A Hybrid DenseNet-LSTM Model for Epileptic Seizure Prediction
Abstract
:1. Introduction
2. Related Work
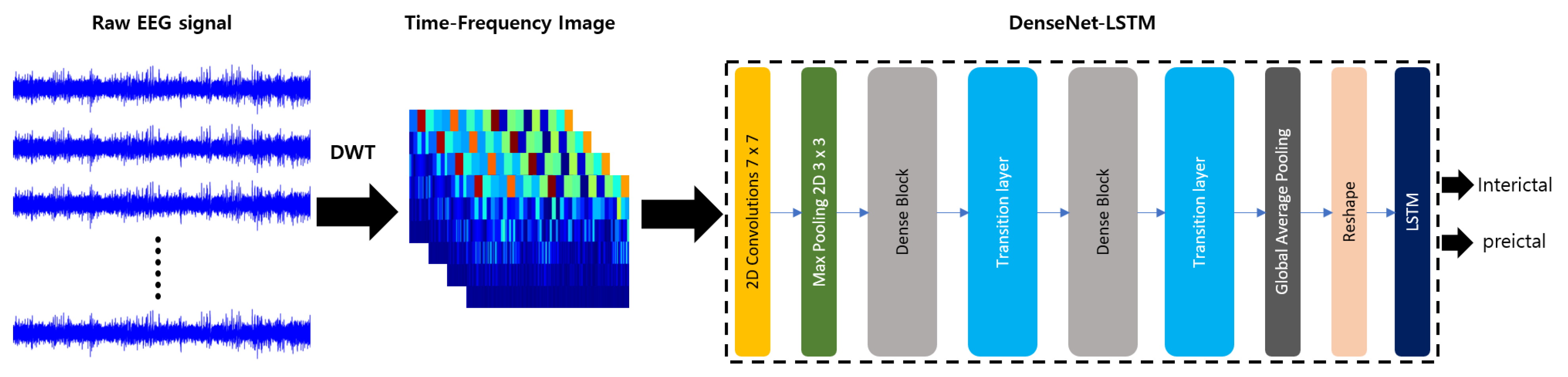
3. Proposed Method: DenseNet-LSTM
3.1. System Model
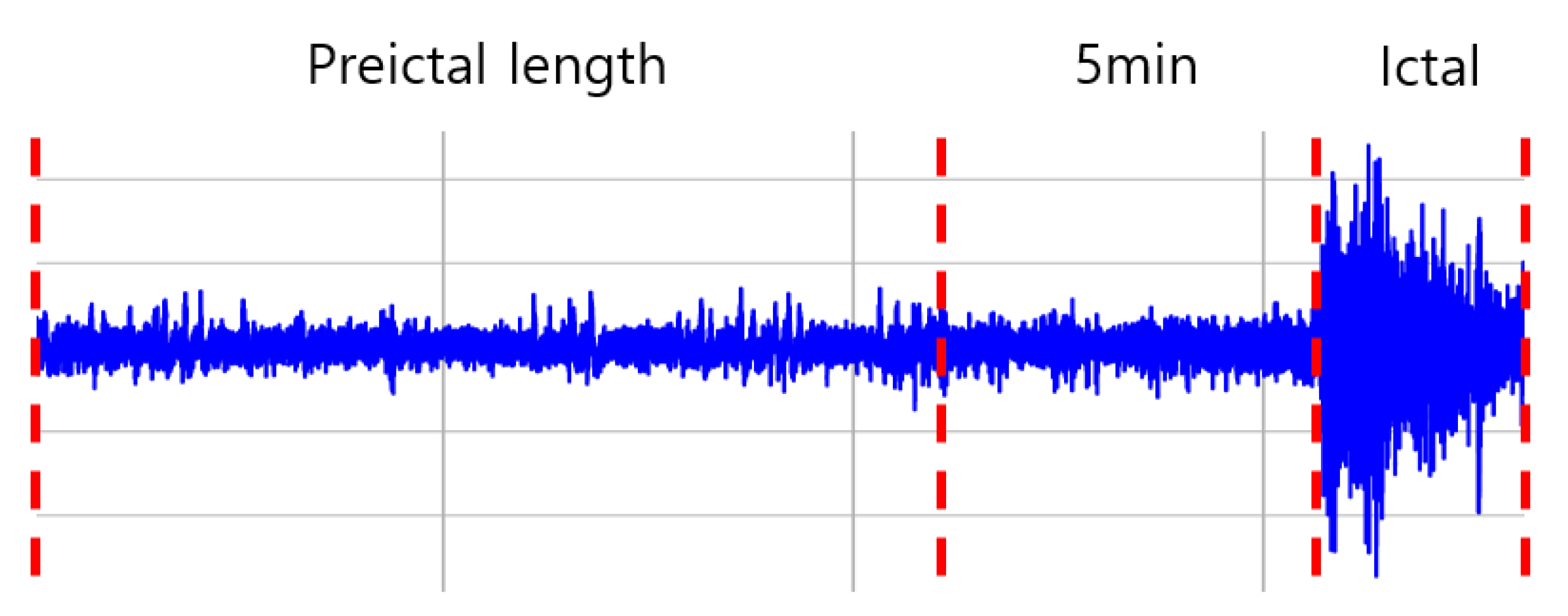
3.2. Dataset and Preprocessing
3.2.1. Dataset
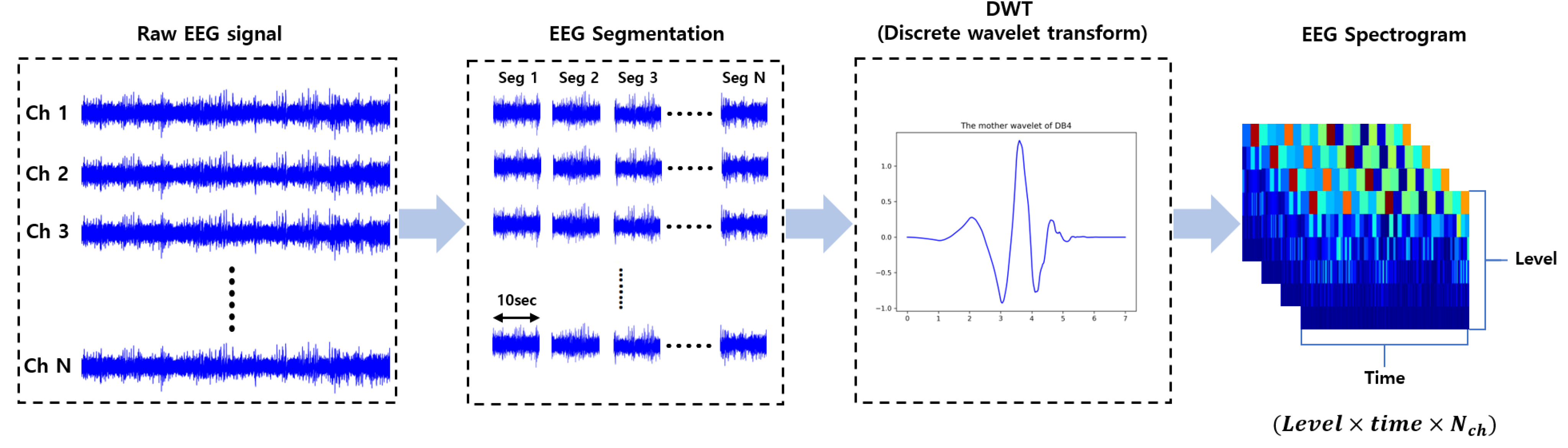
3.2.2. Preprocessing
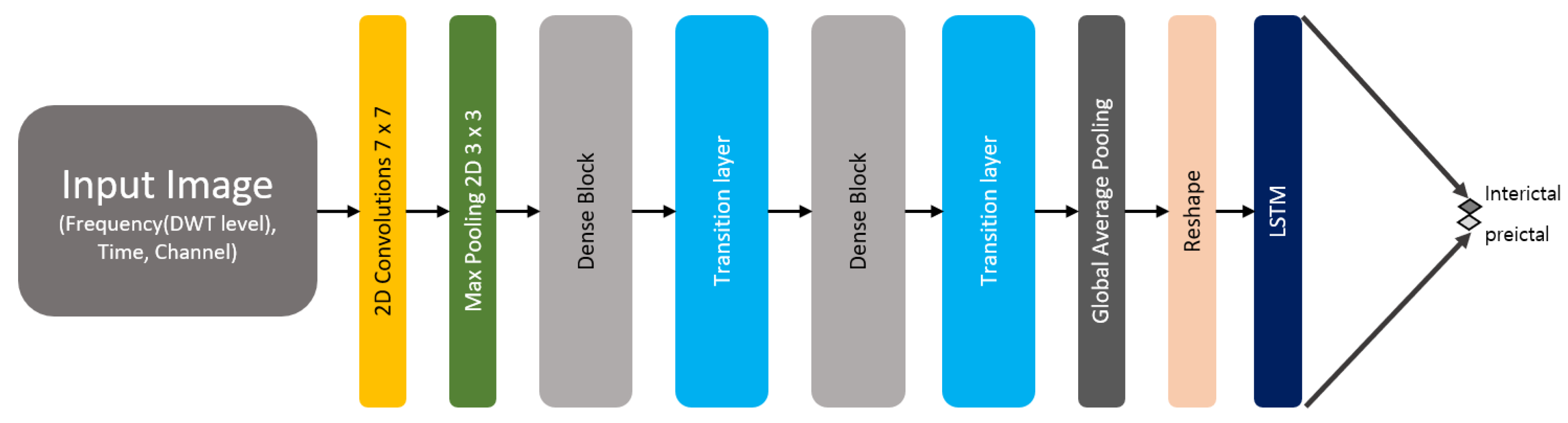
3.3. Deep Learning Architecture
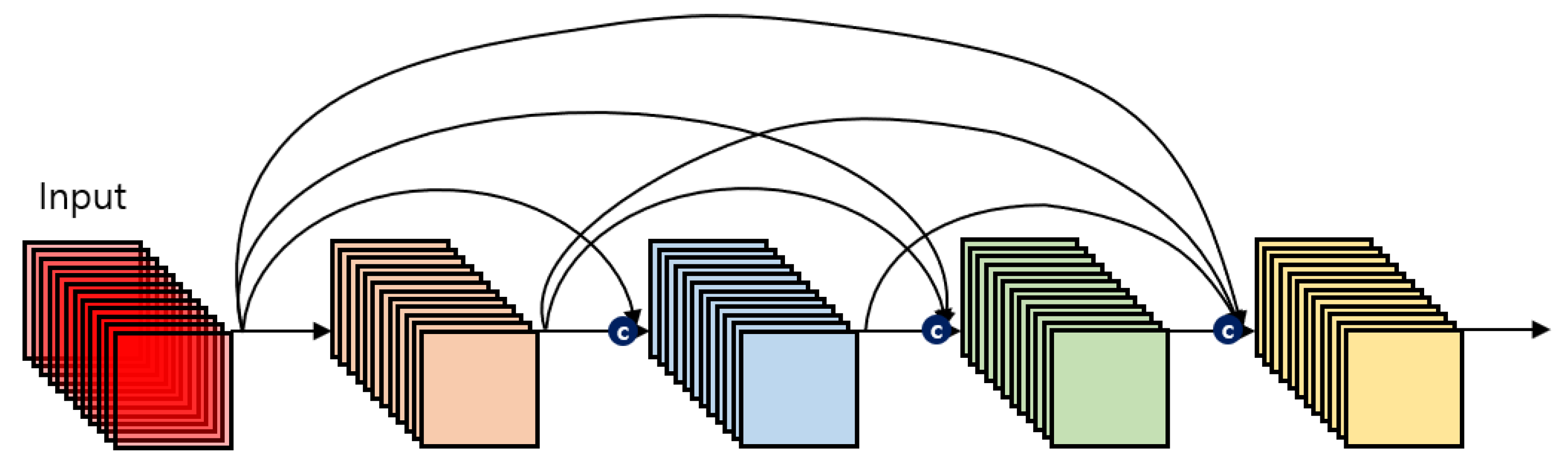
3.3.1. DenseNet
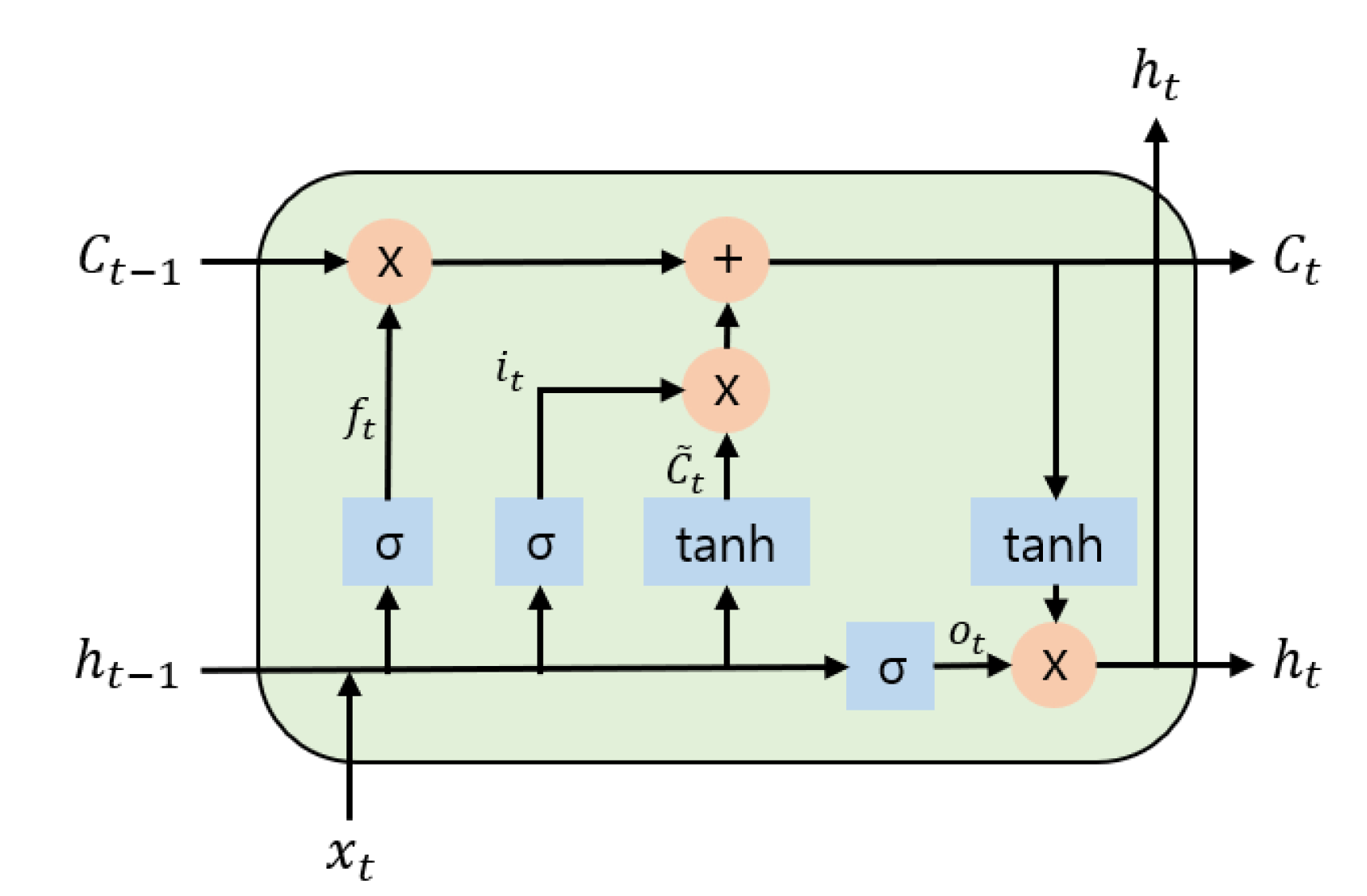
3.3.2. LSTM
3.3.3. Hybrid Model
4. Performance Evaluation
4.1. Experimental Setup
4.2. Experimental Results
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fisher, R.S.; Acevedo, C.; Arzimanoglou, A.; Bogacz, A.; Cross, J.H.; Elger, C.E.; Engel, J., Jr.; Forsgren, L.; French, J.A.; Glynn, M.; et al. ILAE official report: A practical clinical definition of epilepsy. Epilepsia 2014, 55, 475–482. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Neurological Disorders: Public Health Challenges; World Health Organization: Geneva, Switzerland, 2006. [Google Scholar]
- Chiang, C.Y.; Chang, N.F.; Chen, T.C.; Chen, H.H.; Chen, L.G. Seizure prediction based on classification of EEG synchronization patterns with on-line retraining and post-processing scheme. In Proceedings of the 2011 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Boston, MA, USA, 30 August–3 September 2011; pp. 7564–7569. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef] [PubMed]
- Maiwald, T.; Winterhalder, M.; Aschenbrenner-Scheibe, R.; Voss, H.U.; Schulze-Bonhage, A.; Timmer, J. Comparison of three nonlinear seizure prediction methods by means of the seizure prediction characteristic. Phys. D Nonlinear Phenom. 2004, 194, 357–368. [Google Scholar] [CrossRef]
- Winterhalder, M.; Schelter, B.; Maiwald, T.; Brandt, A.; Schad, A.; Schulze-Bonhage, A.; Timmer, J. Spatio-temporal patient–individual assessment of synchronization changes for epileptic seizure prediction. Clin. Neurophysiol. 2006, 117, 2399–2413. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhou, W.; Yuan, Q.; Liu, Y. Seizure prediction using spike rate of intracranial EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2013, 21, 880–886. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, G.; Li, K.; Bao, G.; Wang, J. Epileptic seizure prediction using phase synchronization based on bivariate empirical mode decomposition. Clin. Neurophysiol. 2014, 125, 1104–1111. [Google Scholar] [CrossRef] [PubMed]
- Eftekhar, A.; Juffali, W.; El-Imad, J.; Constandinou, T.G.; Toumazou, C. Ngram-derived pattern recognition for the detection and prediction of epileptic seizures. PLoS ONE 2014, 9, e96235. [Google Scholar] [CrossRef] [PubMed]
- Elgohary, S.; Eldawlatly, S.; Khalil, M.I. Epileptic seizure prediction using zero-crossings analysis of EEG wavelet detail coefficients. In Proceedings of the 2016 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), Chiang Mai, Thailand, 5–7 October 2016; pp. 1–6. [Google Scholar]
- Tsiouris, K.M.; Pezoulas, V.C.; Koutsouris, D.D.; Zervakis, M.; Fotiadis, D.I. Discrimination of preictal and interictal brain states from long-term EEG data. In Proceedings of the 2017 IEEE 30th International Symposium on Computer-Based Medical Systems (CBMS), Thessaloniki, Greece, 22–24 June 2017; pp. 318–323. [Google Scholar]
- Sharif, B.; Jafari, A.H. Prediction of epileptic seizures from EEG using analysis of ictal rules on Poincaré plane. Comput. Methods Programs Biomed. 2017, 145, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Akut, R. Wavelet based deep learning approach for epilepsy detection. Health Inf. Sci. Syst. 2019, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Boonyakitanont, P.; Lek-uthai, A.; Chomtho, K.; Songsiri, J. A Comparison of Deep Neural Networks for Seizure Detection in EEG Signals. bioRxiv 2019, 702654. [Google Scholar] [CrossRef]
- Karim, A.M.; Karal, Ö.; Çelebi, F. A new automatic epilepsy serious detection method by using deep learning based on discrete wavelet transform. In Proceedings of the 3rd International Conference on Engineering Technology and Applied Sciences (ICETAS), Skopje, North Macedonia, 17–21 July 2018; Volume 4, pp. 15–18. [Google Scholar]
- Liang, W.; Pei, H.; Cai, Q.; Wang, Y. Scalp EEG epileptogenic zone recognition and localization based on long-term recurrent convolutional network. Neurocomputing 2020, 396, 569–576. [Google Scholar] [CrossRef]
- Park, Y.; Luo, L.; Parhi, K.K.; Netoff, T. Seizure prediction with spectral power of EEG using cost-sensitive support vector machines. Epilepsia 2011, 52, 1761–1770. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Parhi, K.K. Low-complexity seizure prediction from iEEG/sEEG using spectral power and ratios of spectral power. IEEE Trans. Biomed. Circuits Syst. 2015, 10, 693–706. [Google Scholar] [CrossRef] [PubMed]
- Cho, D.; Min, B.; Kim, J.; Lee, B. EEG-based prediction of epileptic seizures using phase synchronization elicited from noise-assisted multivariate empirical mode decomposition. IEEE Trans. Neural Syst. Rehabil. Eng. 2016, 25, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Truong, N.D.; Nguyen, A.D.; Kuhlmann, L.; Bonyadi, M.R.; Yang, J.; Ippolito, S.; Kavehei, O. Convolutional neural networks for seizure prediction using intracranial and scalp electroencephalogram. Neural Netw. 2018, 105, 104–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, H.; Marcuse, L.; Fields, M.; Swann, K.; Yener, B. Focal onset seizure prediction using convolutional networks. IEEE Trans. Biomed. Eng. 2017, 65, 2109–2118. [Google Scholar] [CrossRef] [Green Version]
- Ozcan, A.R.; Erturk, S. Seizure prediction in scalp EEG using 3D convolutional neural networks with an image-based approach. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 2284–2293. [Google Scholar] [CrossRef]
- Shoeb, A.H. Application of Machine Learning to Epileptic Seizure Onset Detection and Treatment. Ph.D. Thesis, Massachusetts Institute of Technology, Cambridge, MA, USA, 2009. [Google Scholar]
- Choi, G.; Park, C.; Kim, J.; Cho, K.; Kim, T.J.; Bae, H.; Min, K.; Jung, K.Y.; Chong, J. A Novel Multi-scale 3D CNN with Deep Neural Network for Epileptic Seizure Detection. In Proceedings of the 2019 IEEE International Conference on Consumer Electronics (ICCE), Las Vegas, NV, USA, 11–13 January 2019; pp. 1–2. [Google Scholar]
- Heil, C.E.; Walnut, D.F. Continuous and discrete wavelet transforms. SIAM Rev. 1989, 31, 628–666. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity mappings in deep residual networks. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 8–16 October 2016; pp. 630–645. [Google Scholar]


| Layers | Feature Map Size | Configuration |
|---|---|---|
| Convolution Layer | 3 × 1280 × 64 | , stride 2 |
| Dense Block 1 | 2 × 640 × 256 | |
| Transition Layer 1 | 2 × 640 × 128 | |
| 1 × 320 × 128 | 2 × 2 average pooling, stride2 | |
| Dense Block 2 | 1 × 320 × 512 | |
| Transition Layer 2 | 1 × 320 × 256 | |
| 1 × 160 × 256 | 2 × 2 average pooling, stride2 | |
| LSTM Layer | 1 × 256 | global average pooling |
| 4 × 64 | reshape | |
| 1 × 128 | LSTM layer | |
| Classification Layer | 1 × 1 | sigmoid |
| Software or Hardware | Specification |
|---|---|
| CPU | AMD Ryzen 7 3700X |
| GPU | GeForce RTX 2080 Ti |
| RAM | DDR4 64 GB |
| Python | 3.6 |
| Tensorflow | 1.14 |
| Keras | 2.2.4 |
| Hyperparameters | Values |
|---|---|
| Growth rate | 32 |
| Compression factor | 0.5 |
| Activation function | ReLU |
| Optimizer | Adam |
| Learning rate | 0.001 |
| Performance Indicator | Formula |
|---|---|
| Accuracy | (TP + TN)/(TP + TN + FP + FN) |
| Sensitivity (Recall) | TP/(TP + FN) |
| Specificity | TN/(TN + FP) |
| Precision | TP/(TP + FP) |
| False Positive Rate (FPR) | FP/(TN + FP) |
| F1-Score | 2 × ((Precision × Recall)/(Precision + Recall)) |
| Patient | Preictal Length: 5 min | Preictal Length: 10 min | Preictal Length: 15 min | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | FPR | F1-Score | Accuracy | Sensitivity | Specificity | FPR | F1-Score | Accuracy | Sensitivity | Specificity | FPR | F1-Score | |
| chb01 | 100% | 100% | 100% | 0 | 1 | 100% | 100% | 100% | 0 | 1 | 99.97% | 99.95% | 100% | 0 | 0.999 |
| chb02 | 86.94% | 87.97% | 85.91% | 0.141 | 0.869 | 89.89% | 80.79% | 98.98% | 0.01 | 0.877 | 91.47% | 82.94% | 100% | 0 | 0.897 |
| chb03 | 96.82% | 96.3% | 97.33% | 0.026 | 0.967 | 86.86% | 74.49% | 99.23% | 0.007 | 0.808 | 93.66% | 88.77% | 98.54% | 0.014 | 0.929 |
| chb04 | 78.26% | 65.46% | 91.06% | 0.089 | 0.687 | 90.46% | 89.8% | 91.11% | 0.089 | 0.9 | 90.78% | 83.61% | 97.95% | 0.02 | 0.894 |
| chb05 | 94.32% | 97.82% | 90.83% | 0.091 | 0.946 | 97.29% | 96.56% | 98.02% | 0.02 | 0.972 | 98.76% | 98.54% | 98.99% | 0.01 | 0.987 |
| chb06 | 94.2% | 88.61% | 99.78% | 0.002 | 0.902 | 96.6% | 95.41% | 97.79% | 0.022 | 0.963 | 87.34% | 86.9% | 87.78% | 0.122 | 0.861 |
| chb07 | 100% | 100% | 100% | 0 | 1 | 99.4% | 98.81% | 100% | 0 | 0.993 | 100% | 100% | 100% | 0 | 1 |
| chb08 | 100% | 100% | 100% | 0 | 1 | 100% | 100% | 100% | 0 | 1 | 100% | 100% | 100% | 0 | 1 |
| chb09 | 99.82% | 99.65% | 100% | 0 | 0.998 | 99.64% | 99.28% | 100% | 0 | 0.996 | 99.9% | 99.97% | 99.83% | 0.001 | 0.999 |
| chb10 | 90.52% | 94.11% | 86.94% | 0.13 | 0.916 | 91.58% | 90.45% | 92.72% | 0.072 | 0.913 | 90.78% | 89.48% | 92.09% | 0.079 | 0.904 |
| chb11 | 100% | 100% | 100% | 0 | 1 | 100% | 100% | 100% | 0 | 1 | 99.58% | 99.21% | 99.94% | 0 | 0.995 |
| chb12 | 93.07% | 86.99% | 99.16% | 0.008 | 0.879 | 95.91% | 94.39% | 97.43% | 0.025 | 0.953 | 96.46% | 95.06% | 97.86% | 0.021 | 0.961 |
| chb13 | 92.05% | 94.41% | 89.69% | 0.103 | 0.922 | 91.05% | 88.19% | 93.9% | 0.06 | 0.901 | 89.62% | 86.61% | 92.62% | 0.073 | 0.889 |
| chb14 | 89.66% | 93.27% | 86.06% | 0.139 | 0.901 | 85.79% | 80.66% | 90.93% | 0.09 | 0.831 | 83.52% | 81.16% | 85.87% | 0.141 | 0.824 |
| chb15 | 89.41% | 95.46% | 83.36% | 0.166 | 0.902 | 74.97% | 77.12% | 72.82% | 0.272 | 0.74 | 80.54% | 81.97% | 79.12% | 0.208 | 0.817 |
| chb16 | 81.03% | 71.2% | 90.86% | 0.091 | 0.778 | 81.33% | 71.4% | 91.27% | 0.087 | 0.77 | 87.16% | 86.53% | 87.79% | 0.122 | 0.872 |
| chb17 | 100% | 100% | 100% | 0 | 1 | 99.8% | 100% | 99.6% | 0.004 | 0.998 | 100% | 100% | 100% | 0 | 1 |
| chb18 | 92.35% | 91.06% | 93.64% | 0.063 | 0.92 | 93.23% | 95.72% | 90.73% | 0.092 | 0.936 | 86.39% | 92.53% | 80.24% | 0.197 | 0.877 |
| chb19 | 100% | 100% | 100% | 0 | 1 | 100% | 100% | 100% | 0 | 1 | 100% | 100% | 100% | 0 | 1 |
| chb20 | 99.96% | 100% | 99.93% | 0 | 0.999 | 99.86% | 100% | 99.72% | 0.002 | 0.998 | 99.88% | 100% | 99.77% | 0.002 | 0.998 |
| chb21 | 95.4% | 93.81% | 96.99% | 0.03 | 0.952 | 93.36% | 91.87% | 94.83% | 0.051 | 0.932 | 90.81% | 88.8% | 92.81% | 0.071 | 0.906 |
| chb22 | 81.61% | 93.24% | 69.98% | 0.3 | 0.836 | 81.61% | 88.43% | 74.79% | 0.252 | 0.828 | 87.78% | 87.87% | 87.69% | 0.123 | 0.876 |
| chb23 | 96.66% | 96.01% | 97.32% | 0.026 | 0.966 | 91.01% | 99.05% | 82.97% | 0.17 | 0.933 | 93.86% | 99.61% | 88.1% | 0.119 | 0.95 |
| chb24 | 86.86% | 84.76% | 88.96% | 0.11 | 0.825 | 84.93% | 77.47% | 92.38% | 0.076 | 0.755 | 83.7% | 72.57% | 94.83% | 0.051 | 0.758 |
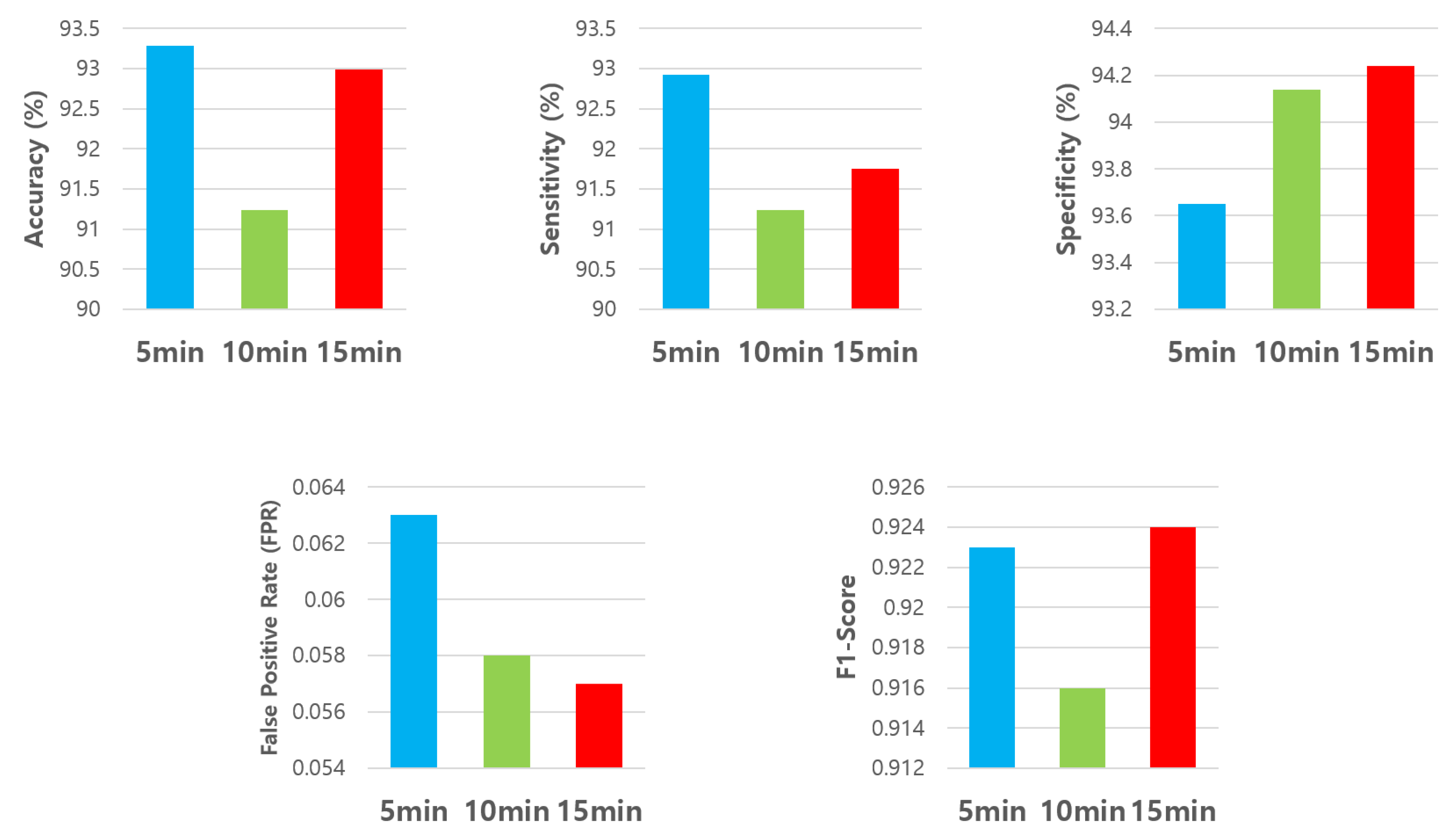
| Average | 93.28% | 92.92% | 93.65% | 0.063 | 0.923 | 92.69% | 91.24% | 94.13% | 0.058 | 0.916 | 92.99% | 91.75% | 94.24% | 0.057 | 0.924 |
| Authors | Year | Datasts | Features | Classifier | Acc (%) | Sen (%) | Spec (%) | FPR (H) | F1-Score |
|---|---|---|---|---|---|---|---|---|---|
| Khan et al. [23] | 2017 | CHB-MIT, 15 patients | Continuous wavelet transform | CNN | - | 87.8 | - | 0.147 | - |
| Truong et al. [22] | 2018 | CHB-MIT, 13 patients | Short-time Fourier transform | CNN | - | 81.2 | - | 0.16 | - |
| Ozcan et al. [24] | 2019 | CHB-MIT, 16 patients | Hjorth parameters | 3D CNN | - | 85.71 | - | 0.096 | - |
| This work | 2021 | CHB-MIT, 24 patients | Discrete wavelet transform | DenseNet-LSTM | 93.28 | 92.92 | 93.65 | 0.063 | 0.923 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ryu, S.; Joe, I. A Hybrid DenseNet-LSTM Model for Epileptic Seizure Prediction. Appl. Sci. 2021, 11, 7661. https://doi.org/10.3390/app11167661
Ryu S, Joe I. A Hybrid DenseNet-LSTM Model for Epileptic Seizure Prediction. Applied Sciences. 2021; 11(16):7661. https://doi.org/10.3390/app11167661
Chicago/Turabian StyleRyu, Sanguk, and Inwhee Joe. 2021. "A Hybrid DenseNet-LSTM Model for Epileptic Seizure Prediction" Applied Sciences 11, no. 16: 7661. https://doi.org/10.3390/app11167661
APA StyleRyu, S., & Joe, I. (2021). A Hybrid DenseNet-LSTM Model for Epileptic Seizure Prediction. Applied Sciences, 11(16), 7661. https://doi.org/10.3390/app11167661







