Characterization of Lactic Acid Bacteria in Pecorino di Farindola Cheese and Manufacturing with a Lacticaseibacillus paracasei Autochthonous Culture
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. DNA Extraction
2.3. PCR Reactions
2.4. Sequencing
2.5. Test of LAB Adhesion to CaCo-2 Cells
2.6. Experimental Production of Pecorino di Farindola Cheese
2.7. Statistical Analyses
3. Results
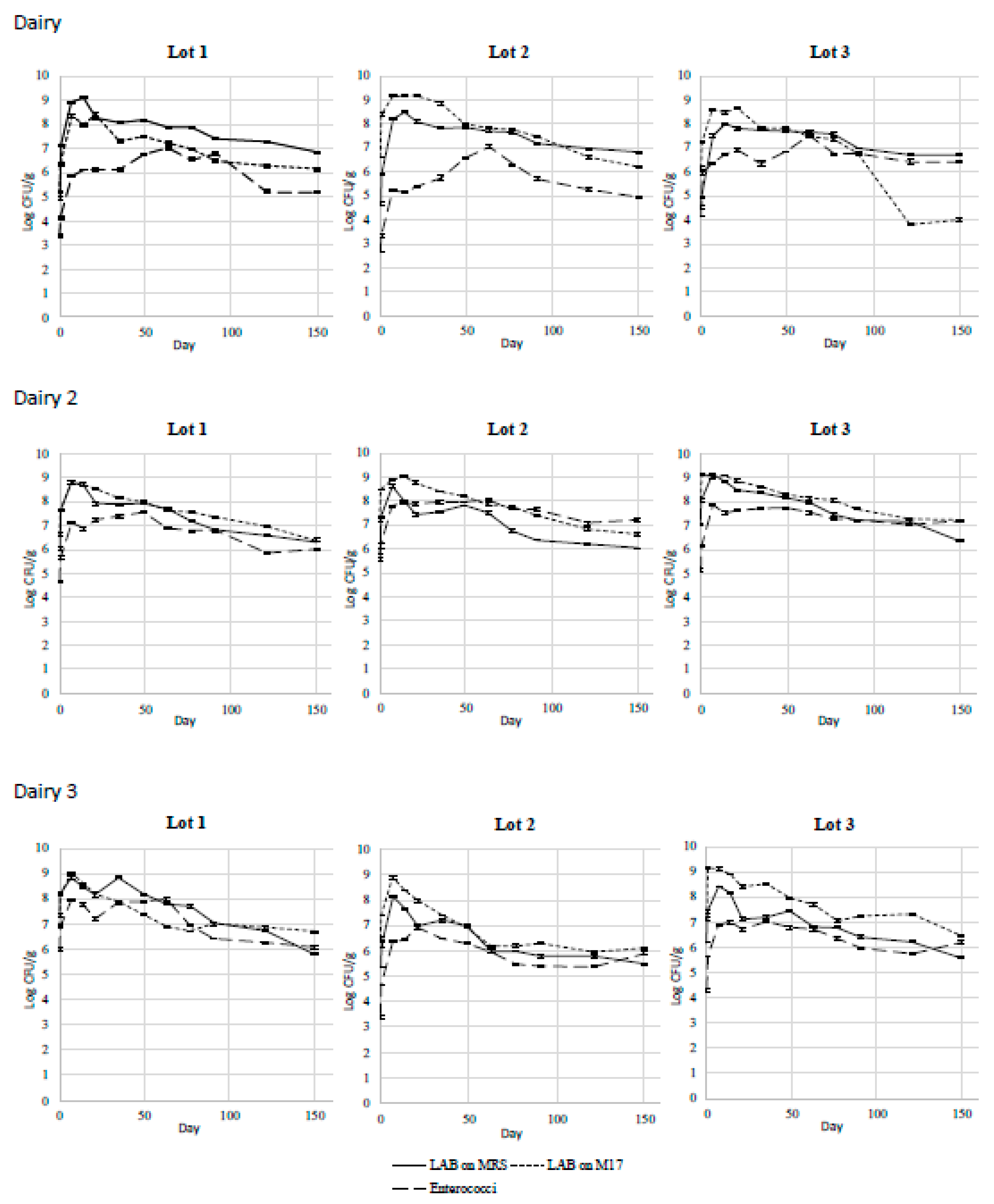
3.1. LAB Dynamics in Pecorino di Farindola Cheese
3.2. Identification of Dominant LAB in Pecorino di Farindola Cheese
3.3. Evaluation of the Prerequisites for Survival in GIT
3.4. LAB Activity against Pathogenic Bacteria
3.5. Cheese Making Trial with the Addition of L. paracasei L11121 Isolate
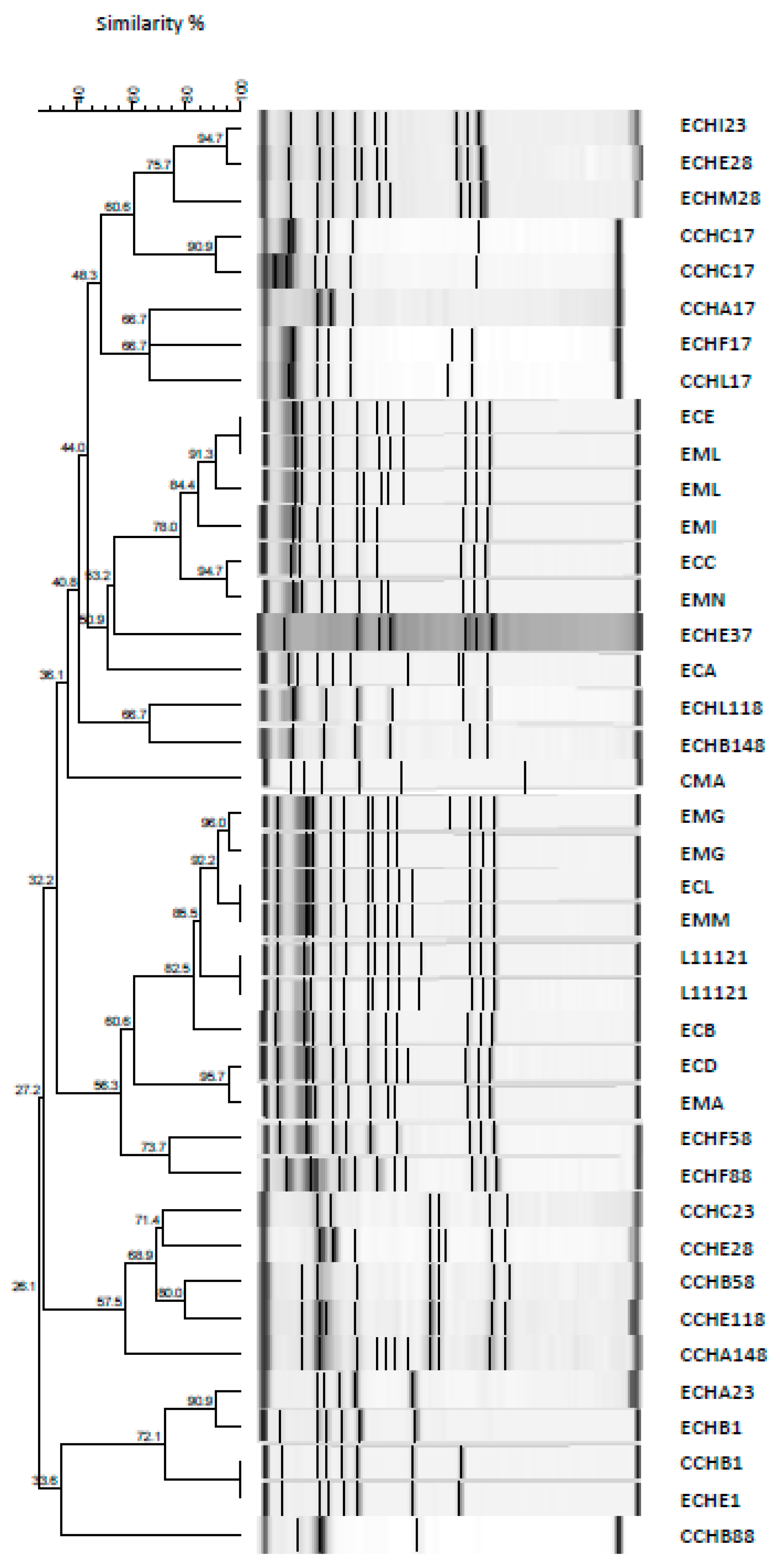
3.6. Rep PCR for L. paracasei L11121 Tracing during Cheese Maturation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Braghieri, A.; Girolami, A.; Riviezzi, A.M.; Piazzolla, N.; Napolitano, F. Liking of traditional cheese and consumer willingness to pay. Ital. J. Anim. Sci. 2014, 13, 155–162. [Google Scholar] [CrossRef]
- European Community. European Guide for Good Hygiene Practices in the Production of Artisanal Cheese and Dairy Products. 2017. Available online: https://ec.europa.eu/food/system/files/2017-12/biosafety_fh_guidance_artisanal-cheese-and-dairy-products_en.pdf (accessed on 12 July 2021).
- Italian Ministry of Agriculture, Food and Forestry (MIPAAF). Ventunesima revisione dell’elenco dei prodotti agroalimentari tradizionali. GU-Ser. Gen. 2021, 48, ord n 15. [Google Scholar]
- Consorzio di Produzione del Pecorino di Farindola. Disciplinare di Produzione del PECORINO di Farindola. Available online: https://www.pecorinodifarindola.it/disciplinare/ (accessed on 12 July 2021).
- Schirone, M.; Tofalo, R.; Mazzone, G.; Corsetti, A.; Suzzi, G. Biogenic amine content and microbiological profile of Pecorino di Farindola cheese. Food Microbiol. 2011, 28, 128–136. [Google Scholar] [CrossRef] [PubMed]
- Tofalo, R.; Perpetuini, G.; Battistelli, N.; Pepe, A.; Ianni, A.; Martino, G.; Suzzi, G. Accumulation γ-aminobutyric acid and biogenic amines in a traditional raw milk ewe’s cheese. Foods 2019, 8, 401. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Velázquez, R.; Salvador-Figueroa, M.; Adriano-Anaya, L.; DeGyves–Córdova, G.; Vázquez-Ovando, A. Use of starter culture of native lactic acid bacteria for producing an artisanal Mexican cheese safe and sensory acceptable. CyTA—J. Food 2018, 16, 460–468. [Google Scholar] [CrossRef]
- Amadoro, C.; Rossi, F.; Pallotta, M.L.; Gasperi, M.; Colavita, G. Traditional dairy products can supply beneficial microorganisms able to survive in the gastrointestinal tract. LWT 2018, 93, 376–383. [Google Scholar] [CrossRef]
- van Staden, A.D.P.; van Zyl, W.F.; Trindade, M.; Dicks, L.M.T.; Smith, C. Therapeutic application of lantibiotics and other lanthipeptides: Old and new findings. Appl. Environ. Microbiol. 2021, 87, AEM.00186-21. [Google Scholar] [CrossRef]
- European Community. Commission Regulation (EC). No 2073/2005 of 15 November 2005 on Microbiological Criteria for Food-Stuffs (Text with EEA Relevance). Off. J. Eur. Union L 2005, 338, 1–26. Available online: https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A32005R2073 (accessed on 10 July 2021).
- Costanzo, N.; Ceniti, C.C.; Santoro, A.; Clausi, M.T.; Casalinuovo, F. Foodborne pathogen assessment in raw milk cheeses. Int. J. Food Sci. 2020, 2020, 3616713. [Google Scholar] [CrossRef]
- ISO 707:2008 [IDF 50:2008]. Milk and Milk Products—Guidance on Sampling; International Organization for Standardization: Geneva, Switzerland, 2008.
- ISO 18787:2017. Foodstuffs—Determination of Water Activity; International Organization for Standardization: Geneva, Switzerland, 2017.
- Del Matto, I.; Rossi, F.; Iannitto, G.; Petrone, D.; Mastrodomenico, M.T.; Alessiani, A.; Sacchini, L.; Amadoro, C.; Tucci, P.; Marino, L. Variability of the microbiota in traditional Caciocavallo, Scamorza and Caciotta cheeses manufactured with raw milk and natural cultures. Int. J. Dairy Technol. 2021, 74, 564–574. [Google Scholar] [CrossRef]
- ISO 6888-2:1999. Microbiology of Food and Animal Feeding Stuffs. Horizontal Method for the Enumeration of Coagulase-Positive Staphylococci (Staphylococcus Aureus and Other Species)—Part 2: Technique Using Rabbit Plasma Fibrinogen Agar Medium; International Organization for Standardization: Geneva, Switzerland, 1999.
- UNI EN ISO 16654:2017. Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Detection of Escherichia Coli O157; International Organization for Standardization: Geneva, Switzerland, 2017.
- UNI EN ISO 11290-1:2017. Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes and of Listeria spp.—Part 1: Detection Method; International Organization for Standardization: Geneva, Switzerland, 2017.
- ISO 16649-1:2001. Microbiology of Food and Animal Feeding Stuffs—Horizontal Method for the Enumeration of Beta-Glucuronidase-Positive Escherichia Coli—Part 1: Colony-Count Technique at 44 Degrees C Using Membranes and 5-Bromo-4-Chloro-3-Indolyl Beta-D-Glucuronide; International Organization for Standardization: Geneva, Switzerland, 2001.
- UNI EN ISO 11290-2:2017. Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes and of LISTERIA spp.—Part 2: Enumeration Method; International Organization for Standardization: Geneva, Switzerland, 2017.
- Ward, L.J.H.; Timmins, M.J. Differentiation of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus by polymerase chain reaction. Lett. Appl. Microbiol. 1999, 29, 90–92. [Google Scholar] [CrossRef]
- Li, H.; O’Sullivan, D.J. Identification of a nisI promoter within the nisABCTIP operon that may enable establishment of nisin immunity prior to induction of the operon via signal transduction. J. Bacteriol. 2006, 188, 8496–8503. [Google Scholar] [CrossRef]
- Kariyama, R.; Mitsuhata, R.; Chow, J.W.; Clewell, D.B.; Kumon, H. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J. Clin. Microbiol. 2000, 38, 3092–3095. [Google Scholar] [CrossRef]
- Versalovic, J.; Schneider, M.; de Bruijn, F.J.; Lupski, J.R. Genomic fingerprinting of bacteria using repetitive sequence-based Polymerase Chain Reaction. Met. Mol. Cell. Biol. 1994, 5, 25–40. [Google Scholar]
- Ren, D.; Li, C.; Qin, Y.; Yin, R.; Du, S.; Ye, F.; Liu, C.; Liu, H.; Wang, M.; Li, Y.; et al. In vitro evaluation of the probiotic and functional potential of Lactobacillus strains isolated from fermented food and human intestine. Anaerobe 2014, 30, 1–10. [Google Scholar] [CrossRef] [PubMed]
- European Union Reference Laboratory for Listeria monocytogenes (EURL Lm). Technical Guidance Document for Conducting Shelf Life Studies on Listeria Monocytogenes in Ready to Eat Foods. Version 3–6 June. 2014, pp. 1–47. Available online: https://www.fsai.ie/uploadedFiles/EURL%20Lm_Technical%20Guidance%20Document%20Lm%20shelf-life%20studies_V3_2014-06-06%20(2).pdf (accessed on 10 July 2021).
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. Past: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. Available online: https://palaeo-electronica.org/2001_1/past/past.pdf (accessed on 11 August 2021).
- Rossi, F.; Veneri, G. Use of bacteriocinogenic cultures without inhibiting cheese associated Nonstarter Lactic Acid Bacteria; a trial with Lactobacillus plantarum. Challenges 2016, 7, 4. [Google Scholar] [CrossRef]
- Ladero, V.; Fernández, M.; Calles-Enríquez, M.; Sánchez-Llana, E.; Cañedo, E.; Martín, M.C.; Alvarez, M.A. Is the production of the biogenic amines tyramine and putrescine a species-level trait in enterococci? Food Microbiol. 2012, 30, 132–138. [Google Scholar] [CrossRef]
- Tuomola, E.M.; Salminen, S.J. Adhesion of some probiotic and dairy Lactobacillus strains to Caco-2 cell cultures. Int. J. Food Microbiol. 1998, 41, 45–51. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Rodríguez-Aparicio, L.; Rúa, J.; Martínez-Blanco, H.; Navasa, N.; García-Armesto, M.R.; Ferrero, M.Á. In vitro evaluation of physiological probiotic properties of different lactic acid bacteria strains of dairy and human origin. J. Funct. Foods 2012, 4, 531–541. [Google Scholar] [CrossRef]
- EFSA Panel on Biological Hazards (BIOHAZ); Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; et al. Update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA 12: Suitability of taxonomic units notified to EFSA until March 2020. EFSA J. 2020, 18, e06174. [Google Scholar] [CrossRef]
- Rasschaert, G.; Houf, K.; Imberechts, H.; Grijspeerdt, K.; De Zutter, L.; Heyndrickx, M. Comparison of five repetitive-sequence-based PCR typing methods for molecular discrimination of Salmonella enterica isolates. J. Clin. Microbiol. 2005, 43, 3615–3623. [Google Scholar] [CrossRef]
- Tzora, A.; Nelli, A.; Voidarou, C.; Fthenakis, G.; Rozos, G.; Theodorides, G.; Bonos, E.; Skoufos, I. Microbiota “Fingerprint” of Greek Feta Cheese through Ripening. Appl. Sci. 2021, 11, 5631. [Google Scholar] [CrossRef]
- Kant, R.; Uroić, K.; Hynönen, U.; Kos, B.; Šušković, J.; Palva, A. Genome sequence of Lactobacillus brevis strain D6, isolated from smoked fresh cheese. Genome Announc. 2016, 4, e00264-16. [Google Scholar] [CrossRef] [PubMed]
- Kariyawasam, K.M.G.M.M.; Yang, S.J.; Lee, N.-K.; Paik, H.-D. Probiotic properties of Lactobacillus brevis KU200019 and synergistic activity with fructooligosaccharides in antagonistic activity against foodborne pathogens. Food Sci. Anim. Resour. 2020, 40, 297–310. [Google Scholar] [CrossRef]
- Rossi, F.; Rizzotti, L.; Felis, G.E.; Torriani, S. Horizontal gene transfer among microorganisms in food: Current knowledge and future perspectives. Food Microbiol. 2014, 42, 232–243. [Google Scholar] [CrossRef]
- Dapkevicius, M.d.L.E.; Sgardioli, B.; Câmara, S.P.A.; Poeta, P.; Malcata, F.X. Current trends of enterococci in dairy products: A comprehensive review of their multiple roles. Foods 2021, 10, 821. [Google Scholar] [CrossRef]
- Albayrak, Ç.B.; Duran, M. Isolation and characterization of aroma producing lactic acid bacteria from artisanal white cheese for multifunctional properties. LWT 2021, 150, 112053. [Google Scholar] [CrossRef]
- Cocconcelli, P.S.; Cattivelli, D.; Gazzola, S. Gene transfer of vancomycin and tetracycline resistances among Enterococcus faecalis during cheese and sausage fermentations. Int. J. Food Microbiol. 2003, 88, 315–323. [Google Scholar] [CrossRef]
- Gazzola, S.; Fontana, C.; Bassi, D.; Cocconcelli, P.S. Assessment of tetracycline and erythromycin resistance transfer during sausage fermentation by culture-dependent and -independent methods. Food Microbiol. 2012, 30, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Malek, A.; De la Hoz, A.; Gomez-Villegas, S.I.; Nowbakht, C.; Arias, C.A. Lactococcus garvieae, an unusual pathogen in infective endocarditis: Case report and review of the literature. BMC Infect. Dis. 2019, 19, 301. [Google Scholar] [CrossRef]
- Gboko, K.D.T.; Traoré, S.G.; Sanhoun, A.R.; Kirioua, J.; Otaru, N.; Kurt, F.; Jaeger, F.N.; Isenring, J.; Kaindi, D.W.M.; Kreikemeyer, B.; et al. Risk factors for the carriage of Streptococcus infantarius subspecies infantarius isolated from African fermented dairy products. PLoS ONE 2019, 14, e0225452. [Google Scholar] [CrossRef]
- Abdelfatah, E.N.; Mahboub, H.H.H. Studies on the effect of Lactococcus garvieae of dairy origin on both cheese and Nile tilapia (O. niloticus). Int. J. Vet. Sci. Med. 2018, 6, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Özkan, E.R.; Öztürk, H.İ.; Demirci, T.; Akın, N. Detection of biofilm formation, virulence factors genes, antibiotic-resistance, adherence properties and some beneficial properties of cheese-related S. infantarius, S. gallolyticus, and S. lutetiensis strains belonging to the S. bovis/S. equinus complex. LWT 2021, 150, 112077. [Google Scholar] [CrossRef]
- Stefanovic, E.; Kilcawley, K.N.; Roces, C.; Rea, M.C.; O’Sullivan, M.; Sheehan, J.J.; McAuliffe, O. Evaluation of the potential of Lactobacillus paracasei adjuncts for flavor compounds development and diversification in short-aged Cheddar cheese. Front. Microbiol. 2018, 9, 1506. [Google Scholar] [CrossRef] [PubMed]
- Centorotola, G.; Salini, R.; Sperandii, A.F.; Neri, D.; Tucci, P.; Santarelli, G.A.; Di Marzio, V.; Romantini, R.; Candeloro, L.; Conte, A.; et al. Validation via challenge test of a dynamic growth-death model for the prediction of Listeria monocytogenes kinetics in Pecorino di Farindola cheese. Int. J. Food Microbiol. 2020, 329, 108690. [Google Scholar] [CrossRef] [PubMed]
- Iannetti, L.; Visciano, O.; Marfoglia, C.; Iannitto, G.; Parisciani, G.; Sericola, M.; Petrone, D.; Mangieri, M.S.; Pomilio, F.; Schirone, M. Coagulase positive staphylococci enumeration and enterotoxins detection in milk and dairy products from Central Italy. Ital. J. Food Sci. 2019, 31, 3. [Google Scholar] [CrossRef]
- Langa, S.; Peirotén, A.; Gaya, P.; Escudero, C.; Rodríguez-Mínguez, E.; Landete, J.M.; Arqués, J.L. Development of multi-strain probiotic cheese: Nisin production in food and gut. LWT 2021, 148, 111706. [Google Scholar] [CrossRef]

| Species, Detected Genes, Source and Time of Isolation | |
|---|---|
| Manufacturer 1 | |
| Lactobacilli | Lacticaseibacillus paracasei: Lot 1, days 49, 63, 77, 121, 150; Lot 2, days 14, 49, 63, 91, 121, 150; Lot 3, days 49, 77, 150; Lactiplantibacillus plantarum: Lot 1, days 14, 21, 49; Lot 2, days 14, 21, 35; Levilactobacillus brevis: Lot 1, days 7, 35, 91; Lot 3, day 121; L. brevis (odc): Lot 2, day 77; Leuconostoc mesenteroides: Lot 2, day 7; L. pseudomesenteroides: Lot 3, days 7, 14. |
| Lactic cocci | Lactococcus lactis subsp. Lactis: Lot 1, milk, curd; L. lactis subsp. lactis (nisA): Lot 2, milk, curd; Lot 3, milk, curd; Streptococcus infantarius: Lot 3, day 35; S. lutetiensis: Lot 1, day 91. |
| Enterococci | Enterococcus durans: Lot 3, day 150; E. faecalis (tdcA): Lot 2, day 77; E. faecalis (ace, tdcA): Lot 1, days 77, 121, 150; Lot 3, day 150; E. faecalis (gelE, tdcA): Lot 2, day 35; E. faecalis (gelE, ace, tdcA): Lot 1, milk, day 49; Lot 2, curd; E. faecalis (gelE, ace, efaA): Lot 1, day 63; E. faecium: Lot 1, days 28, 35; Lot 2, days 49, 63, 77, 150; Lot 3, days 7, 14, 21, 35, 63, 91; E. faecium (entB): Lot 1, day 49; E. faecium (entP): Lot 1, day 77; Lot 2, day 91; E. faecium (gelE): Lot 3, day 121; Lot 2, day 121; E. faecium (gelE, tdcA): Lot 1, day 14; E. gallinarum: Lot 2, milk. |
| Manufacturer 2 | |
| Lactobacilli | L. plantarum: Lot 1, days 7, 21, 49, 77, 91; Lot 2, days 14, 21, 35, 77, 91, 150; Lot 3, day 77; L. brevis: Lot 1, days 121, 150; Lot 2, day 121; Lot 3, days 14, 49, 91, 121, 150; L. mesenteroides: Lot 1, day 14; Loigolactobacillus coryniformis: Lot 1, day 35. |
| Lactic cocci | L. lactis subsp. Cremoris: Lot 1, milk, curd; Lot 2, milk, curd, day 7; Lot 3, milk, curd, day 14; L. lactis subsp. lactis: Lot 3, day 7. |
| Enterococci | E. casseliflavus: L1, day 63; E. durans: Lot 3, day 121; E. faecalis (gelE, tdcA): Lot 1, milk, day 35; Lot 2, milk, day 35; Lot 3, day 35; E. faecalis (ace, tdcA): Lot 2, day 21; E. faecalis (gelE, ace, tdcA): Lot 3, day 21; E. faecium: Lot 2, days 21, 49, 63, 91; Lot 3, days 14, 21, 49, 91, 121, 150; E. faecium (gelE): Lot 3, day 63; E. faecium (entB): Lot 1, days 35, 63, 121, 150; Lot 2, days 35, 77; Lot 3, days 7, 63, 121; E. faecium (entL50AB): Lot 3, day 14; Lot 2, day 150; E. faecium (entP): Lot 1, days 21, 49, 63, 121, 150; Lot 3, day 91. |
| Manufacturer 3 | |
| Lactobacilli | L. paracasei: Lot 1, days 63, 77, 91; Lot 2, days 14, 77, 91, 121, 150; Lot 3, days 21, 35, 77, 91, 150; L. plantarum: Lot 3, days 14, 49; L. brevis: Lot 1, days 35, 121, 150; Lot 2, day 121; Lot 3, day 121; L. mesenteroides: Lot 3, curd. |
| Lactic cocci | L. garviae: Lot 1, days 7, 49; L. lactis subsp. lactis: Lot 2, milk, curd; Lot 3, milk, curd; L. lactis subsp. lactis (nisI): Lot 1, milk, curd; Lot 2, day 7; Lot 3, day 7 |
| Enterococci | E. faecalis: Lot 3, day 14; E. faecalis (tdcA): Lot 1, day 49; Lot 2, day 21; E. faecalis (ace, tdcA): Lot 1, days 14, 21, 49, 150; Lot 2, days 7, 14; Lot 3, curd, days 7, 21, 49; E. faecalis (asa1): Lot 3, day 14; E. faecalis (gelE, ace, tdcA): Lot 1, day 49; Lot 2, day 35; E. faecalis (ace, tdcA, asa1): Lot 1, day 35; Lot 2, curd, day 35; Lot 3, curd, day 21; E. faecium: Lot 3, day 63; E. faecium (entL50AB): Lot 1, days 77, 121, 150; Lot 2, days 21, 49, 63, 77; Lot 3, days 21, 35, 63, 150. |
| Isolate | pH Tolerance (% Survival after 2 h) | Bile Tolerance (% Growth vs. Control) | Adhesion to CaCo-2 Cells (%) | |||
|---|---|---|---|---|---|---|
| 2.5 | 3 | 0.3 | 0.5 | 1 | ||
| Lacticaseibacillus casei ATCC 393 | 0 | 94.60 ± 2.12 | 27.00 ± 0.71 | 12.00 ± 0.97 | 5.00 ± 1.22 | 0.42 ± 0.04 |
| Enterococcus faecium E12150 | 47.22 ± 1.34 | 90.82 ± 1.45 | 96.00 ± 3.48 | 95.00 ± 4.37 | 80.01 ± 2.43 | 0.52 ± 0.07 |
| E. durans E13150 | 0 | 93.76 ± 2.67 | 63.42 ± 1.34 | 35.03 ± 2.14 | 20.46 ± 1.76 | 0.22 ± 0.02 |
| E. faecium E21121a * | 0 | 92.80 ± 2.13 | 31.93 ± 4.22 | 23.39 ± 1.36 | 12.79 ± 1.46 | 0.34 ± 0.09 |
| E. faecium E21121b † | 0 | 85.10 ± 1.17 | 41.50 ± 3.12 | 32.81 ± 1.44 | 12.71 ± 1.24 | 0.35 ± 0.11 |
| E. faecium E21150a * | 42.62 ± 1.25 | 83.96 ± 2.46 | 93.01 ± 2.33 | 78.79 ± 2.65 | 73.74 ± 1.96 | 0.61 ± 0.08 |
| E. faecium E21150b † | 0 | 95.40 ± 2.21 | 51.69 ± 4.32 | 32.14 ± 1.87 | 22.19 ± 1.47 | 0.71 ± 0.02 |
| E. faecium E22150 ‡ | 0 | 87.08 ± 1.27 | 41.37 ± 1.22 | 32.31 ± 2.98 | 12.33 ± 0.98 | 0.97 ± 0.18 |
| E. durans E23121 | 0 | 91.67 ± 1.65 | 39.66 ± 2.09 | 35.43 ± 3.16 | 33.34 ± 1.33 | 0.30 ± 0.05 |
| E. faecium E23121 * | 0 | 77.59 ± 2.14 | 74.87 ± 1.76 | 57.04 ± 2.56 | 52.16 ± 2.06 | 0.52 ± 0.07 |
| E. faecium E23150 | 0 | 96.52 ± 2.56 | 35.63 ± 1.09 | 24.41 ± 2.16 | 20.68 ± 1.67 | 0.38 ± 0.03 |
| E. faecium E31121 | 0 | 90.38 ± 1.68 | 39.10 ± 1.99 | 38.76 ± 1.03 | 29.18 ± 2.12 | 0.21 ± 0.01 |
| E. faecium E31150 | 0 | 98.05 ± 1.35 | 34.63 ± 1.47 | 30.90 ± 1.76 | 26.05 ± 1.09 | 0.59 ± 0.02 |
| E. faecium E33150 ‡ | 0 | 90.96 ± 2.22 | 36.76 ± 1.76 | 17.24 ± 1.33 | 16.56 ± 1.23 | 0.47 ± 0.03 |
| L. paracasei L11121 | 0 | 95.44 ± 2.13 | 80.23 ± 2.46 | 45.88 ± 2.46 | 16.32 ± 1.76 | 0.03 ± 0.01 |
| Levilactobacillus brevis L21121 | 0 | 80.37 ± 2.97 | 48.02 ± 1.96 | 24.79 ± 1.22 | 23.36 ± 1.33 | 0.45 ± 0.02 |
| L. brevis L31121 | 0 | 94.15 ± 4.22 | 97.00 ± 2.65 | 72.83 ± 2.09 | 65.28 ± 2.95 | 3.20 ± 0.12 |
| L. paracasei L12121 | 0 | 97.00 ± 1.76 | 52.75 ± 2.06 | 28.72 ± 1.67 | 13.16 ± 1.22 | 0.03 ± 0.01 |
| L. brevis L22121 | 73.75 ± 2.11 | 92.62 ± 2.56 | 58.92 ± 1.99 | 19.10 ± 1.09 | 9.28 ± 0.97 | 1.22 ± 0.03 |
| L. brevis L32121 | 0 | 80.28 ± 1.48 | 83.62 ± 2.55 | 33.85 ± 2.98 | 29.59 ± 1.96 | 1.05 ± 0.02 |
| L. brevis L13121 | 48.07 ± 1.63 | 93.33 ± 2.77 | 51.87 ± 2.46 | 24.83 ± 1.24 | 14.78 ± 1.03 | 0.96 ± 0.02 |
| L. brevis L23121 | 42.68 ± 1.25 | 97.00 ± 2.55 | 43.55 ± 1.33 | 11.54 ± 1.76 | 9.93 ± 0.98 | 1.00 ± 0.01 |
| L. brevis L33121 | 0 | 92.45 ± 3.15 | 74.58 ± 2.16 | 51.55 ± 1.99 | 49.43 ± 2.16 | 1.58 ± 0.07 |
| L. paracasei L11150 | 0 | 95.59 ± 3.11 | 17.71 ± 1.34 | 18.14 ± 0.98 | 10.02 ± 1.22 | 0.03 ± 0.00 |
| L. brevis L21150 | 0 | 97.36 ± 1.47 | 8.73 ± 1.03 | 6,89 ± 0.03 | 3.38 ± 0.04 | 0.28 ± 0.03 |
| L. brevis L31150 | 0 | 91.62 ± 1.27 | 80.06 ± 2.67 | 41.84 ± 2.05 | 26.30 ± 1.33 | 1.54 ± 0.02 |
| L. paracasei L12150 | 33.09 ± 1.34 | 98.00 ± 1.03 | 36.69 ± 1.22 | 18.81 ± 1.33 | 8.34 ± 0.96 | 0.08 ± 0.02 |
| Lactiplantibacillus plantarum L22150 | 90.87 ± 1.98 | 97.81 ± 2.19 | 50.42 ± 2.09 | 30.04 ± 1.47 | 21.77 ± 1.99 | 1.33 ± 0.07 |
| L. paracasei L32150 | 42.18 ± 1.55 | 95.00 ± 4.21 | 46.72 ± 2.06 | 34.56 ± 2.16 | 14.48 ± 1.67 | 0.03 ± 0.01 |
| L. paracasei L13150 | 78.57 ± 1.58 | 94.22 ± 2.23 | 9.56 ± 0.98 | 4.61 ± 0.78 | 4.78 ± 0.06 | 0.03 ± 0.01 |
| L. brevis L23150 | 54.09 ± 1.44 | 73.40 ± 2.67 | 53.30 ± 1.34 | 45.00 ± 1.33 | 12.00 ± 1.24 | 0.02 ± 0.00 |
| L. paracasei L33150 | 45.05 ± 1.77 | 72.05 ± 1.33 | 97.42 ± 2.55 | 96.13 ± 3.13 | 68.46 ± 2.05 | 0.01 ± 0.00 |
| Batch | Milk | Curd | Day 1 | Day 7 | Day 17 | Day 23 | Day 28 | Day 37 | Day 58 | Day 88 | Day 118 | Day 148 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aw | C | 0.994 ± 0.004 | 0.965 ± 0.000 | 0.952 ± 0.000 | 0.916 ± 0.001 | 0.867 ± 0.000 | 0.859 ± 0.001 | 0.856 ± 0.002 | 0.828 ± 0.004 | 0.822 ± 0.000 | 0.827 ± 0.001 | 0.819 ± 0.001 | |
| E | 0.992 ± 0.005 | 0.957 ± 0.000 | 0.951 ± 0.001 | 0.916 ± 0.001 | 0.895 ± 0.003 | 0.868 ± 0.002 | 0.861 ± 0.001 | 0.827 ± 0.002 | 0.820 ± 0.02 | 0.822 ± 0.000 | 0822 ± 0.001 | ||
| pH | C | 6.85 ± 0.05 | 6.74 ± 0.01 | 6.60 ± 0.01 | 6.01 ± 0.08 | 5.80 ± 0.01 | 5.71 ± 0.01 | 5.67 ± 0.02 | 5.74 ± 0.01 | 5.43 ± 0.02 | 5.38 ± 0.02 | 5.41 ± 0.01 | 5.46 ± 0.03 |
| E | 6.85 ± 0.05 | 6.74 ± 0.01 | 6.60 ± 0.01 | 6.01 ± 0.08 | 5.80 ± 0.01 | 5.65 ± 0.04 | 5.58 ± 0.02 | 5.75 ± 0.06 | 5.45 ± 0.05 | 5.43 ± 0.05 | 5.39 ± 0.00 | 5.26 ± 0.01 | |
| LAB on MRS (Log CFU/g) | C a | 5.33 ± 0.02 | 5.44 ± 0.04 | 7.63 ± 0.02 | 7.53 ± 0.03 | 7.54 ± 0.04 | 7.85 ± 0.02 | 7.63 ± 0.02 | 7.85 ± 0.02 | 6.48 ± 0.03 | 6.41 ± 0.05 | 4.72 ± 0.09 | 4.58 ± 0.06 |
| E a | 6.18 ± 0.05 | 6.33 ± 0.04 | 8.73 ± 0.02 | 8.71 ± 0.03 | 8.66 ± 0.03 | 8.56 ± 0.02 | 8.03 ± 0.00 | 7.89 ± 0.02 | 6.56 ± 0.08 | 6.42 ± 0.08 | 5.14 ± 0.05 | 5.11 ± 0.03 | |
| LAB on M17 (Log CFU/g) | C c | 5.55 ± 0.03 | 5.61 ± 0.02 | 8.72 ± 0.02 | 8.72 ± 0.01 | 8.55 ± 0.03 | 8.51 ± 0.02 | 7.69 ± 0.01 | 7.97 ± 0.02 | 6.41 ± 0.04 | 6.40 ± 0.10 | 5.35 ± 0.06 | 5.27 ± 0.07 |
| E c | 6.33 ± 0.01 | 6.53 ± 0.02 | 8.64 ± 0.02 | 8.70 ± 0.02 | 8.73 ± 0.02 | 8.60 ± 0.02 | 8.78 ± 0.05 | 7.96 ± 0.03 | 6.51 ± 0.07 | 6.21 ± 0.01 | 5.74 ± 0.07 | 5.68 ± 0.06 | |
| Enterococci (Log CFU/g) | C | 3.62 ± 0.02 | 3.77 ± 0.03 | 3.80 ± 0.02 | 4.94 ± 0.01 | 5.42 ± 0.05 | 5.37 ± 0.02 | 5.96 ± 0.03 | 5.91 ± 0.03 | 5.37 ± 0.03 | 4.86 ± 0.08 | 4.66 ± 0.14 | 4.45 ± 0.10 |
| E | 3.35 ± 0.03 | 3.49 ± 0.03 | 3.91 ± 0.01 | 5.03 ± 0.02 | 5.77 ± 0.02 | 5.93 ± 0.02 | 5.88 ± 0.02 | 5.87 ± 0.02 | 5.58 ± 0.01 | 4.74 ± 0.09 | 4.86 ± 0.08 | 4.50 ± 0.15 | |
| Coagulase-positive staphylococci (Log CFU/g) | C | 4.48 ± 0.00 | 4.51 ± 0.04 | 5.02 ± 0.03 | 5.01 ± 0.02 | 4.93 ± 0.01 | 4.81 ± 0.02 | 4.72 ± 0.01 | 4.63 ± 0.02 | 3.45 ± 0.04 | 3.47 ± 0.04 | 2.34 ± 0.12 | nd |
| E | 4.42 ± 0.03 | 4.51 ± 0.04 | 5.02 ± 0.03 | 5.01 ± 0.02 | 4.93 ± 0.01 | 4.81 ± 0.02 | 4.63 ± 0.02 | 4.40 ± 0.07 | 3.60 ± 0.03 | 3.30 ± 0.08 | 2.54 ± 0.11 | nd | |
| E. coli O157 (Log CFU/g) | C | 3.77 ± 0.02 | 3.49 ± 0.04 | 4.03 ± 0.01 | 4.04 ± 0.01 | 4.02 ± 0.01 | 3.92 ± 0.01 | 3.65 ± 0.11 | 3.38 ± 0.02 | 3.38 ± 0.08 | 2.62 ± 0.04 | nd | nd |
| E | 3.63 ± 0.02 | 3.40 ± 0.05 | 4.01 ± 0.02 | 4.00 ± 0.00 | 3.64 ± 0.03 | 3.06 ± 0.05 | 3.08 ± 0.05 | 3.06 ± 0.05 | 3.05 ± 0.05 | nd | nd | nd | |
| L. monocytogenes (Log CFU/g) | C b | 3.32 ± 0.02 | 3.34 ± 0.06 | 3.95 ± 0.02 | 3.94 ± 0.01 | 3.37 ± 0.08 | 3.31 ± 0.03 | 3.30 ± 0.03 | 3.32 ± 0.02 | 3.27 ± 0.03 | 2.25 ± 0.06 | 2.03 ± 0.11 | nd |
| E b | 3.44 ± 0.05 | 3.37 ± 0.06 | 3.47 ± 0.03 | 3.48 ± 0.04 | 3.36 ± 0.04 | 3.33 ± 0.02 | 1.88 ± 0.03 | 1.71 ± 0.05 | 1.68 ± 0.05 | nd | nd | nd |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aprea, G.; Alessiani, A.; Rossi, F.; Sacchini, L.; Boni, A.; D’Angelantonio, D.; Scattolini, S.; Sperandii, A.F.; Centorotola, G.; Neri, D.; et al. Characterization of Lactic Acid Bacteria in Pecorino di Farindola Cheese and Manufacturing with a Lacticaseibacillus paracasei Autochthonous Culture. Appl. Sci. 2021, 11, 7897. https://doi.org/10.3390/app11177897
Aprea G, Alessiani A, Rossi F, Sacchini L, Boni A, D’Angelantonio D, Scattolini S, Sperandii AF, Centorotola G, Neri D, et al. Characterization of Lactic Acid Bacteria in Pecorino di Farindola Cheese and Manufacturing with a Lacticaseibacillus paracasei Autochthonous Culture. Applied Sciences. 2021; 11(17):7897. https://doi.org/10.3390/app11177897
Chicago/Turabian StyleAprea, Giuseppe, Alessandra Alessiani, Franca Rossi, Lorena Sacchini, Arianna Boni, Daniela D’Angelantonio, Silvia Scattolini, Anna Franca Sperandii, Gabriella Centorotola, Diana Neri, and et al. 2021. "Characterization of Lactic Acid Bacteria in Pecorino di Farindola Cheese and Manufacturing with a Lacticaseibacillus paracasei Autochthonous Culture" Applied Sciences 11, no. 17: 7897. https://doi.org/10.3390/app11177897
APA StyleAprea, G., Alessiani, A., Rossi, F., Sacchini, L., Boni, A., D’Angelantonio, D., Scattolini, S., Sperandii, A. F., Centorotola, G., Neri, D., Pomilio, F., Di Giannatale, E., Del Matto, I., Tucci, P., & Migliorati, G. (2021). Characterization of Lactic Acid Bacteria in Pecorino di Farindola Cheese and Manufacturing with a Lacticaseibacillus paracasei Autochthonous Culture. Applied Sciences, 11(17), 7897. https://doi.org/10.3390/app11177897







