Dietary Intervention Induced Distinct Repercussions in Response to the Individual Gut Microbiota as Demonstrated by the In Vitro Fecal Fermentation of Beef
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Vitro Gastrointestinal Digestion and Fecal Fermentation
2.2. Quantitative Analysis of SCFAs
2.3. DNA Extraction and 16S rRNA Gene Sequencing
2.4. Fecal Microbiome Data Analysis
2.5. Statistical Analysis and Visualization
3. Results
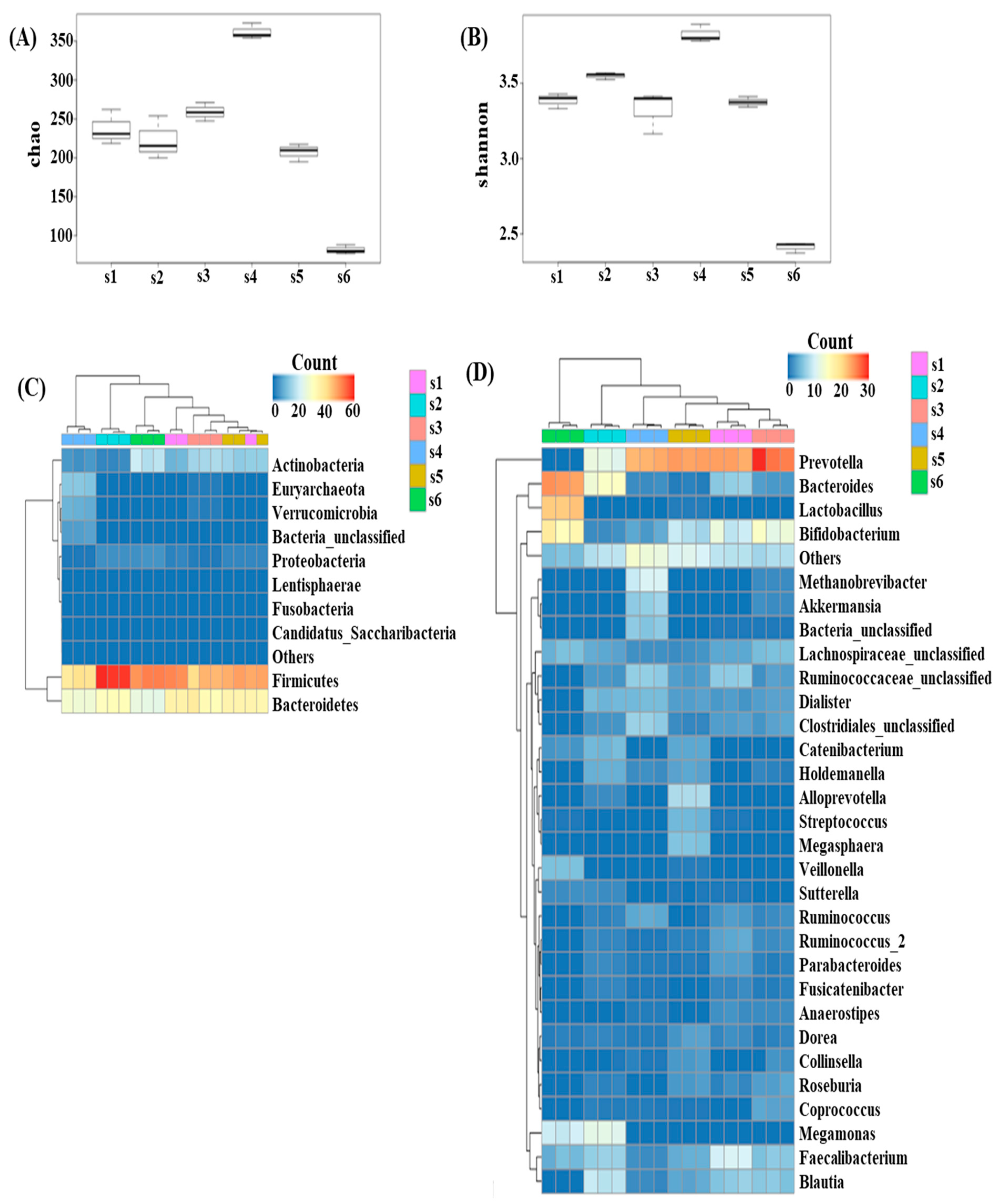
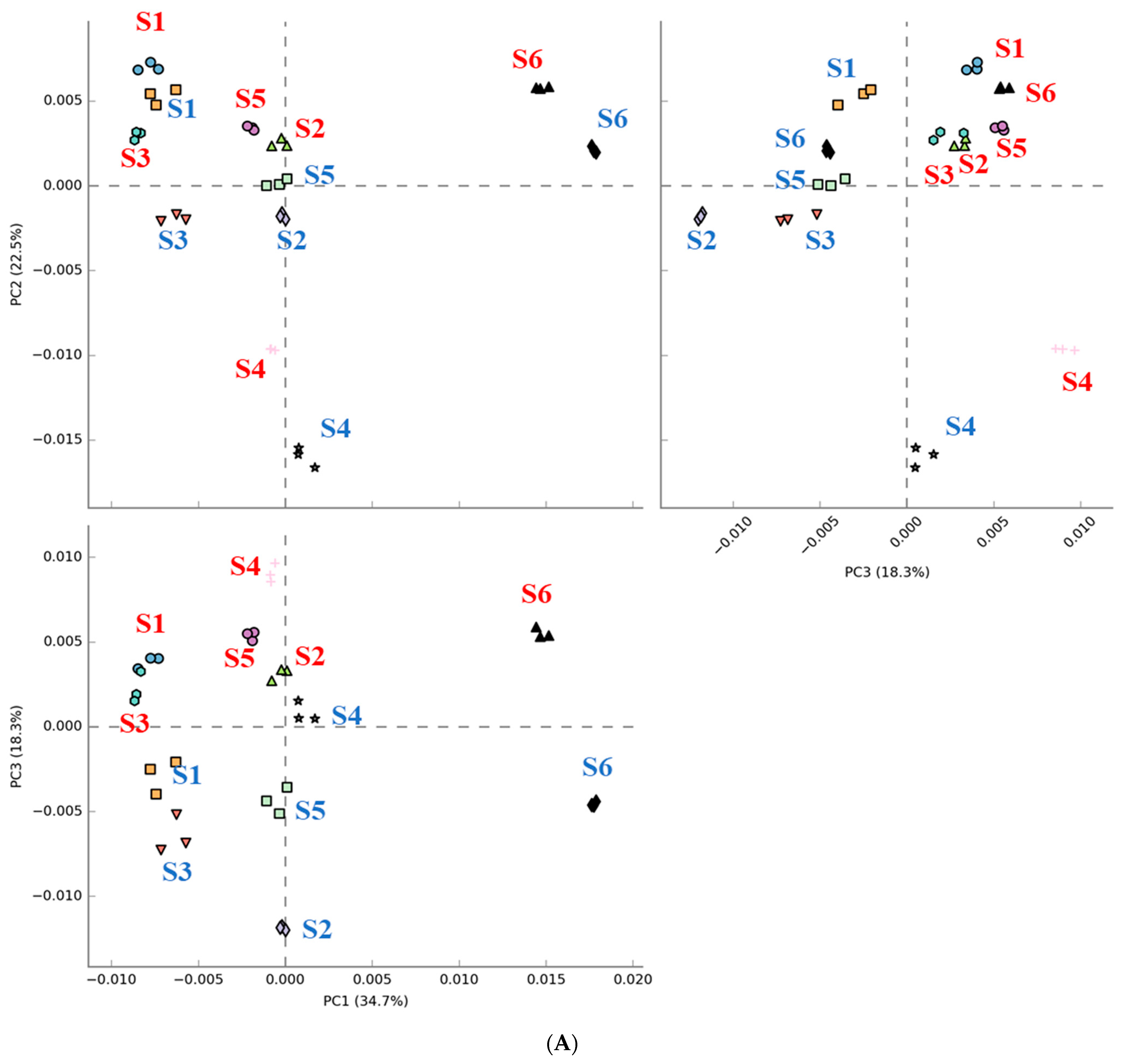
3.1. Diversity in the Fecal Microbiota of Different Fecal Donors
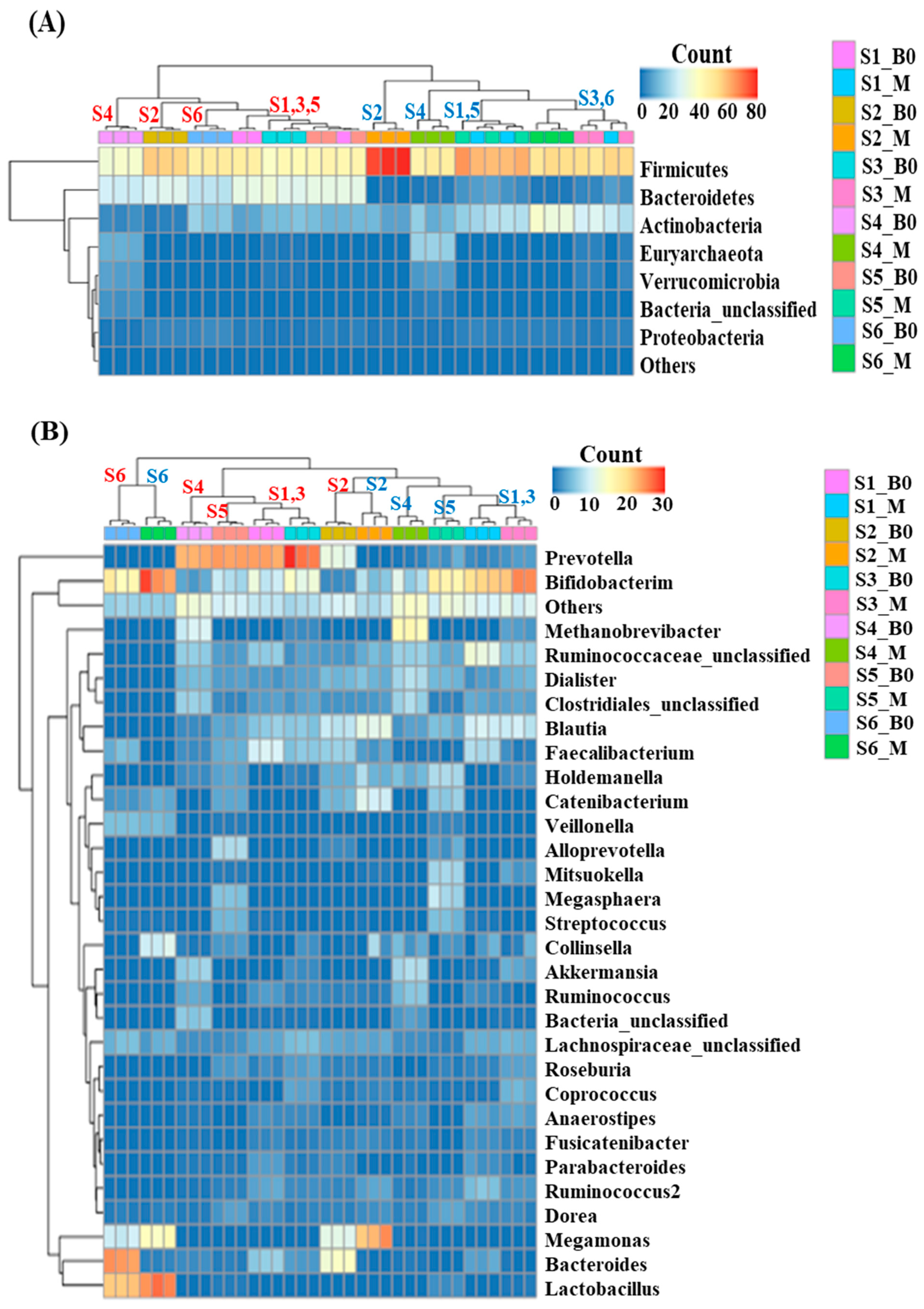
3.2. Effects of In Vitro Condition on Fecal Microbiota
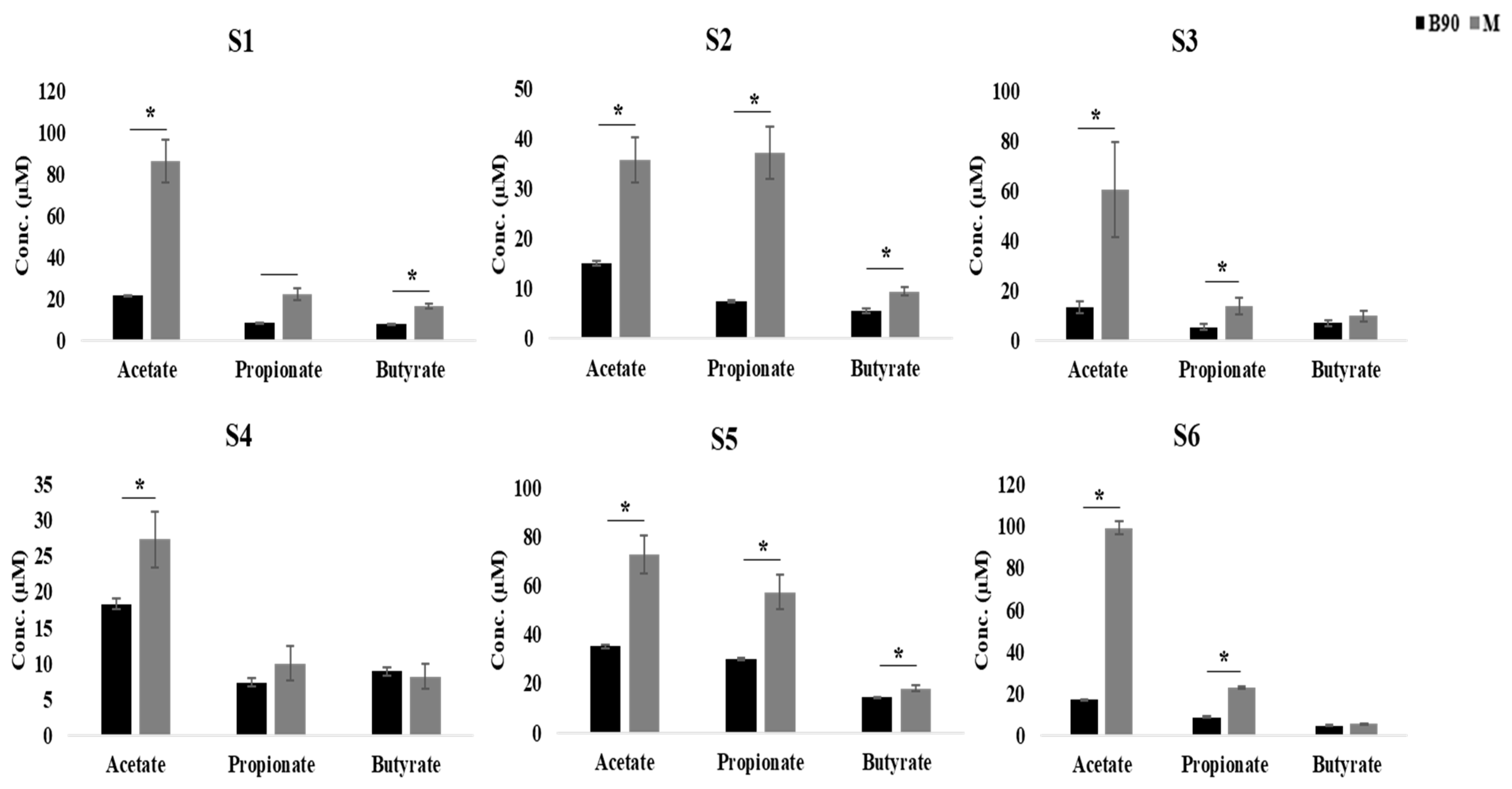
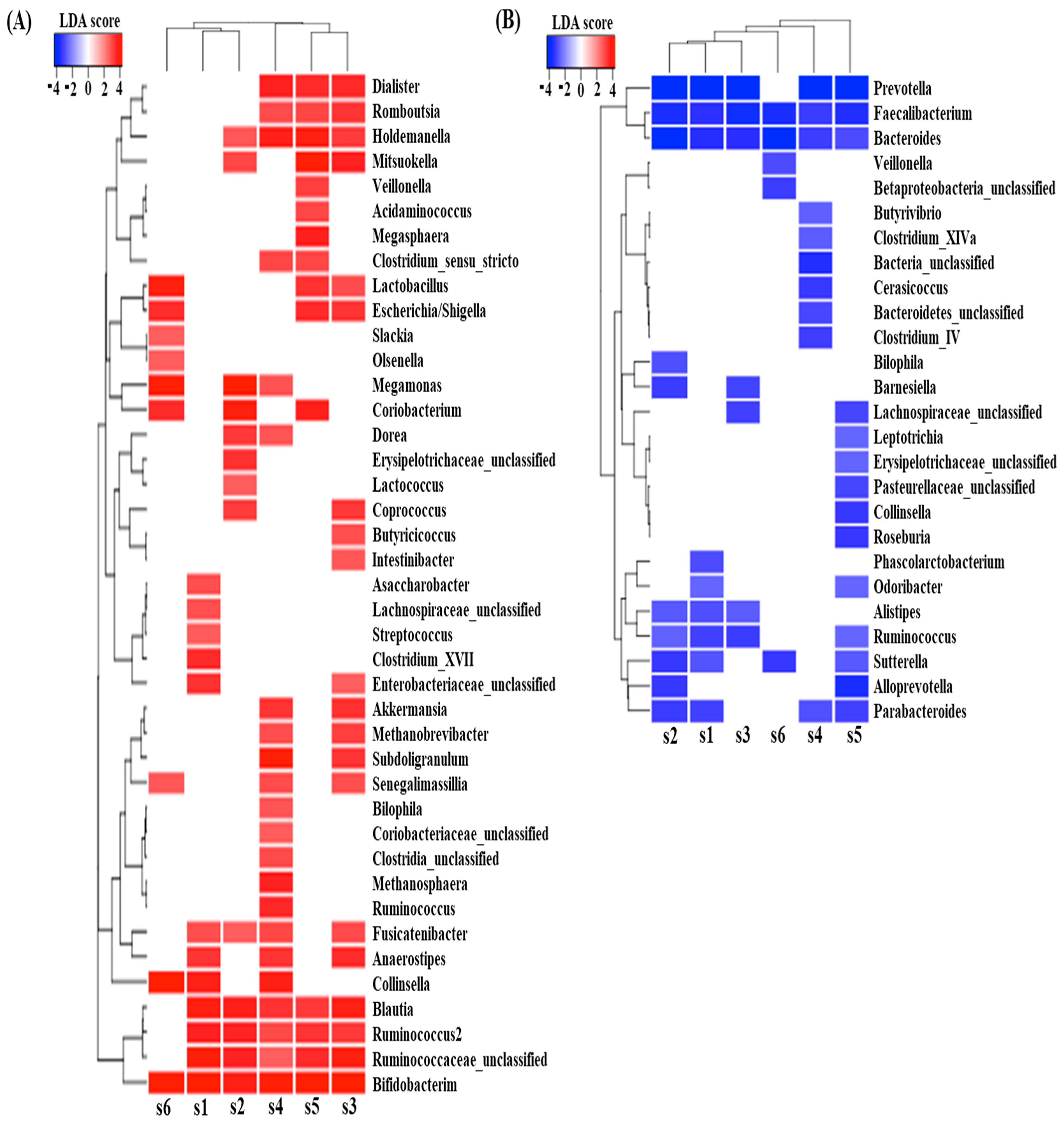
3.3. Effect of Digested Beef on SCFA Content and the Fecal Microbiome
3.4. Effect of Digested Beef on Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lepage, P.; Leclerc, M.C.; Joossens, M.; Mondot, S.; Blottière, H.M.; Raes, J.; Ehrlich, D.; Doré, J. A metagenomic insight into our gut’s microbiome. Gut 2013, 62, 146–158. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Chang, H.-W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 1–17. [Google Scholar] [CrossRef] [Green Version]
- De Filippis, F.; Pellegrini, N.; Vannini, L.; Jeffery, I.; La Storia, A.; Laghi, L.; Serrazanetti, D.I.; Di Cagno, R.; Ferrocino, I.; Lazzi, C.; et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 2015, 65, 1812–1821. [Google Scholar] [CrossRef] [PubMed]
- Tomova, A.; Bukovsky, I.; Rembert, E.; Yonas, W.; Alwarith, J.; Barnard, N.D.; Kahleova, H. The Effects of Vegetarian and Vegan Diets on Gut Microbiota. Front. Nutr. 2019, 6, 47. [Google Scholar] [CrossRef] [Green Version]
- Oliphant, K.; Allen-Vercoe, E. Macronutrient metabolism by the human gut microbiome: Major fermentation by-products and their impact on host health. Microbiome 2019, 7, 1–15. [Google Scholar] [CrossRef]
- Scott, K.P.; Gratz, S.W.; Sheridan, P.O.; Flint, H.J.; Duncan, S.H. The influence of diet on the gut microbiota. Pharmacol. Res. 2013, 69, 52–60. [Google Scholar] [CrossRef]
- Yang, J.; Yu, J. The association of diet, gut microbiota and colorectal cancer: What we eat may imply what we get. Protein Cell 2018, 9, 474–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brodkorb, A.; Egger, C.; Alminger, M.; Alvito, P.; Assunção, R.; Ballance, S.; Bohn, T.; Bourlieu-Lacanal, C.; Boutrou, R.; Carrière, F.; et al. INFOGEST static in vitro simulation of gastrointestinal food digestion. Nat. Protoc. 2019, 14, 991–1014. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food—An international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Peyron, M.-A.; Mishellany, A.; Woda, A. Particle Size Distribution of Food Boluses after Mastication of Six Natural Foods. J. Dent. Res. 2004, 83, 578–582. [Google Scholar] [CrossRef]
- Jalabert-Malbos, M.-L.; Mishellany-Dutour, A.; Woda, A.; Peyron, M.-A. Particle size distribution in the food bolus after mastication of natural foods. Food Qual. Prefer. 2007, 18, 803–812. [Google Scholar] [CrossRef]
- Moon, J.S.; Li, L.; Bang, J.; Han, N.S. Application of in vitro gut fermentation models to food components: A review. Food Sci. Biotechnol. 2016, 25, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Cole, J.R.; Wang, Q.; Fish, J.A.; Chai, B.; McGarrell, D.M.; Sun, Y.; Brown, C.T.; Porras-Alfaro, A.; Kuske, C.R.; Tiedje, J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2013, 42, D633–D642. [Google Scholar] [CrossRef] [Green Version]
- Clarke, K.R.; Somerfield, P.J.; Chapman, M.G. On resemblance measures for ecological studies, including taxonomic dissimilarities and a zero-adjusted Bray–Curtis coefficient for denuded assemblages. J. Exp. Mar. Biol. Ecol. 2006, 330, 55–80. [Google Scholar] [CrossRef]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and extensible approach for metagenome inference. bioRxiv 2019, 672295. [Google Scholar] [CrossRef] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, I.; Harris, K.; Quince, C. Dirichlet Multinomial Mixtures: Generative Models for Microbial Metagenomics. PLoS ONE 2012, 7, e30126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parks, D.H.; Tyson, G.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [Green Version]
- Shen, Q.; Chen, Y.A.; Tuohy, K.M. A comparative in vitro investigation into the effects of cooked meats on the human faecal microbiota. Anaerobe 2010, 16, 572–577. [Google Scholar] [CrossRef]
- Reddy, B.S.; Weisburger, J.H.; Wynder, E.L. Effects of High Risk and Low Risk Diets for Colon Carcinogenesis on Fecal Microflora and Steroids in Man. J. Nutr. 1975, 105, 878–884. [Google Scholar] [CrossRef]
- Biddle, A.; Stewart, L.; Blanchard, J.L.; Leschine, S. Untangling the Genetic Basis of Fibrolytic Specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities. Diversity 2013, 5, 627–640. [Google Scholar] [CrossRef]
- Albracht-Schulte, K.; Islam, T.; Johnson, P.; Moustaid-Moussa, N. Systematic Review of Beef Protein Effects on Gut Microbiota: Implications for Health. Adv. Nutr. 2020, 12, 102–114. [Google Scholar] [CrossRef]
- Adamberg, K.; Tomson, K.; Talve, T.; Pudova, K.; Puurand, M.; Visnapuu, T.; Alamäe, T.; Adamberg, S. Levan Enhances Associated Growth of Bacteroides, Escherichia, Streptococcus and Faecalibacterium in Fecal Microbiota. PLoS ONE 2015, 10, e0144042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandberg, J.; Kovatcheva-Datchary, P.; Björck, I.; Bäckhed, F.; Nilsson, A. Abundance of gut Prevotella at baseline and metabolic response to barley prebiotics. Eur. J. Nutr. 2018, 58, 2365–2376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Long, W.; Zhang, C.; Liu, S.; Zhao, L.; Hamaker, B.R. Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Sci. Rep. 2017, 7, 1–7. [Google Scholar] [CrossRef]
- Parveez, G.K.A.; Rasid, O.A.; Hashim, A.T.; Ishak, Z.; Rosli, S.K.; Sambanthamurthi, R. 4—Tissue Culture and Genetic Engineering of Oil Palm. In Palm Oil; Lai, O.-M., Tan, C.-P., Akoh, C.C., Eds.; AOCS Press: Champaign, IL, USA, 2012; pp. 87–135. [Google Scholar]
- Lopez, S.; Bermudez, B.; la Paz, S.M.; Pacheco, Y.M.; Ortega-Gomez, A.; Varela, L.M.; Lemus-Conejo, A.; Millan-Linares, M.C.; Rosillo, M.A.; Abia, R.; et al. Oleic Acid: The Main Component of Olive Oil on Postprandial Metabolic Processes. In Olives and Olive Oil in Health and Disease Prevention; Preedy, V.R., Watson, R.R., Eds.; Academic Press: San Diego, CA, USA, 2010; Chapter 154; pp. 1385–1393. [Google Scholar]
- Senyilmaz-Tiebe, D.; Pfaff, D.H.; Virtue, S.; Schwarz, K.V.; Fleming, T.; Altamura, S.; Muckenthaler, M.U.; Okun, J.G.; Vidal-Puig, A.; Nawroth, P.; et al. Dietary stearic acid regulates mitochondria in vivo in humans. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mota, J.S.D.O.; Boué, G.; Guillou, S.; Pierre, F.; Membré, J.-M. Estimation of the burden of disease attributable to red meat consumption in France: Influence on colorectal cancer and cardiovascular diseases. Food Chem. Toxicol. 2019, 130, 174–186. [Google Scholar] [CrossRef]
- Bohn, T.; Carriere, F.; Day, L.; Deglaire, A.; Egger, L.; Freitas, D.; Golding, M.; le Feunteun, S.; Macierzanka, A.; Menard, O.; et al. Correlation between in vitro and in vivo data on food digestion. What can we predict with static in vitro digestion models? Crit. Rev. Food Sci. Nutr. 2018, 58, 2239–2261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mulet-Cabero, A.; Egger, L.; Portmann, R.; Ménard, O.; Marze, S.; Minekus, M.; le Feunteun, S.; Sarkar, A.; Grundy, M.M.-L.; Carrière, F.; et al. A standardised semi-dynamic in vitro digestion method suitable for food—An international consensus. Food Funct. 2020, 11, 1702–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Metabolic Pathways (MetaCyc) | I/D * | Description |
|---|---|---|
| PWY-7664 | D | Oleate biosynthesis IV (anaerobic) |
| PWYG-321 | D | Mycolate biosynthesis |
| PWY-6282 | D | Palmitoleate biosynthesis I (from (5z)-dodec-5-enoate) |
| PWY-5989 | D | Stearate biosynthesis II (bacteria and plants) |
| FASYN-ELONG-PWY | D | Fatty acid elongation—saturated |
| PWY0-862 | D | (5z)-dodec-5-enoate biosynthesis |
| FASYN-INITIAL-PWY | D | Super pathway of fatty acid biosynthesis initiation (E. Coli) |
| RHAMCAT-PWY | D | L-rhamnose degradation I |
| PWY-6163 | I | Chorismite biosynthesis from 3-dehydroquinate |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, V.; Ryu, Y.-C.; Unno, T. Dietary Intervention Induced Distinct Repercussions in Response to the Individual Gut Microbiota as Demonstrated by the In Vitro Fecal Fermentation of Beef. Appl. Sci. 2021, 11, 6841. https://doi.org/10.3390/app11156841
Singh V, Ryu Y-C, Unno T. Dietary Intervention Induced Distinct Repercussions in Response to the Individual Gut Microbiota as Demonstrated by the In Vitro Fecal Fermentation of Beef. Applied Sciences. 2021; 11(15):6841. https://doi.org/10.3390/app11156841
Chicago/Turabian StyleSingh, Vineet, Youn-Chul Ryu, and Tatsuya Unno. 2021. "Dietary Intervention Induced Distinct Repercussions in Response to the Individual Gut Microbiota as Demonstrated by the In Vitro Fecal Fermentation of Beef" Applied Sciences 11, no. 15: 6841. https://doi.org/10.3390/app11156841
APA StyleSingh, V., Ryu, Y.-C., & Unno, T. (2021). Dietary Intervention Induced Distinct Repercussions in Response to the Individual Gut Microbiota as Demonstrated by the In Vitro Fecal Fermentation of Beef. Applied Sciences, 11(15), 6841. https://doi.org/10.3390/app11156841








