammad Umar, Joel J. P. C. Rodrigues, and Samy R. Mahmoud. 2021. "Soft Computing Paradigms to Find the Numerical Solutions of a Nonlinear Influenza Disease Model" Applied Sciences 11, no. 18: 8549. https://doi.org/10.3390/app11188549
APA StyleSabir, Z., Ag Ibrahim, A. A., Raja, M. A. Z., Nisar, K., Umar, M., Rodrigues, J. J. P. C., & Mahmoud, S. R. (2021). Soft Computing Paradigms to Find the Numerical Solutions of a Nonlinear Influenza Disease Model. Applied Sciences, 11(18), 8549. https://doi.org/10.3390/app11188549








