Abstract
Classification and regression are the major applications of machine learning algorithms which are widely used to solve problems in numerous domains of engineering and computer science. Different classifiers based on the optimization of the decision tree have been proposed, however, it is still evolving over time. This paper presents a novel and robust classifier based on a decision tree and tabu search algorithms, respectively. In the aim of improving performance, our proposed algorithm constructs multiple decision trees while employing a tabu search algorithm to consistently monitor the leaf and decision nodes in the corresponding decision trees. Additionally, the used tabu search algorithm is responsible to balance the entropy of the corresponding decision trees. For training the model, we used the clinical data of COVID-19 patients to predict whether a patient is suffering. The experimental results were obtained using our proposed classifier based on the built-in sci-kit learn library in Python. The extensive analysis for the performance comparison was presented using Big O and statistical analysis for conventional supervised machine learning algorithms. Moreover, the performance comparison to optimized state-of-the-art classifiers is also presented. The achieved accuracy of 98%, the required execution time of 55.6 ms and the area under receiver operating characteristic (AUROC) for proposed method of 0.95 reveals that the proposed classifier algorithm is convenient for large datasets.
1. Introduction
Classification plays a vital role in machine leaning, pattern recognition and data analytics. There are several classifications and prediction algorithms, proposed in recent solutions to provide intelligent decision making by extracting the relevant information from historical/large data [1,2,3,4]. Moreover, the exploration of machine learning algorithms and techniques has grown enormously due to the considerable progress made in the information storage and processing capabilities of computers [3,5,6]. Therefore, the machine learning algorithms can be classified into four categories: (1) supervised learning; (2) unsupervised learning; (3) semi-supervised learning; and (4) reinforcement learning [7,8]. Concerning supervised machine learning, there are several prediction algorithms (or) approaches in the literature such as the kth nearest neighbor, naïve Bayes, decision trees, support vector machines, logic regression and random forest, etc. [9]. All these classification algorithms are first trained using historical data. Then, the trained prediction model is employed in the designated application environment.
The decision tree algorithm is one of the prominent techniques for machine learning, data mining and pattern recognition. These algorithms construct classification or regression models exploiting the partitioning of data based on regular recursion [10]. Generally, a decision tree algorithm starts with the complete dataset, breaks the dataset into two or more subsets on the basis of one or more than one attribute and then this process repeats until an appropriate number of finer subsets are generated. The modeled process could be represented as a tree structure and generally, it is summarized as a set of “if-else” statements [11]. The decision tree algorithm is easy to elucidate and it can handle noisy data. In the discipline of computer science and engineering, a decision tree is used for the diagnosis of medical diseases, electrical troubleshooting, image recognition, signal processing, etc. [10,12]. Concerning its industrial applications, the decision tree algorithm is widely used for fraud detection, the management of customer data, payment evaluations, etc. Moreover, a decision tree is also employed in different data mining applications and it can aid in large dataset evaluation.
We note that the aforementioned classification algorithms have the inability to adapt their dynamic behaviors or vary the conditions of different learning models [10,13,14,15,16,17]. Therefore, there is a real need for robust algorithms, (or) designs, (or) approaches with the capability to accommodate the dynamic behaviors of different learning models. Furthermore, the behavioral/functional improvement of the aforementioned prediction algorithms in terms of different parameters, i.e., performance, accuracy, etc., is essential. As compared to other aforementioned prediction algorithms, the decision tree algorithm (DT) provides higher computational accuracy [11,18,19]. Therefore, by considering the robustness, the main objective of this work was to provide performance and an accuracy-oriented classifier using both the decision tree and tabu search algorithms at the same time.
Our Contributions
The contributions of this work are given as follows:
- We proposed a performance-oriented classifier algorithm for training supervised ML models over large datasets using decision tree and tabu search algorithms, respectively. We termed the proposed classifier algorithm “tabu search oriented decision tree (TSODT)” (see Section 4).
- We provided a functional setup for the proposed TSODT classifier algorithm to perform several experimentations (as can be seen in Section 5).
- The linear and logarithmic Big O evaluation of our proposed classifier is presented (as can be seen in Section 6.4.3).
- We provided a statistical analysis of the proposed TSODT classifier algorithm (see Section 6.4.3).
The experimental results were performed using the TSODT classifier which exploits the built-in sci-kit-learn library in Python. The achieved 98% accuracy and the required 55.6 ms execution time reveals that the proposed classifier algorithm is convenient for large datasets.
The remainder of this work is structured as follows: The required background pertaining to the decision tree and tabu search algorithms is presented in Section 3. The proposed classifier algorithm is described in Section 4. The functional setup to perform this study is presented in Section 5. The results and comparison with state-of-the-art methods’ performances are given in Section 6. Finally, the article is concluded in Section 7.
2. Existing Classifiers and Their Limitations
Classifiers based on the DT algorithm. Initially, the DT algorithm was proposed to achieve higher accuracy with a reduced computational cost [11,20,21]. The accuracy varies with the robustness of the computational algorithm over the targeted data. The earlier DT algorithm, termed ID3, is more convenient for making simple decision trees. It results in a decrease in accuracy when the computational complexity increases. The most recent solutions for the accuracy improvement of the DT algorithm is described in [10,20,22,23,24,25].
In [10], an intelligent classifier based on a decision tree algorithm was structured for diverse applications. Recently, the solution given in [20] proposed an optimized structure of the decision tree to improve the efficiency and to reduce the error rate by simplifying 19 nodes to five node structures. The results reveal that the accuracy of ID3 was 6% to 7% higher as compared to conventional supervised machine learning classifiers. The solution described in [22] proposed an optimized genetic algorithm by merging the DT algorithm with a parallel genetic decision tree.
Towards performance improvement, the solution given in [23] has utilized a linear regression decision tree algorithm for data classification. To evaluate the effectiveness of their approach, a student evaluation and zoo datasets from the UCI Machine Learning Repository were considered. In [24], several classification models were trained to extract the relevant features from the targeted clinical or laboratory data. To achieve higher performance, relevant features from the different datasets were merged to construct a new dataset to train the learning model. A scalable decision tree algorithm was employed in [25] to classify the large dataset with improved efficiency as compared with other methods (such as SLIQ, SPRINT and RainForest). The efficiency was improved by reducing the sorting cost of the decision tree.
Classifiers based on combined DT and tabu search algorithms. Limited solutions are available in the literature where both DT and tabu search (TS) algorithms are utilized at the same time to improve the performance and accuracy of the learning classifiers [26,27,28,29,30].
In [26], a new algorithm was proposed to optimize the feature selection of hierarchical classification trees. To achieve this, a binary hierarchical classifier (BHC), also named the decision tree, was employed in this work. The result shows an improved accuracy when compared to traditional classifiers. Similarly, the solution described in [27], proposed a new classifier (they termed this “ad hoc”) for logistic regression and discriminant analysis. This results in better accuracy as compared to conventional regression methods (i.e., stepwise, backward and forward).
The authors in [28] proposed an algorithm for classifying large datasets. Their classifier outperformed as compared to nominal classifiers in terms of accuracy. Moreover, the achieved accuracy was validated by employing a well-known dataset containing information relevant to different chronic diseases. An interesting classifier was proposed in [29] where better accuracy was achieved when compared to multivariate decision trees. In fact, the execution time linearly increased with the increase in the size of a dataset. Finally, the authors in [30] described a new algorithm (that they termed “cooperative-tabu-search”) for classifying important information into different categories. It iteratively reduces the leaf nodes and transforms the data into new space. The results after several experiments reveal that their classifier achieves better accuracy as compared to conventional classifiers. In a nutshell, the work given in [26,27,28,30] reports improved accuracy while the solution proposed in [29] outperformed in terms of both accuracy and execution time.
Classifiers have also been employed for the detection and prediction of COVID-19. In December 2019, a number of people started suffering from the idiopathic disease in Wuhan, China, displaying symptoms similar to pneumonia. A number of samples were collected from the affected pneumonia patients to determine the specific health issues. The published works were reported in [31,32,33,34,35,36], where the occurrence of unbiased sequences was discovered among the affected pneumonia patients. These studies employed different classifiers to either detect or predict COVID-19.
As summarized, machine learning and data mining techniques are getting more important in analyzing large datasets using different classifiers. Moreover, extracting important information or making a decision from the trained model over-employed datasets are additional benefits. Therefore, in the aforesaid solutions for medical-related applications (published in [10,20,22,23,24,25,26,27,28,29,30]), there are various implementations of supervised machine learning algorithms or approaches, either to detect or predict hazardous diseases, which has recently included the COVID-19 [31,32,34,35,37,38]. These solutions are based on standard supervised machine learning algorithms and tend to be specifically tailored for a single algorithm. These results decrease in accuracy and require higher computational time (especially for the provided datasets on COVID-19 patients). Consequently, there should be an intuitive way to obtain an early prediction of health issues with less computational time and higher accuracy.
3. Preliminaries
This section provides the essential background related to the decision tree algorithm in Section 3.1.
3.1. Decision Tree Algorithm
The hierarchy of the DT algorithm is presented in Figure 1. It belongs to a list of supervised machine learning algorithms. This is frequently employed for the classification of large datasets. Additionally, the DT algorithm was efficiently used to solve regression problems. The required computations for the implementation of the DT algorithm were based on a training model. The training model was required to predict the class (or) value of the given variable based on the learning rules. The learning rules were derived from the training data.
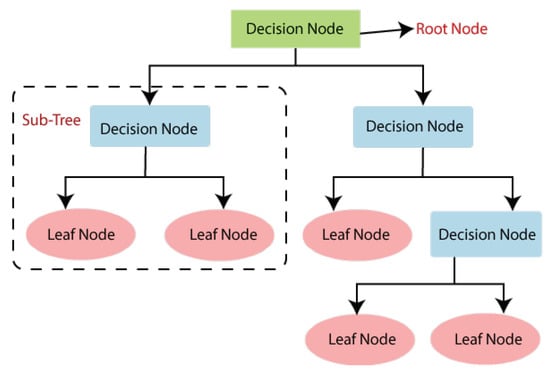
Figure 1.
Structure of the decision tree (DT) algorithm.
As shown in Figure 1, the structure of the DT algorithm consists of several nodes, i.e., root, decision and leaf. The root node initiates the tree while the decision nodes are responsible for decision-making, i.e., switching from one node to another. The leaf nodes act as an output from decision nodes. In other words, the DT algorithm starts from the root of the tree (root node, shown in green color in the figure) to predict a class label. It compares the root attribute with the data attributes. Based on this comparison, it follows the corresponding branch and moves to the next node (decision nodes, shown in blue color in the figure). Concerning a certain set of rules, the decision nodes are responsible for moving on the corresponding leaf node (shown with pink color in the figure). In short, the entire tree contains several sub-trees, as presented in Figure 1. Each sub-tree consists of decision and leaf nodes.
4. Proposed Classifier Algorithm
The general idea of the DT algorithm is presented in Section 3.1. Therefore, the proposed TSODT algorithm goes recursively to divide the given data using the “greedy depth-first approach”. This approach is also utilized in our work to optimize the traditional decision tree algorithm. The data flow and global overview of our proposed algorithm is given in Figure 2. It constructs a single decision tree (T) from a given dataset and then the tabu search selects the root node as a current node to start the optimization of the decision tree. It is noteworthy that our proposed algorithm was flexible while creating new decision trees and this feature also aids TSODT to optimize the performance—regardless of the size of the dataset. We assumed N = 3 to explain the working principle of TDOST and this means that TSODT will construct three decision trees (e.g., DT1, DT2, DT3) and the tabu search extracts the neighbor nodes (V*) from the standard decision tree (T). This generates the best solution which is a leaf node (S*). Then, it creates the N number of decision trees (N = 3 for our case), extracting the best solution (S*) and then it appends into previously created decision trees. The place of the appended leaf node (S*) was decided using the entropy of the decision trees. In what follows, the tabu search checks the aspiration criterion and continues the same process until it achieves the aspiration criterion. Consequently, it builds an optimized decision tree that is robust as compared to the traditional decision tree (T).
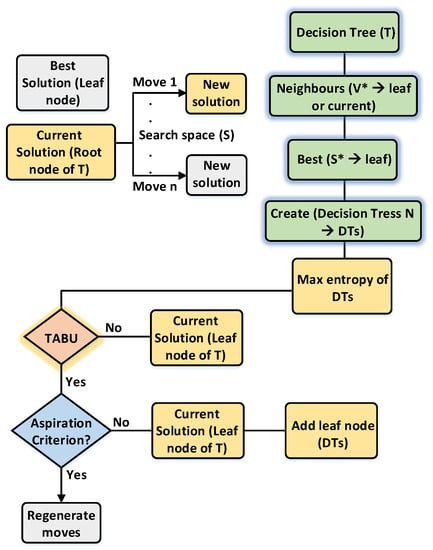
Figure 2.
Data flow of the proposed TSODT algorithm.
The proposed algorithm for the TSODT classifier is illustrated in Algorithm 1. Initially, it builds a tree (leaf nodes and decision nodes) and then it prunes so that the tree makes a lists [S, TABU] from the training instances. In what follows, it checks the lists [S, TABU], and if it is not empty, then it selects a node from the list and flags it as a current node J. It involves three steps (or) decision trees: (step-1) call tree classifies the extracted options as traditional or abnormal; (step-2) the identification of the abnormal options that contain signs of tuberculosis; and (step-3) the calculation of the likelihood of the malady from the results, obtained in step-2. Considering the aforementioned steps (step-1–step-3), it defines a splitting rule for the instances of the current node J. Moreover, it applies a select-split from J and forms child nodes. These child nodes are not a part of the decision trees. Once the resultant node is the only one, then J will be added to leaf nodes. The position of the current node (J) in the leaf nodes is defined on the entropy of the decision trees. On the other hand, the resultant nodes are added to the defined decision trees (DT1, DT2, DT3). The probability of each decision tree is based on the fraction of entropy, calculated using Equation (1). It is important to note that the proposed TSODT classifier algorithm is capable of dealing with big datasets. Moreover, it is more flexible than other classification algorithms because the user can select the number of decision trees which will be sued to build a final decision tree as explained in the Algorithm 1:
In Equation (1), determines the percentage of each class present in the child node (node for a specific value of the feature). The term, i.e., DTi presents the decision trees (as mentioned earlier in step-1–step-3) while k shows the number of classes.
| Algorithm 1: Proposed Tabu Search Oriented Decision Tree (TSODT) Algorithm |
Input: TD (training data), R (target aspiration), N (number of decision trees), RN( rootnode), A (attributes of trainig data) |
Output: CLo ← f (TD, R, N, RN) |
 |
5. Experimental Setup for the Evaluation of Tsodt
The block diagram of settings to perform different experiments is illustrated in Figure 3. The dataset was collected from clinical sources based on the symptoms of COVID-19. The preprocessing of the clinical data was accomplished by employing different filters (the corresponding details are given in Section 5.2). After preprocessing, the features were extracted based on their importance. Thereafter, the data were split into two parts, i.e., for training and testing. In the selected COVID-19 dataset, there is a class imbalance problem due to the negative class dominating the positive. In order to overcome this issue, we used a synthetic minority oversampling technique (SMOT). Once all the issues related to the selected dataset were resolved, in the following, training and modeling over data were performed by using supervised ML algorithms, i.e., naïve Bayes, KNN, logistic regression, support vector machine, random forest, DT and our proposed TSODT. Subsequently, each model was tested using test data to predict whether the patient suffered from COVID-19 or not. For each ML model, the performance accuracy was calculated using different accuracy measures (as shown in Figure 3). Finally, the performance comparison for selected ML models was provided.
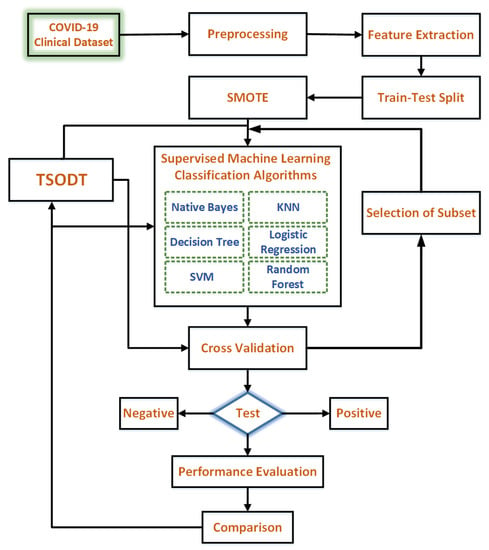
Figure 3.
Complete hierarchy of the proposed methodology.
Therefore, the relevant details for each block of Figure 3 was provided in the text that follows:
5.1. Clinical Data
The dataset of COVID-19 was collected from the GitHub repository of Carbon Health [39]. They compiled a repository named “Coronavirus Disease 2019 (COVID-19) Clinical Data Repository”, having the clinical data characteristics of patients who had taken a COVID-19 test. The repository was composed of CSV files and maintained its batches. Each batch contained the weekly test data of the patients from Carbon and Braid Health. The data include the clinical characteristics and symptoms for radiological and laboratory findings. Consequently, for our evaluations, we considered 11 weeks of data starting from the 7th of January to the 25th of April 2021. In short, all clinical data files contain 46 features and 11,168 records of patients.
5.2. Preprocessing
For training, preprocessing is essential to make the data readable for our model. Therefore, we considered: (1) the CSV files of 11 weeks of data for transformation into a single CSV file; (2) the exclusion of irrelevant columns; (3) transformation from categorical or text data into numerical data; (4) handled missing data problem; and (5) solved class imbalance problem.
5.2.1. Categorical Data into Numeric Data
To make the data machine-readable, these must be converted into machine language, as numeric data are very easy to understand for the training algorithms. Figure 4 shows the conversion of data categorical data into numerical data. The numbers used for converting categorical data into numeric data are “0” for false and “1” for true, “0” for negative and “1” for positive, “0” for mild, “1” for moderate and “2” for severe.
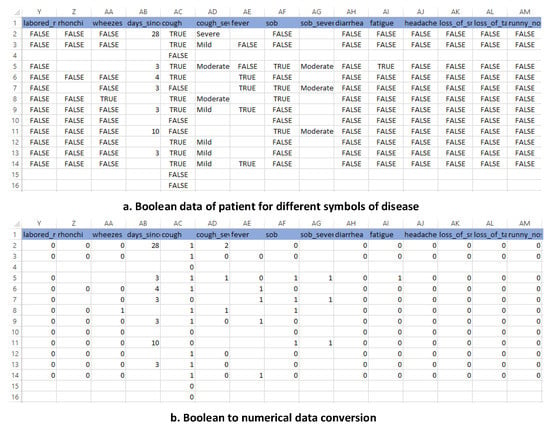
Figure 4.
Categorical data into numerical data.
5.2.2. Missing Data
The data which do not store properly for their respective variables are the missing data. The machine learning classifier does not train the data with missing values. Therefore, it is essential to handle these first. There are a few techniques to handle the missing data, i.e., imputation, a model-based technique, (or) simply ignore the missing data [40]. Initially, the filling rate of the data is to be visualized and then analyzed. Figure 5 represents the filling data rate for the selected COVID-19 dataset. The x axis of Figure 5 shows the symptoms and vitals of the patients and the y axis illustrates the corresponding filling rate for the symptoms and vitals. After filtration, the result of the data versus symptoms is 52.49 %. From Figure 5 and from previously obtained values, we can analyze how many data are filled and how many are missing for each symptom of COVID-19. Therefore, in this work, we used the following criteria to discard the missing data:
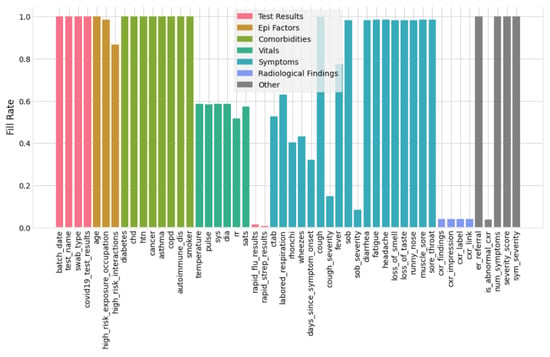
Figure 5.
Filling rate of the dataset.
- The columns with ≥50% missing data are dropped from the dataset. Consequently, after removing irrelevant and more than 50% missing data columns, the remaining columns are 31 (shown in Figure 5).
- For those features having less than 50% missing data, respective records/rows are dropped. After dropping missing data rows, 3616 records of patients were left for training and testing.
5.3. Feature Extraction
Feature extraction has great importance for the classification model to achieve accuracy, avoid overfitting and reduce training time. Therefore, we extracted 15 features out of 31 for the classification algorithms, as shown in Figure 6. This was achieved by using the built-in class, i.e., the ExtraTreeClassifier classifier.
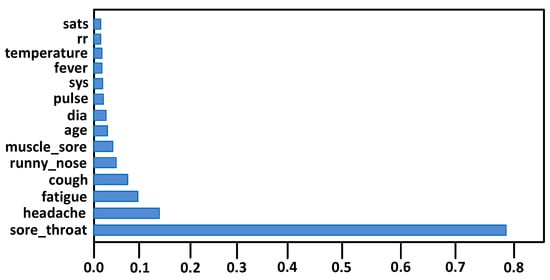
Figure 6.
Extracted features.
5.4. Classification Algorithms
All classification algorithms are implemented by using the sci-kit-learn library of machine learning in Python, which basically features various classification models. The implemented supervised ML models are (1) naïve Bayes (NB); (2) logistic regression (LR); (3) K-nearest neighbors (KNN); (4) decision tree (DT); (5) support vector machine (SVM); (5) random forest (RF); and (6) the proposed TSODT (see Algorithm 1). For mathematical details over these ML models, we turn readers to [41,42,43,44].
5.5. Test–Train Split
Prior to training the classifier model for prediction, we need to split our dataset into two parts: (1) to train the classification model, and (2) to test the performance of the model after training. In fact, the training data should be more than testing data. Each model is trained with 5-fold cross validation to tune the hyper-parameters. Normally, the 75–25 train–test ratio is used for classification [45]. Consequently, we used 70% of the data for training and 30% for testing.
5.6. Class Imbalance
Class imbalance is a common problem for classification algorithms. In binary classification, an imbalanced class problem means that one class dominates another, in which the first class is in the minority and the other is in majority [46]. In the selected COVID-19 dataset (covid19_test_results), there are two categories/labels, i.e., “1” (means positive) and “0” (means negative). Moreover, it contains a total of 3616 records. Among these 3616 records, 315 are positive, while the remaining are negative. This dominates 91.3% to 8.7% with the minority class of “positive”. There are numerous methods to resolve the class imbalance problem, including adaptive synthetic (ADASYN), random oversampling, synthetic minority oversampling technique (SMOTE), borderline SMOTE, SMOTE nominal and continuous (NC) [47,48]. The most frequently utilized techniques to reduce this issue are upsampling and downsampling. Each method has advantages and disadvantages, for example, random oversampling reduces the variance of the dataset, SMOTE can increase the overlapping of classes and it can create noise in the data. SMOTE is useful for large datasets and it alleviates the overfitting introduced due to oversampling [49]. Consequently, we employed a synthetic minority oversampling technique (SMOT) to reduce the aforementioned described class imbalance problem.
6. Experimental Results and Performance Evaluations
This section provides the criterion to investigate the achieved results, the calculated results and performance comparisons in Section 6.1–Section 6.4, respectively.
6.1. Parameters for Classifiers
As explained in Section 5.6, 5-fold cross validation is used to obtain the results. During training, the F1-score is maximized to optimize each classifier. The best learning model is extracted using the tuning of the learning rate (α) and the regularization parameter (λ) for LR. A set of nearest neighbors (100, 200…., 1000) is experimented with to obviate bias and overfitting. For the case of traditional decision tree and TDOST, pre-pruning is exploited to prevent overfitting with maximum splits. Considering SVM, Gaussian kernels were used for optimization.
6.2. Criterion to Calculate the Results
The most common performance measurement is based on the construction of a confusion matrix, as shown in Table 1. Column one provides the status of the COVID-19 patients in terms of them being either true or false. The terms, i.e., true and false, determine the COVID-19 patients in terms of sufferer and non-sufferer. Moreover, column two of Table 1 shows the classifier prediction-based on certain conditions, (either positive or negative). Consequently, there are four cases to construct the confusion matrix: (1) the classifier predicts the COVID-19 patient as positive (meaning a true positive, denoted as TP); (2) the classifier predicts the COVID-19 patient as negative (meaning a false negative, termed as FN); (3) the classifier predicts the non-COVID-19 patient as positive (meaning a false positive, denoted as FP); and (4) the classifier predicts the non-COVID-19 patient as negative (meaning a false negative, termed as FN).

Table 1.
Constructed confusion matrix.
6.3. Calculated Results
The experimental results were performed using TSODT and a traditional classifier that exploits the built-in sci-kit-learn library in Python. As stated in the explanation of TSODT, it is flexible for the selection of internal decision trees, so we performed decision tree space exploration. We recorded the execution of TSODT for decision trees (1, 2, 3…, 10) and this resulted in the selection of three decision trees (N = 3, input of Algorithm 1) which provided efficient execution. Hence, we used three decision trees for further computation. The achieved results are given in Table 2. The first column presents the type of classifier while the second column shows the execution time (in ms) required for training the data. Columns three to six give the numerical values for the constructed confusion matrix over each classifier, respectively. The remaining columns (i.e., columns seven to eleven) provide different parameters in terms of accuracy, error rate, precision, recall and F score. Therefore, the corresponding values for these parameters (accuracy, error rate, precision, recall, Fscore and AUROC (area under receiver operating characteristic)) in Table 2 are calculated using Equations (A1)–(A5), respectively, as given in Appendix A.

Table 2.
Results and performance comparison to conventional state-of-the-art classifiers.
As shown in Table 2, the proposed TSODT algorithm (Algorithm 1) requires 55.6 ms to train the model. The combined utilization of the decision tree and a tabu search algorithms, at the same time, result in 98.1% accuracy. Apart from the higher accuracy and the required execution time, the error rate is only 3.7% (see column eight of Table 2). The additional parameters, i.e., precision, recall, and Fscore, result in more than 97% (as shown in columns nine to eleven in Table 2).
6.4. Performance Comparison
In order to provide a realistic and reasonable performance evaluation, the proposed TSODT classifier (see Algorithm 1) is compared with conventional classifiers (i.e., NB, LR, DT, SVM, RF, and KNN) in Section 6.4.1. The comparison to most recent state-of-the-art classifier algorithms is provided in Section 6.4.2. Finally, the performance validation of the proposed TSODT classifier algorithm is given in Section 6.4.3.
6.4.1. Comparison to Conventional Classifiers
As shown in column two of Table 2, the most efficient classifier with respect to execution time (or performance) is NB as it trains the model in 8.5 ms. This is due to a requirement for the computation of prior probabilities. Once all the probabilities are calculated, they can be stored in a storage memory for efficient use in the next required computations. On the other hand, SVM is shown to slow down as compared to other classifiers in Table 2. This is because it requires to load the whole dataset in a RAM and thereafter, process each data point sequentially. It is important to worth mentioning that the selected clinical dataset for training in this work is quite large. Therefore, the proposed TSODT classifier algorithm (Algorithm 1) requires 1.79, 224.7 and 9.09 times lower execution/computational time to train the model as compared to LR, SVM and RF classifiers, respectively. Both the proposed TSODT and DT classifier algorithms take the same execution time (55.6 ms). On the other hand, the proposed classifier algorithm (Algorithm 1) requires a 6.54 and 3.97 times higher execution time as compared to the conventional NB and KNN classifier algorithms, respectively.
The proposed TSODT classifier algorithm efficiently performs over a large number of features. Therefore, for the same number of features (fifteen), our proposed TSODT classifier algorithm (Algorithm 1) results in higher accuracy (98%, as can be seen in column seven of Table 2) as compared to other classifiers. It is important to worth mentioning that the higher the accuracy, the higher the classifier performance will be. As compared to other classifiers, i.e., NB, LR, DT, SVM, RF and KNN, the proposed TSODT classifier results as 1.01, 1.02, 1.04, 1.09, 1.02 and 1.04 times higher in accuracy. Moreover, the proposed TSODT algorithm aggregates a large number of decision trees, which limit the overfitting and error rate parameters due to certain bias.
As shown in column eight of Table 1, the proposed TSODT algorithm results in an error rate of 3.7%. Therefore, when concerning only the error rate for the comparisons, our proposed classifier provides a 1.45, 2.97, 2.60, 2.97, 1.18 and 2.78 times lower error rate as compared to the NB, LR, DT, SVM, RF and KNN conventional classifier approaches, respectively. As shown in columns nine to eleven in Table 3, our proposed TSODT classifier algorithm results in higher values for other parameters, i.e., precision (97.5%), recall (99.3%) and Fscore (98.4%).

Table 3.
Comparison to state-of-the-art classifiers.
6.4.2. Comparison to State-of-the-Art Classifier Algorithms
The performance comparison with the most recent state-of-the-art classifiers in terms of execution time and accuracy is presented in Table 3. In order to train the model, the classifiers, reported in [34,36,38,50], employed different classification approaches while the classifiers, reported in [23,28,30], consider decision tree optimizations using the tabu search algorithm. Note that we used the “–” in Table 3 where the relevant information is not provided.
Comparison to classifiers given in [34,36,38,50]. In [50], the employed classifiers are NB, SVM, RF and KNN. For these classifiers (NB, SVM, RF and KNN), the reported accuracy in Table 3 are 0.98%, 69.79%, 77.98% and 75.68%. Our proposed TSODT classifier results to be 100 (for NB), 1.40 (for SVM), 1.25 (for RF) and 1.29 (for KNN) times higher in accuracy when compared to [50]. Apart from accuracy, the execution time in [50] is only reported for the NB classifier (0.96 ms). This is comparatively 1.02 times higher than in this work (0.94).
Recently, the most interesting work is described in [36], where utilized classifiers are SVM, RF, and DT. For these classifiers (SVM, RF and DT), the reported values for accuracy in Table 3 are 89%, 94.3% and 0.91%. The proposed TSODT classifier result 1.10 (ratio of 98 over 89 for SVM), 1.03 (ratio of 98 over 94.3 for RF) and 107.69 (ratio of 98 over 0.91 for DT) times higher accuracy as compared to [36]. Similar to [50], the execution time in [36] is only given for the RF classifier (0.93 ms). This is comparatively 1.02 times lower than in this work (0.95).
In [38], the employed classifiers are NB, LR, SVM and KNN. For the corresponding classifiers, the achieved accuracies are 94.3%, 94.41%, 92.4% and 89.2% (shown in Table 3). As compared to [38], our proposed TSODT classifier result 1.03 (for NB), 1.03 (for LR), 1.06 (for SVM) and 1.09 (for KNN) times higher accuracy. Similarly to [38], the work reported in [34] utilizes different classifiers (i.e., NB, LR, SVM, RF, and DT) for the detection of COVID-19. For the performance comparisons, only the values for accuracy are provided, as shown in Table 3. Comparatively, the proposed TSODT classifier algorithm is 1.01, 1.01, 1.08, 1.03 and 106.52 times faster (in terms of accuracy) than NB, LR, SVM, RF and DT algorithms, respectively.
Comparison over the optimized decision tree using the tabu search algorithm [23,28,30]. As shown in Table 3, the authors in [23] achieved 92.5% accuracy for their proposed optimized algorithm. This is comparatively 1.05 times lower than in this work. Moreover, there is a linear relationship between the execution time with datapoints. The classifier reported in [28] tested their algorithm for different datasets of chronic diseases. A maximum accuracy of 96% was achieved, which is comparatively 1.02 times lower than in our work (98%). The authors in [30] achieved 95.5% accuracy, which is comparatively 1.02 times lower than in this work. Similarly to [23], there is a linear increase in execution time with the increase in datapoints.
6.4.3. Performance Validation of the Proposed TSODT Classifier Algorithm
After providing comparisons in Section 6.4.1 and Section 6.4.2, we validated the performance of our proposed TSODT algorithm for different data points, as shown in Figure 7. The x axis in the figure shows the number of datasets with different data points (in ) while the y axis, shows the execution time (in ms). Consequently, the trend (presented in Figure 7) explicitly shows that there is a linear increase in the execution time as with the increase in the number of data points.
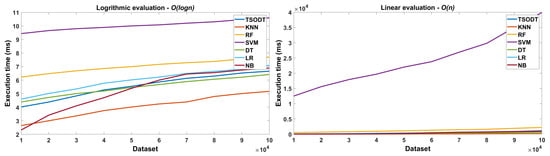
Figure 7.
Performance comparison different classifiers for logarithmic and linear Big O.
Evaluation of Big O. We evaluated our classifier with other classifiers for linear and logarithmic Big O. Figure 8 illustrates the evaluation of logarithmic and linear Big O for TSODT and other traditional classifiers. The x axis of this figure shows the number of elements in a dataset and y axis shows the complexity of Big O in terms of execution time. It is clear that the algorithm complexity of TSODT is linear O(n) and this trend is depicted in algorithmic and logarithmic complexity.
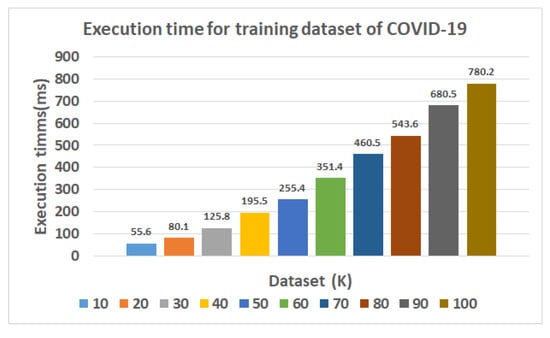
Figure 8.
Performance validation (based on execution time) of our proposed TSODT classifier.
Statistical analysis. Statistical analysis was performed to compare the proposed classifier and traditional classifiers. For this, the Friedman test and a post hoc analysis were performed [51]. The results for classification algorithms using the dataset are presented in Table 4. This table compares the TSODT and other classifiers with each other in terms of average ranks. The listed numbers illustrate subsequent classification algorithms sorted by the accuracy in ascending order. TSODT is highlighted as a number 1, the sum of average ranks is maximum for TSODT. Consequently, TSODT outperforms as compared to other classifiers. On the other hand, the value of AUROC, as shown in the last column of Table 2, confirms that TSODT has the highest probability as compared to the other classifiers. The results shows that NB and LR also offer better performance which shows high sensitivity and specificity.

Table 4.
Results using post hoc analysis.
Analysis of AUROC. We also evaluated the performance of our classifier by comparing the AUROC of traditional and TSODT. Figure 9 illustrates the ROC curves for different classifiers. The ROC curve is generated by considering sensitivity and specificity. The x axis of this figure shows the false positive rate (FPR) and the y axis shows the true positive rate (TPR) of the corresponding classifier. The numeric values of AUROC are listed in the last column of Table 2. The AUROC for TSODT is higher as compared to other classifiers. On the other hand, the AUROC of KNN is the lowest among all classifiers. The evaluation of results is based on the COVID-19 dataset, which proves that our classifiers outperform. The values for AUROC might be different for other datasets but the comparative value will be higher. Generally speaking, the evaluation also shows that this classifier is well suited for medical applications.
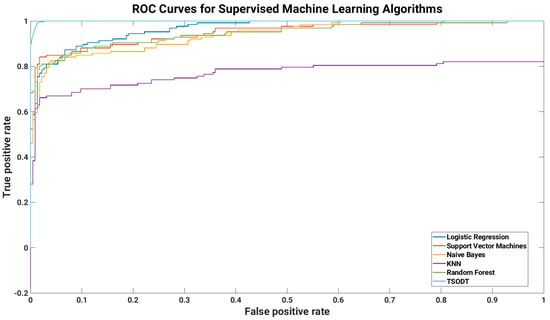
Figure 9.
Performance validation (based on AUROC) of our proposed TSODT classifier.
7. Conclusions
Machine learning algorithms are extensively being in used for the prediction of numerous diseases, e.g., diabetic retinopathy, COVID-19. The prediction/diagnosis is achieved with the aid of a classifier. In this work, a new classifier based on DT and tabu search is presented to optimize the performance and accuracy at the same time. Half a dozen supervised machine learning (supervised ML) techniques (including TSODT) were exploited to diagnose the accustomed COVID-19 illness victimization using the clinical information of the patients. The experimental results reveal that the proposed classifier algorithm provides a higher performance for the medical applications with an accuracy of 98%. Additionally, the execution time required for the training is 55.6 ms. Consequently, the proposed classifier will help the relevant community solve different medical, social and statistical problems with an elevated accuracy and performance parameters.
Author Contributions
Conceptualization, H.T. and M.R.; methodology, M.A.H.; software, M.A.H. and Z.U.A.; validation, M.A.H., Z.U.A. and M.R.; formal analysis, M.R.; investigation, S.S.A.; resources, S.S.A. and M.H.S.; data curation, M.H.S.; writing—original draft preparation, M.A.H.; writing—review and editing, S.S.A. and M.R.; visualization, Z.U.A.; supervision, S.S.A.; funding acquisition, M.R. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported in part by the King Abdulaziz City for Science and Technology (KACST) under Grant 14-ELE1049-10, and in part by the Science and Technology Unit (STU), Mecca, Saudi Arabia.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We are thankful for the support of King Abdul-Aziz City for Science and Technology (KACST) and Science and Technology Unit (STU), MAKKAH, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. Mathematical Equations
References
- Russell, S. Artificial Intelligence: A Modern Approach, 4th ed.; Pearson: New York, NY, USA, 2019. [Google Scholar]
- Sivasakthi, M. Classification and prediction based data mining algorithms to predict students’ introductory programming performance. In Proceedings of the 2017 International Conference on Inventive Computing and Informatics (ICICI), Coimbatore, India, 23–24 November 2017; pp. 346–350. [Google Scholar] [CrossRef]
- Savla, A.; Israni, N.; Dhawan, P.; Mandholia, A.; Bhadada, H.; Bhardwaj, S. Survey of classification algorithms for formulating yield prediction accuracy in precision agriculture. In Proceedings of the 2015 International Conference on Innovations in Information, Embedded and Communication Systems (ICIIECS), Coimbatore, India, 19–20 March 2015; pp. 1–7. [Google Scholar] [CrossRef]
- Imran, M.; Bashir, F.; Jafri, A.R.; Rashid, M.; ul Islam, M.N. A systematic review of scalable hardware architectures for pattern matching in network security. Comput. Electr. Eng. 2021, 92, 107169. [Google Scholar] [CrossRef]
- Negi, A.; Rajesh, K. A Review of AI and ML Applications for Computing Systems. In Proceedings of the 2019 9th International Conference on Emerging Trends in Engineering and Technology—Signal and Information Processing (ICETET-SIP-19), Nagpur, India, 1–2 November 2019; pp. 1–6. [Google Scholar] [CrossRef]
- Shehzad, F.; Rashid, M.; Sinky, M.H.; Alotaibi, S.S.; Zia, M.Y.I. A Scalable System-on-Chip Acceleration for Deep Neural Networks. IEEE Access 2021, 9, 95412–95426. [Google Scholar] [CrossRef]
- Binkhonain, M.; Zhao, L. A review of machine learning algorithms for identification and classification of non-functional requirements. Expert Syst. Appl. X 2019, 1, 100001. [Google Scholar]
- Hady, M.F.A.; Schwenker, F. Semi-supervised Learning. In Handbook on Neural Information Processing; Bianchini, M., Maggini, M., Jain, L.C., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 215–239. [Google Scholar]
- Rashid, M.; Imran, M.; Jafri, A.R. Exploration of Hardware Architectures for String Matching Algorithms in Network Intrusion Detection Systems. In Proceedings of the 11th International Conference on Advances in Information Technology; Association for Computing Machinery: New York, NY, USA, 2020. [Google Scholar]
- Navada, A.; Ansari, A.N.; Patil, S.; Sonkamble, B.A. Overview of use of decision tree algorithms in machine learning. In Proceedings of the 2011 IEEE Control and System Graduate Research Colloquium, Shah Alam, Malaysia, 27–28 June 2011; pp. 37–42. [Google Scholar] [CrossRef]
- Hao, H.; Chen, T.; Lu, J.; Liu, J.; Ma, X. The Research and Analysis in Decision Tree Algorithm Based on C4.5 Algorithm. In Proceedings of the 2018 IEEE 3rd Advanced Information Technology, Electronic and Automation Control Conference (IAEAC), Chongqing, China, 12–14 October 2018; pp. 1882–1886. [Google Scholar] [CrossRef]
- RMishina, V.; Khomyakova, L. Dedollarization and settlements in national currencies: Eurasian and Latin American experience. Vopr. Ekon. 2020, 9, 61–79. [Google Scholar] [CrossRef]
- Zhang, S.; Li, X.; Zong, M.; Zhu, X.; Wang, R. Efficient kNN Classification With Different Numbers of Nearest Neighbors. IEEE Trans. Neural Netw. Learn. Syst. 2018, 29, 1774–1785. [Google Scholar] [CrossRef]
- Aksoy, G.; Karabatak, M. Performance Comparison of New Fast Weighted Naïve Bayes Classifier with Other Bayes Classifiers. In Proceedings of the 2019 7th International Symposium on Digital Forensics and Security (ISDFS), Barcelos, Portugal, 10–12 June 2019; pp. 1–5. [Google Scholar]
- Junli, C.; Licheng, J. Classification mechanism of support vector machines. In Proceedings of the WCC 2000—ICSP 2000, 2000 5th International Conference on Signal Processing, 16th World Computer Congress, Beijing, China, 21–25 August 2000; Volume 3, pp. 1556–1559. [Google Scholar] [CrossRef]
- Rashid, M.; Shah, S.A.B.; Arif, M. Determination of Worst-Case Data using an Adaptive Surrogate Model for Real-Time System. J. Circuits Syst. Comput. 2020, 29, 2050005. [Google Scholar] [CrossRef]
- Shah, S.A.B.; Rashid, M.; Arif, M. Estimating WCET using prediction models to compute fitness function of a genetic algorithm. Real-Time Syst. 2020, 56, 28–63. [Google Scholar] [CrossRef]
- Al Hamad, M.; Zeki, A.M. Accuracy vs. Cost in Decision Trees: A Survey. In Proceedings of the 2018 International Conference on Innovation and Intelligence for Informatics, Computing, and Technologies (3ICT), Sakhier, Bahrain, 18–20 November 2018; pp. 1–4. [Google Scholar] [CrossRef]
- Alam, F.I.; Bappee, F.K.; Rabbani, M.R.; Islam, M.M. An Optimized Formulation of Decision Tree Classifier. In Advances in Computing, Communication, and Control; Unnikrishnan, S., Surve, S., Bhoir, D., Eds.; Springer: Singapore, 2013; pp. 105–118. [Google Scholar]
- Zhang, H.; Zhou, R. The analysis and optimization of decision tree based on ID3 algorithm. In Proceedings of the 2017 9th International Conference on Modelling, Identification and Control (ICMIC), Kunming, China, 10–12 July 2017; pp. 924–928. [Google Scholar] [CrossRef]
- Jati, W.K.; Kemas Muslim, L. Optimization of Decision Tree Algorithm in Text Classification of Job Applicants Using Particle Swarm Optimization. In Proceedings of the 2020 3rd International Conference on Information and Communications Technology (ICOIACT), Yogyakarta, Indonesia, 24–25 November 2020; pp. 201–205. [Google Scholar] [CrossRef]
- Yuan, F.; Lian, F.; Xu, X.; Ji, Z. Decision tree algorithm optimization research based on MapReduce. In Proceedings of the 2015 6th IEEE International Conference on Software Engineering and Service Science (ICSESS), Beijing, China, 23–25 September 2015; pp. 1010–1013. [Google Scholar] [CrossRef]
- Ahmed, A.M.; Rizaner, A.; Ulusoy, A.H. A Decision Tree Algorithm Combined with Linear Regression for Data Classification. In Proceedings of the 2018 International Conference on Computer, Control, Electrical, and Electronics Engineering (ICCCEEE), Khartoum, Sudan, 12–14 August 2018; pp. 1–5. [Google Scholar] [CrossRef]
- Eman Abdel, M.; Sherif, B.; Mohammed, E. Chapter 9—Medical Images Analysis Based on Multilabel Classification. In Machine Learning in Bio-Signal Analysis and Diagnostic Imaging; Nilanjan, D., Surekha, B., Amira, S.A., Fuqian, S., Eds.; Academic Press: Cambridge, MA, USA, 2019; Chapter 10; pp. 209–245. [Google Scholar]
- Thangaparvathi, B.; Anandhavalli, D. An improved algorithm of decision tree for classifying large data set based on rainforest framework. In Proceedings of the 2010 International Conference on Communication Control and Computing Technologies, Nagercoil, India, 7–9 October 2010; pp. 800–805. [Google Scholar] [CrossRef]
- Donna, K.K. Investigating the Use of Tabu Search to Find Near-Optimal Solutions in Multiclassifier Systems; University of Texas at Austin: Austin, TX, USA, 2013. [Google Scholar]
- Silvia, C.Y.; Joaquín, P.B. Tabu Search for Variable Selection in Classification. In Encyclopedia of Data Warehousing and Mining; IGI Global: Hershey, PA, USA, 2009; pp. 1909–1915. [Google Scholar]
- Orsenigo, C.; Vercellis, C. Discrete support vector decision trees via tabu search. Comput. Stat. Data Anal. 2004, 47, 311–322. [Google Scholar] [CrossRef]
- Li, X.B.; Sweigart, J.; Teng, J.; Donohue, J.; Thombs, L.; Wang, S. Multivariate decision trees using linear discriminants and tabu search. IEEE Trans. Syst. Man Cybern. Part A Syst. Hum. 2003, 33, 194–205. [Google Scholar] [CrossRef]
- Ansari, E.; Sadreddini, M.; Sadeghi Bigham, B.; Alimardani, F. A combinatorial cooperative-tabu search feature reduction approach. Sci. Iran. 2013, 20, 657–662. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Chan, J.F.W.; Yuan, S.; Kok, K.H.; To, K.K.W.; Chu, H.; Yang, J.; Xing, F.; Liu, J.; Yip, C.C.Y.; Poon, R.W.S.; et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet 2020, 395, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Qin, L.; Yang, Y.; Cao, Q.; Cheng, Z.; Wang, X.; Sun, Q.; Yan, F.; Qu, J.; Yang, W. A predictive model and scoring system combining clinical and CT characteristics for the diagnosis of COVID-19. Eur. Radiol. 2020, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Akib Mohi Ud Din, K.; Syed Tanzeel, R.; Qamar Rayees, K.; Nusrat, R.; Masarat, M.U.D. Machine learning based approaches for detecting COVID-19 using clinical text data. Int. J. Inf. Technol. 2020, 12, 731–739. [Google Scholar]
- Mienye, I.D.; Sun, Y.; Wang, Z. Prediction performance of improved decision tree-based algorithms: A review. Procedia Manuf. 2019, 35, 698–703. [Google Scholar] [CrossRef]
- Rustam, F.; Khalid, M.; Aslam, W.; Rupapara, V.; Mehmood, A.; Choi, G.S. A performance comparison of supervised machine learning models for COVID-19 tweets sentiment analysis. PLoS ONE 2021, 16, e0245909. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Mu, L.; Li, J.; Wang, J.; Yin, Z.; Liu, D. Early Prediction of the 2019 Novel Coronavirus Outbreak in the Mainland China Based on Simple Mathematical Model. IEEE Access 2020, 8, 51761–51769. [Google Scholar] [CrossRef] [PubMed]
- Muhammad, L.J.; Algehyne, E.A.; Usman, S.S.; Ahmad, A.; Chakraborty, C.; Mohammed, I.A. Supervised Machine Learning Models for Prediction of COVID-19 Infection using Epidemiology Dataset. SN Comput. Sci. 2020, 2, 11. [Google Scholar] [CrossRef]
- Carbon Health and Braid Health. Coronavirus Disease 2019 (COVID-19) Clinical Data Repository. 2020. Available online: https://github.com/mdcollab/covidclinicaldata (accessed on 10 May 2020).
- Sanagapati, P. A Simple Tutorial—How to Handle Missing Data. 2020. Available online: https://www.kaggle.com/pavansanagapati/simple-tutorial-how-to-handle-missing-data (accessed on 20 April 2020).
- Sudirman, I.; Nugraha, D. Naive bayes classifier for predicting the factors that influence death due to COVID-19 in China. J. Theor. Appl. Inf. Technol. 2020, 98, 1686–1696. [Google Scholar]
- Xu, K.; Zhou, M.; Yang, D.; Ling, Y.; Liu, K.; Bai, T.; Cheng, Z.; Li, J. Application of Ordinal Logistic Regression Analysis to Identify the Determinants of Illness Severity of COVID-19 in China. Epidemiol. Infect. 2020, 148, 1–25. [Google Scholar] [CrossRef]
- Sethy, P.K.; Behera, S.K.; Ratha, P.K.; Biswas, P. Detection of coronavirus Disease (COVID-19) based on Deep Features and Support Vector Machine. Int. J. Math. Eng. Manag. Sci. 2020, 643–651. [Google Scholar] [CrossRef]
- Iwendi, C.; Bashir, A.K.; Peshkar, A.; Sujatha, R.; Chatterjee, J.M.; Pasupuleti, S.; Mishra, R.; Pillai, S.; Jo, O. COVID-19 Patient Health Prediction Using Boosted Random Forest Algorithm. Front. Public Health 2020, 8, 357. [Google Scholar] [CrossRef] [PubMed]
- Bose, A. Cross Validation—Why & How. 2020. Available online: https://towardsdatascience.com/cross-validation-430d9a5fee2 (accessed on 8 May 2012).
- Chawla, N.; Bowyer, K.; Hall, L.; Kegelmeyer, W. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. (JAIR) 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Jiang, Z.; Pan, T.; Zhang, C.; Yang, J. A New Oversampling Method Based on the Classification Contribution Degree. Symmetry 2021, 13, 194. [Google Scholar] [CrossRef]
- Salazar, A.; Vergara, L.; Safont, G. Generative Adversarial Networks and Markov Random Fields for oversampling very small training sets. Expert Syst. Appl. 2021, 163, 113819. [Google Scholar] [CrossRef]
- Izonin, I.; Tkachenko, R.; Shakhovska, N.; Lotoshynska, N. The Additive Input-Doubling Method Based on the SVR with Nonlinear Kernels: Small Data Approach. Symmetry 2021, 13, 612. [Google Scholar] [CrossRef]
- Almansoor, M.; Hewahi, N.M. Exploring the Relation between Blood Tests and COVID-19 Using Machine Learning. In Proceedings of the 2020 International Conference on Data Analytics for Business and Industry: Way Towards a Sustainable Economy (ICDABI), Sakheer, Bahrain, 26–27 October 2020; pp. 1–6. [Google Scholar] [CrossRef]
- Friedman, M. The Use of Ranks to Avoid the Assumption of Normality Implicit in the Analysis of Variance. J. Am. Stat. Assoc. 1937, 32, 675–701. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).