1. Introduction
Two dimensional (2D) Ultrasound (US) is a standard modality in medical imaging [
1,
2,
3,
4,
5]. It has several advantages. First, it does not have harmful effects on the human body. Second, it is relatively low cost compared with other modalities. Third, it provides real-time imaging during surgical operations [
1,
4,
5]. The research community is actively concerned about the advancements made in the area of ultrasound imagery analysis [
1,
4,
5,
6,
7]. This gives physicians the ability to get more information for image-guided intervention and surgery. One of the most tackled tasks in medical imaging analysis is the semantic segmentation of body organs and tissues [
4]. This procedure makes a partitioning of the input image into separate regions. Every region has a particular clinical relevance [
4].
On the other hand, Ultrasound imaging is challenging regards to the application of semantic segmentation algorithms. In fact, it is operator dependent and presents relatively fewer details compared to other modalities like Computerized Tomography scans (CT scan) or Magnetic Resonance Imaging (MRI) [
4]. Furthermore, medically used ultra-sonographic waves do not pass through bones [
5,
6,
7]. Even in the absence of bones, the depth penetration of the ultrasound waves depends on the acoustic impedance of the body organs. This makes an unclear representation of the organ boundaries [
4].
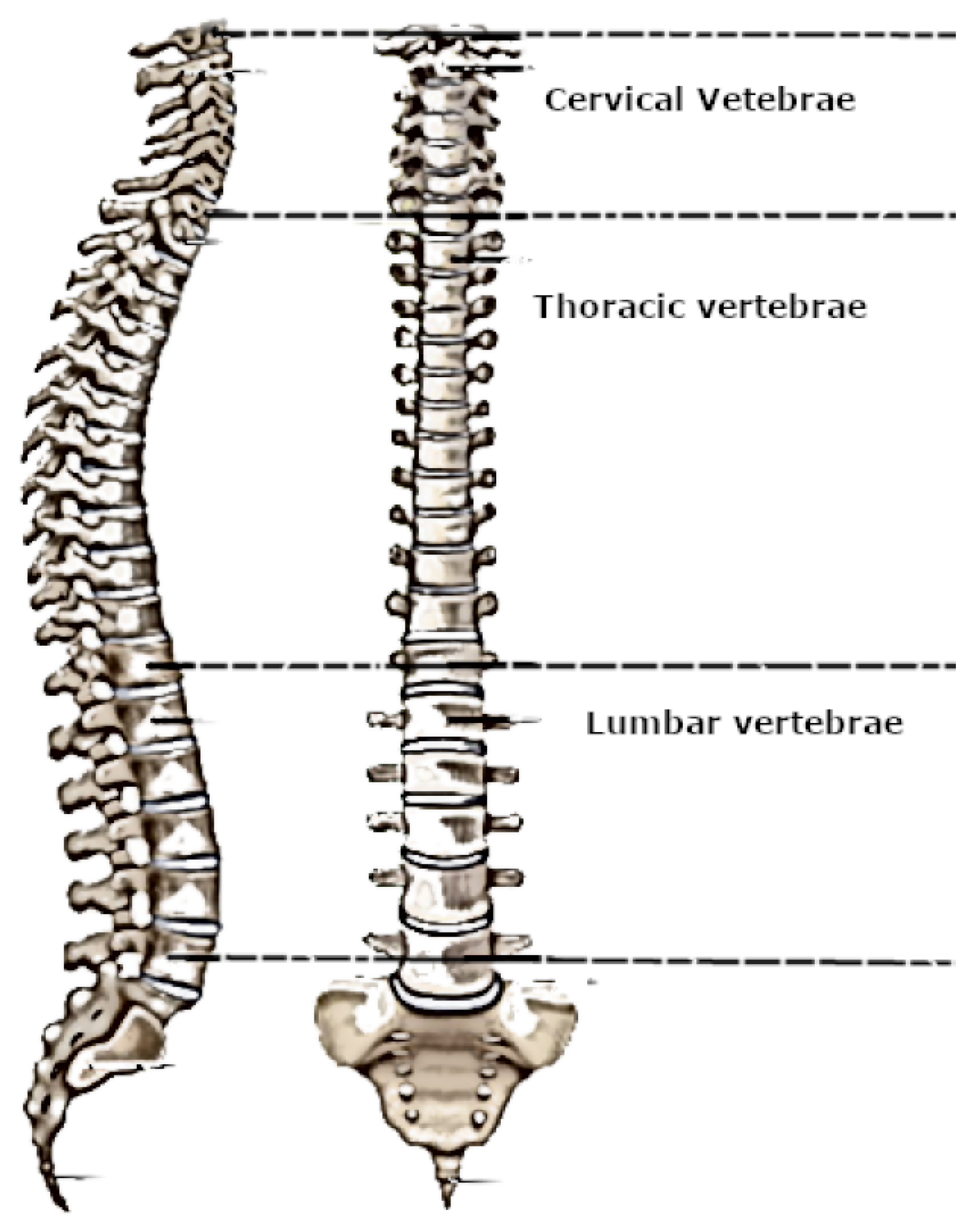
The Spine is also called vertebral column, spinal column, or backbone. It is composed of several bones named vertebrae [
2,
3]. The basic functions of the spine are to protect the spinal cord and to maintain the body alignment. The spinal cord is a part of the central nervous system containing major nerve tracts [
2,
3]. It connects the brain to nerves throughout the body and helps to regulate body functions. The spinal cord descends out of the brain at the base of the skull through the spinal column until it ends at the second lumbar vertebrae. It is enclosed and protected inside the bones of the spinal column. It is approximately about 45 cm (18 inches) long with a varying width from 13 mm (0.5 inch) in the cervical and lumbar regions to 6.4 (0.25 inch) in the thoracic area [
2].
Figure 1 shows a description of the human vertebrae from the anterior and the right lateral view.
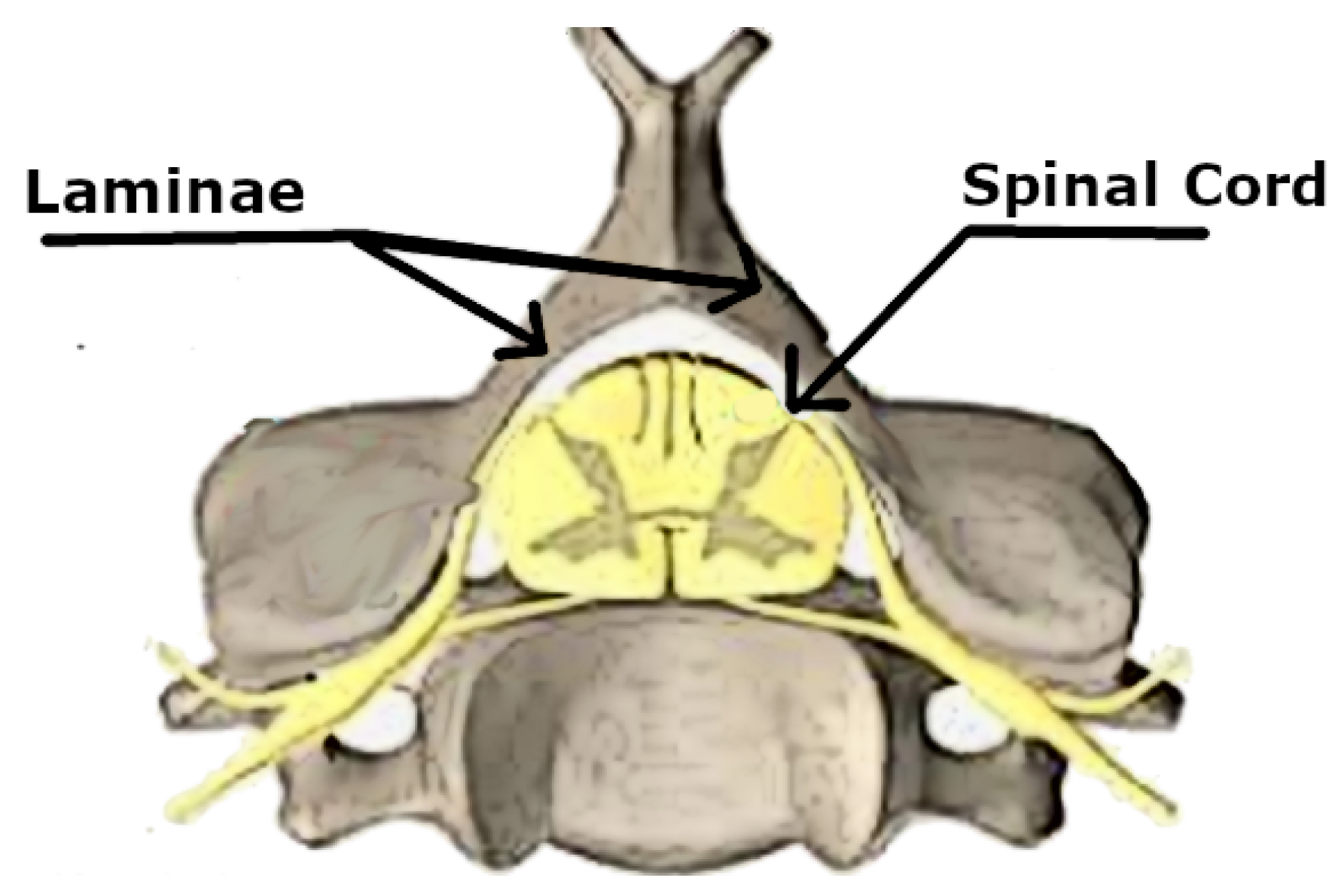
Figure 2 demonstrated the location of the spinal cord inside the spinal canal of the vertebral column. We could realize the location of the lamina on the back part of the vertebra and behind the spinal cord.
The laminectomy is a surgical operation performed by neurosurgeons and orthopedic surgeons to relieve compression on the spinal cord. This pressure may engender mild to severe back pain or difficulty in walking or in controlling limb functions. It can also have other symptoms that can interfere with daily life. The surgical procedure creates more space for the spinal cord and nerve roots to relieve abnormal pressure on the spinal cord by removing the laminae and the intervening ligaments [
3].
Figure 3 shows the laminectomy surgery and the spinal cord.
In normal situations, the spinal cord could not be imaged using ultrasound imaging because ultrasonic waves do not pass well through bones [
8]. In fact, the spinal cord is completely enclosed inside the bones of the spinal column. During laminectomy, ultrasonic waves can pass through the created bone defect to give real-time exploitable images of the spinal cord. Ultrasound imaging can demonstrate the spinal cord and the surrounding structures. This helps greatly to confirm the adequacy of the spinal cord decompression [
8]. The ultrasound is also able to detect spinal cord pulsation and record it in a video format [
8]. The created video can be saved for later use and analysis. It is believed that a decompressed spinal cord tends to have better pulsation. Spinal cord pulsation, however, is variable from one person to another [
8]. Providing an automatic solution for detecting the exact boundaries of the spinal cord is helpful for automatic interpretation and analysis of spinal cord pulsation. This spinal cord pulsation values could be interpreted alongside the electrocardiogram (the electrical activity of the heart) to interpret if both signals are synchronized, or there is some latency to interpret [
8].
But the great challenge here is the quality of the ultrasound image as boundaries could be obscured and some parts could be of foggy appearance [
8]. This is due to the different attenuations applied to the sonographic waves while passing through the human body. This makes the automatic segmentation more difficult to achieve. But the service that gives this solution for neurosurgeons strengthens the interest for further examination to resolve this issue.
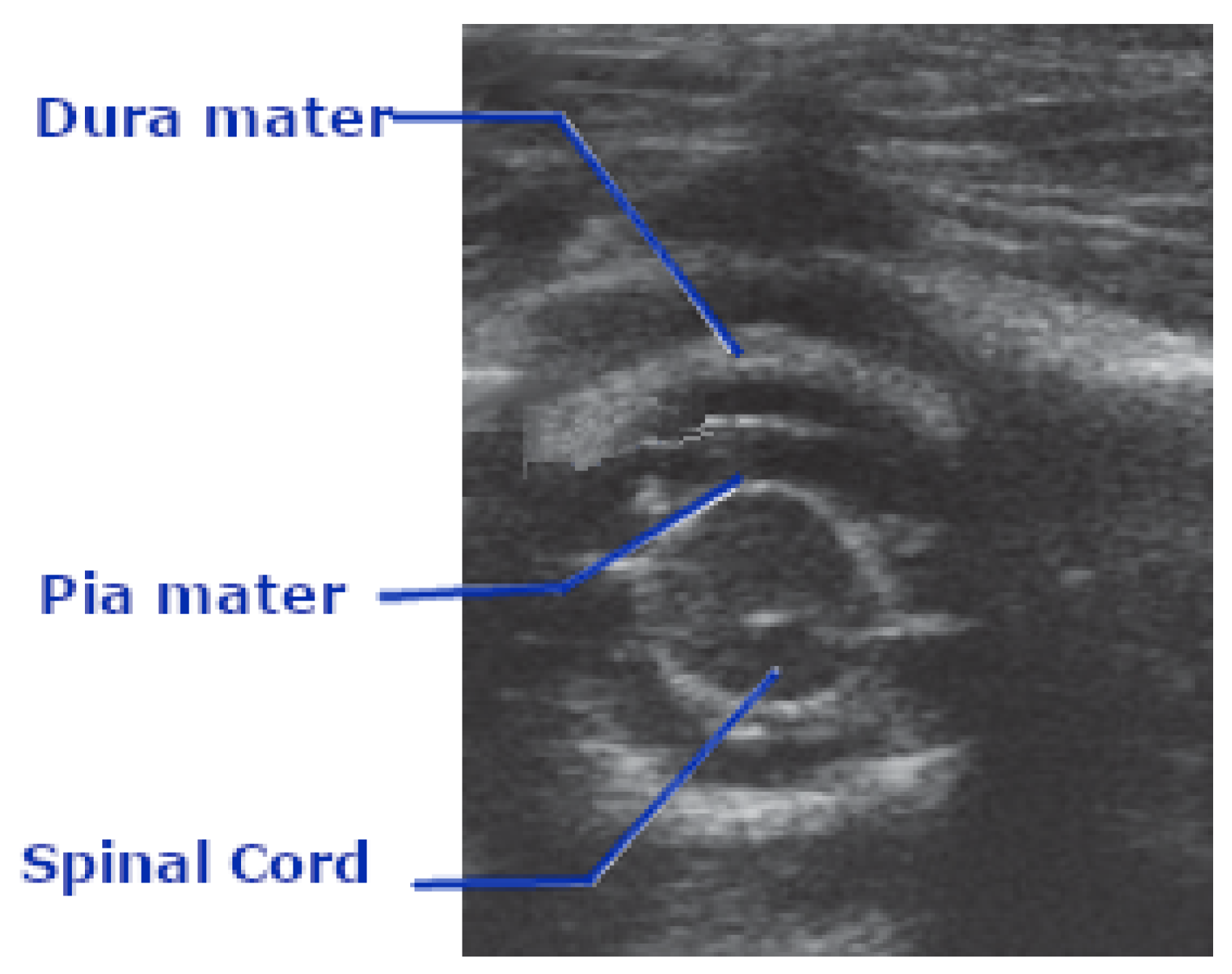
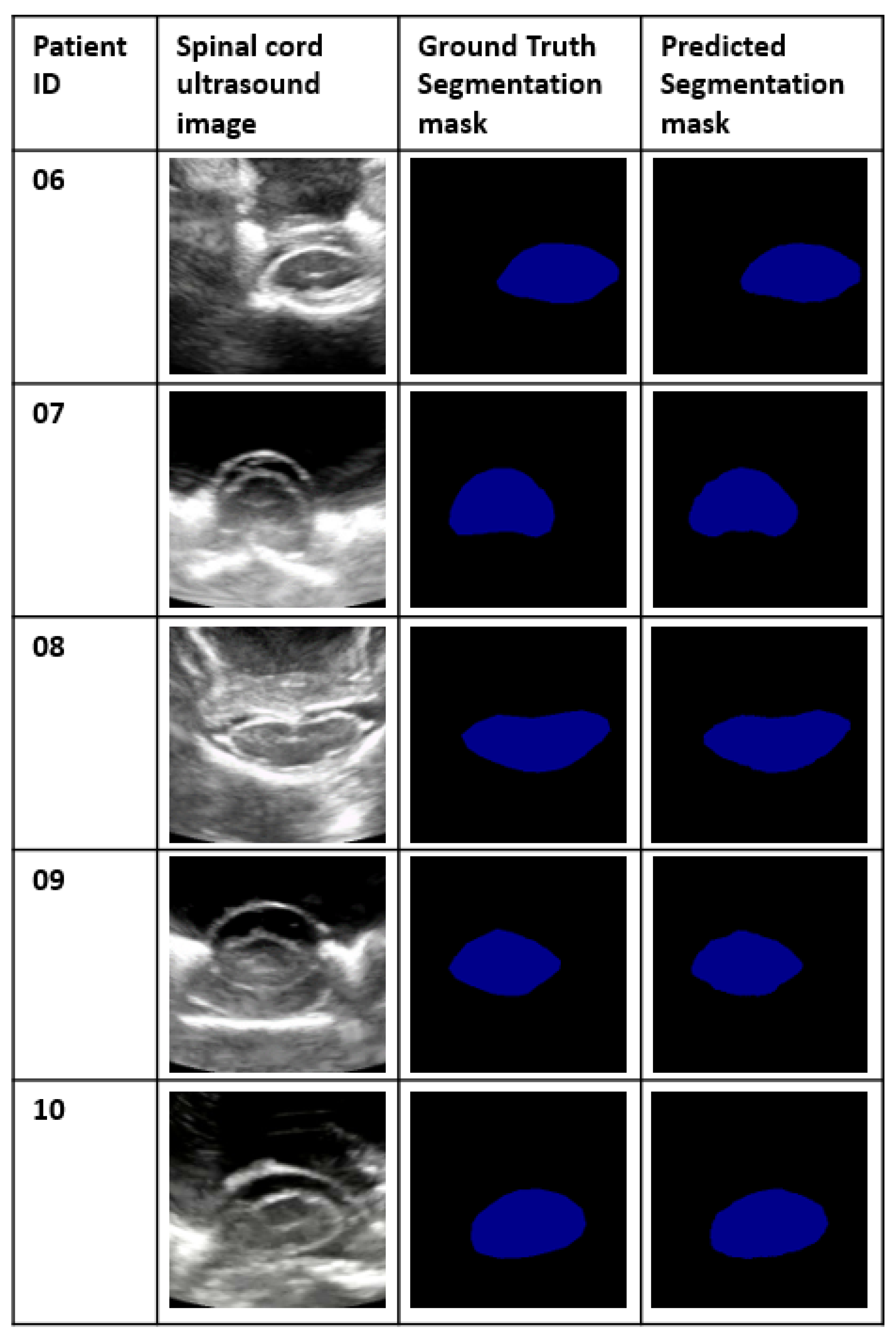
Figure 4 gives a clear representation of how the spinal cord is represented in ultrasound imaging.
Since 2012 [
9] deep learning based approaches have shown an attractive efficiency in object segmentation for multiple types of imaging (RGB imaging, aerial imaging, multi-spectral imaging etc.) [
10,
11,
12,
13,
14]. This was a source of inspiration for the medical imaging community to move their interest towards a great adoption of these approaches for different medical imaging modalities (MRI, CT Scan, 2D Ultrasound, 3D Ultrasound etc.) [
4].
Concerning the ultrasound medical segmentation, different works have been made in the detection of different parts of the body [
4] (breast image segmentation, vessel image segmentation, heart image segmentation etc.). However, as demonstrated in
Section 2, no one has treated the spinal cord image segmentation in 2D ultrasound medical imagery. In this study, we deduced, based on the current state of the art of image semantic segmentation, the best solution to adopt for our specific task. Our objectives in this paper were:
Treating, for the first time, the problem of spinal cord segmentation in ultrasound imaging.
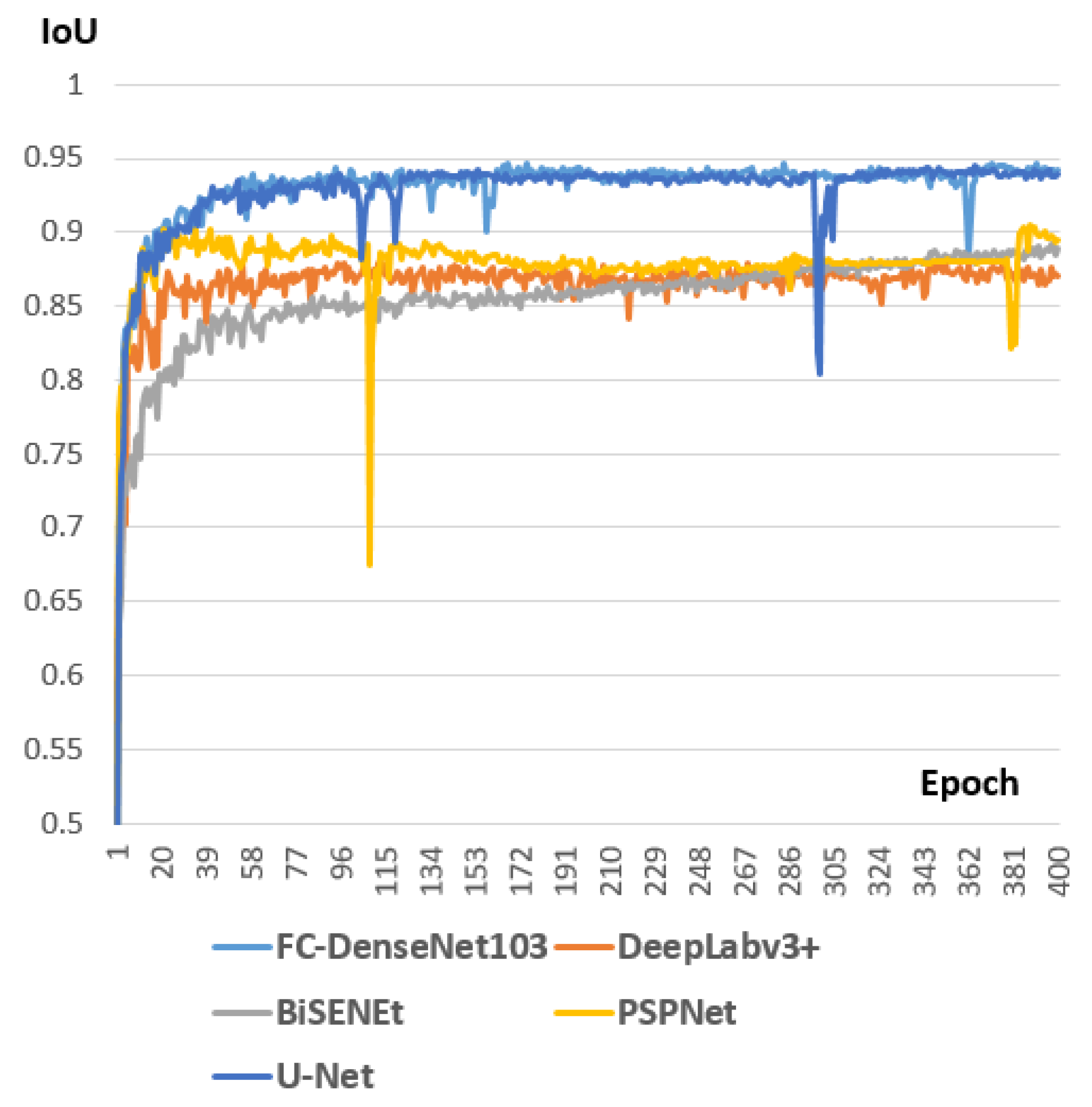
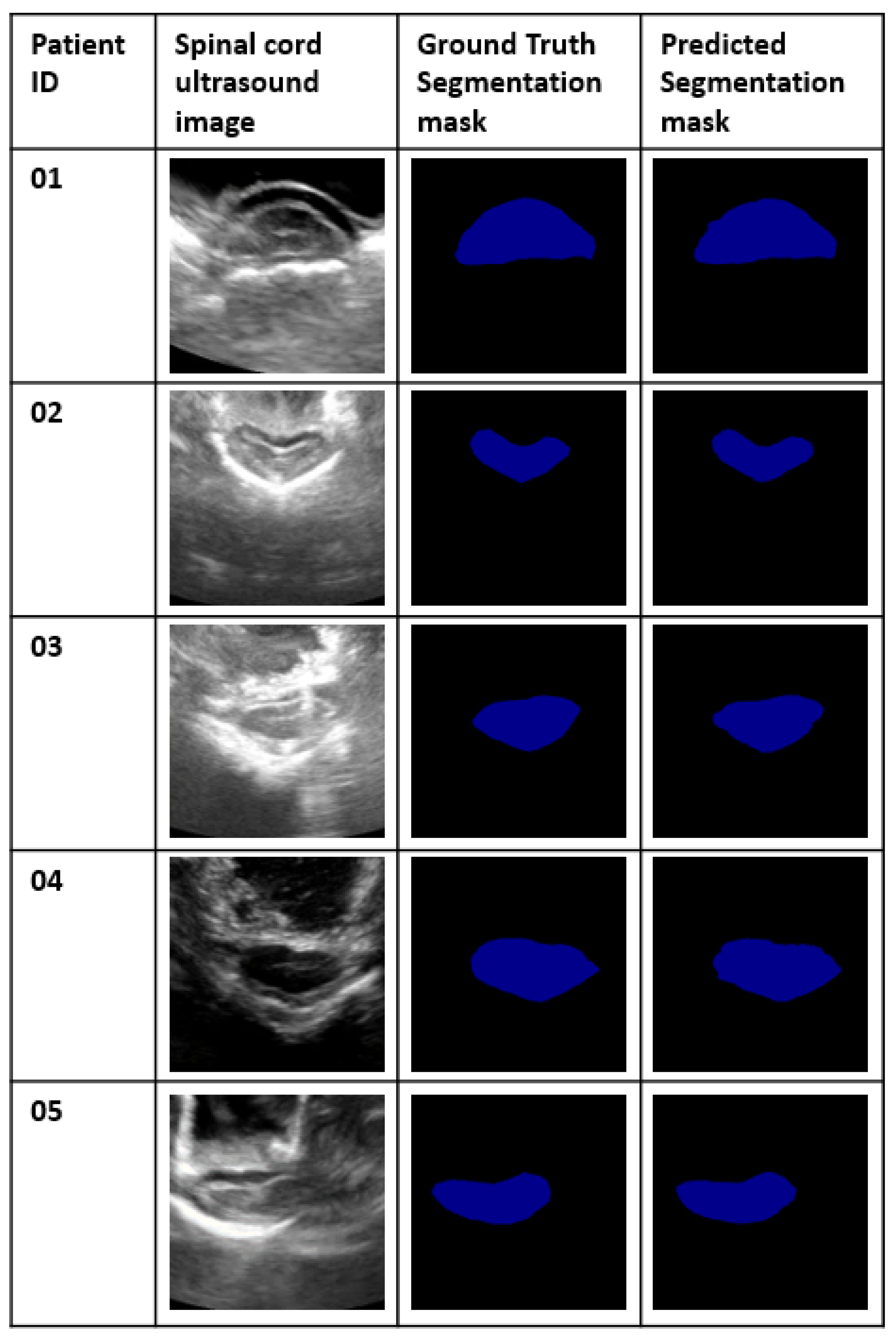
Introducing, based on the state of the art in the area of image semantic segmentation, the best model for two case studies. The first case study is when the spinal cord is already in the train set of the model. The second case study is when the spinal cord does not belong to the train set of the model. We constructed a separate dataset for each scenario and selected the best model in each case study.
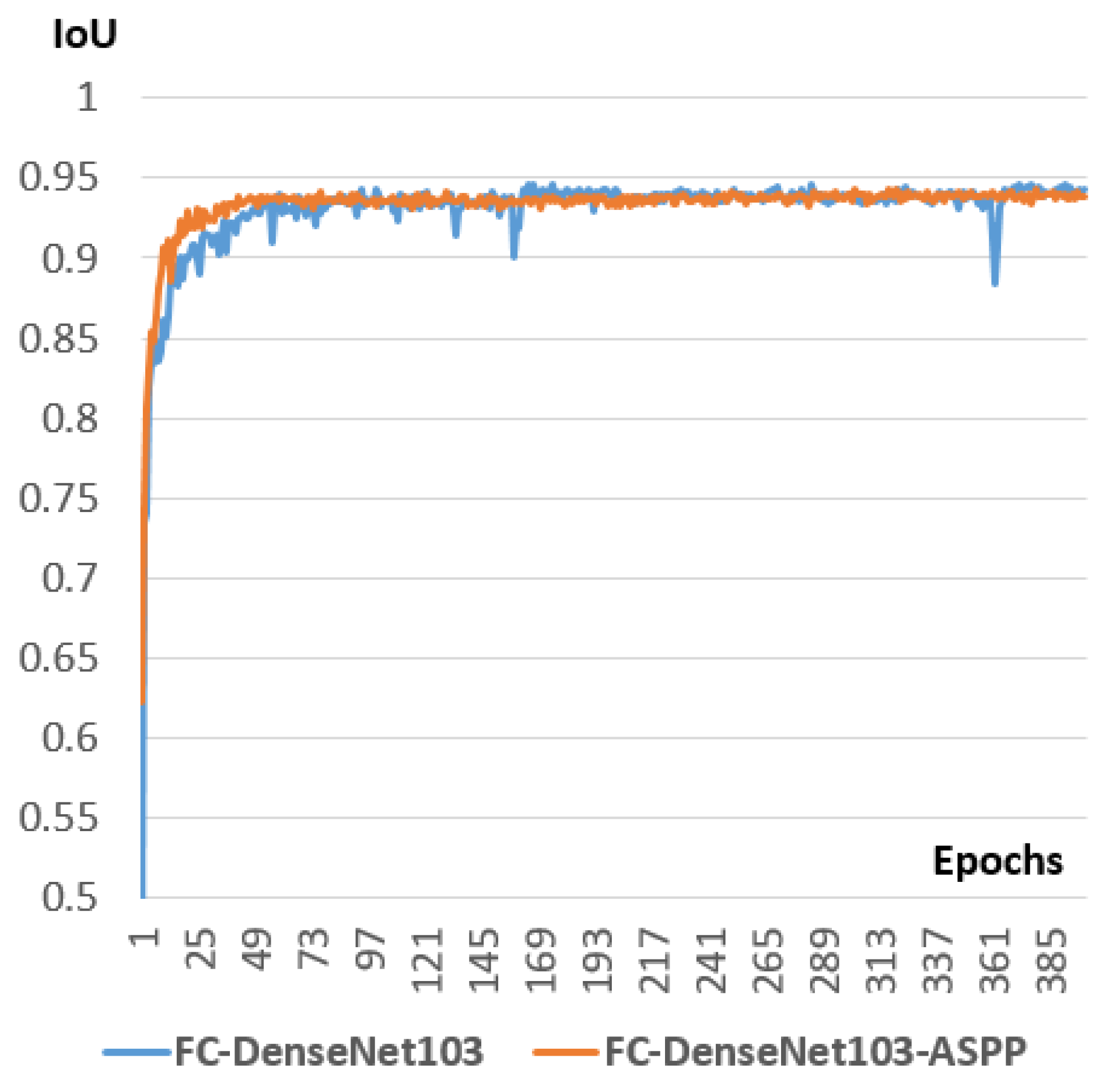
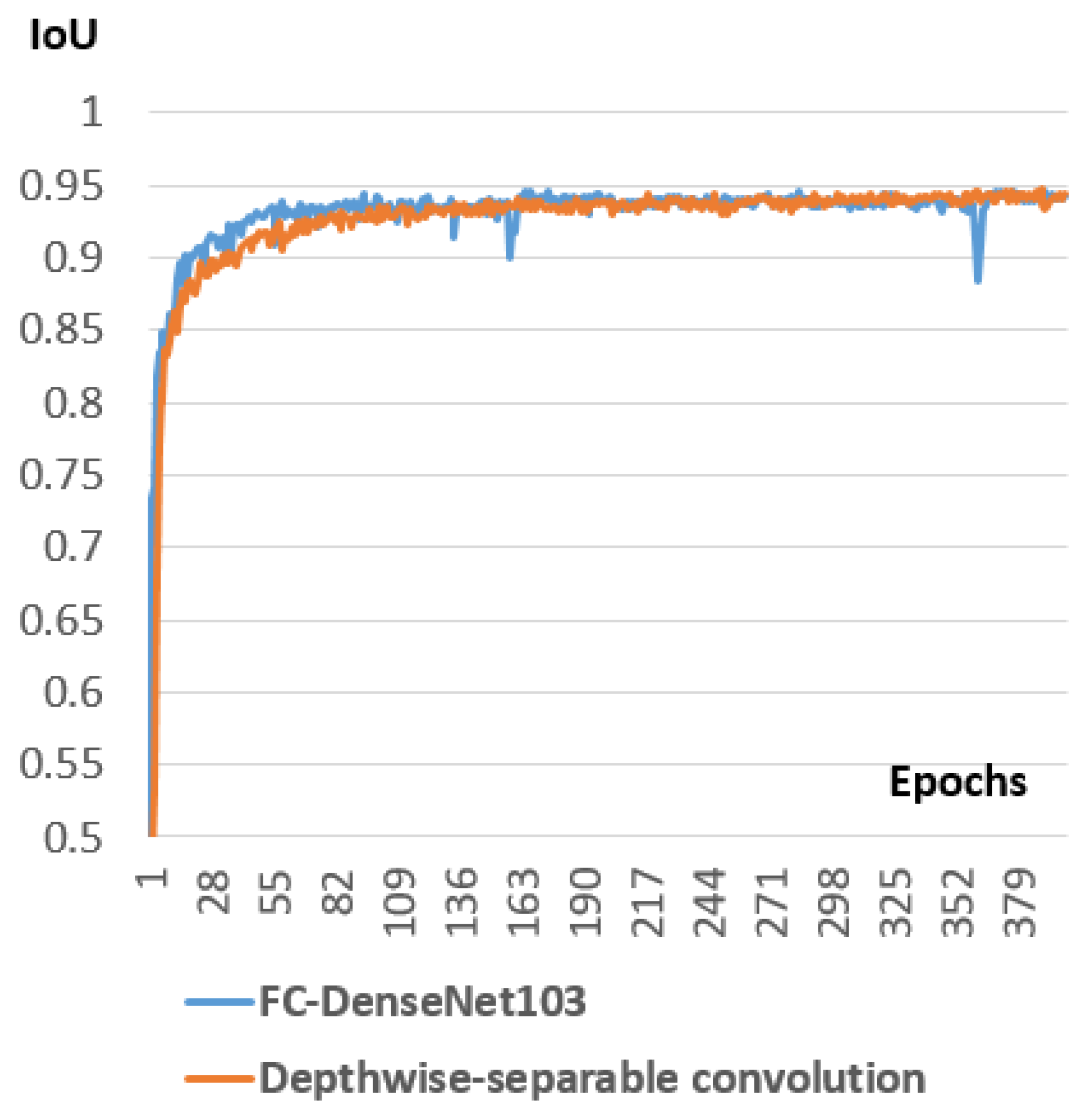
Studying the integration of some successful deep learning components like ASPP (Atrous Spatial Pyramid Pooling) and DSC (Depthwise Separable Convolution) inside the selected models.
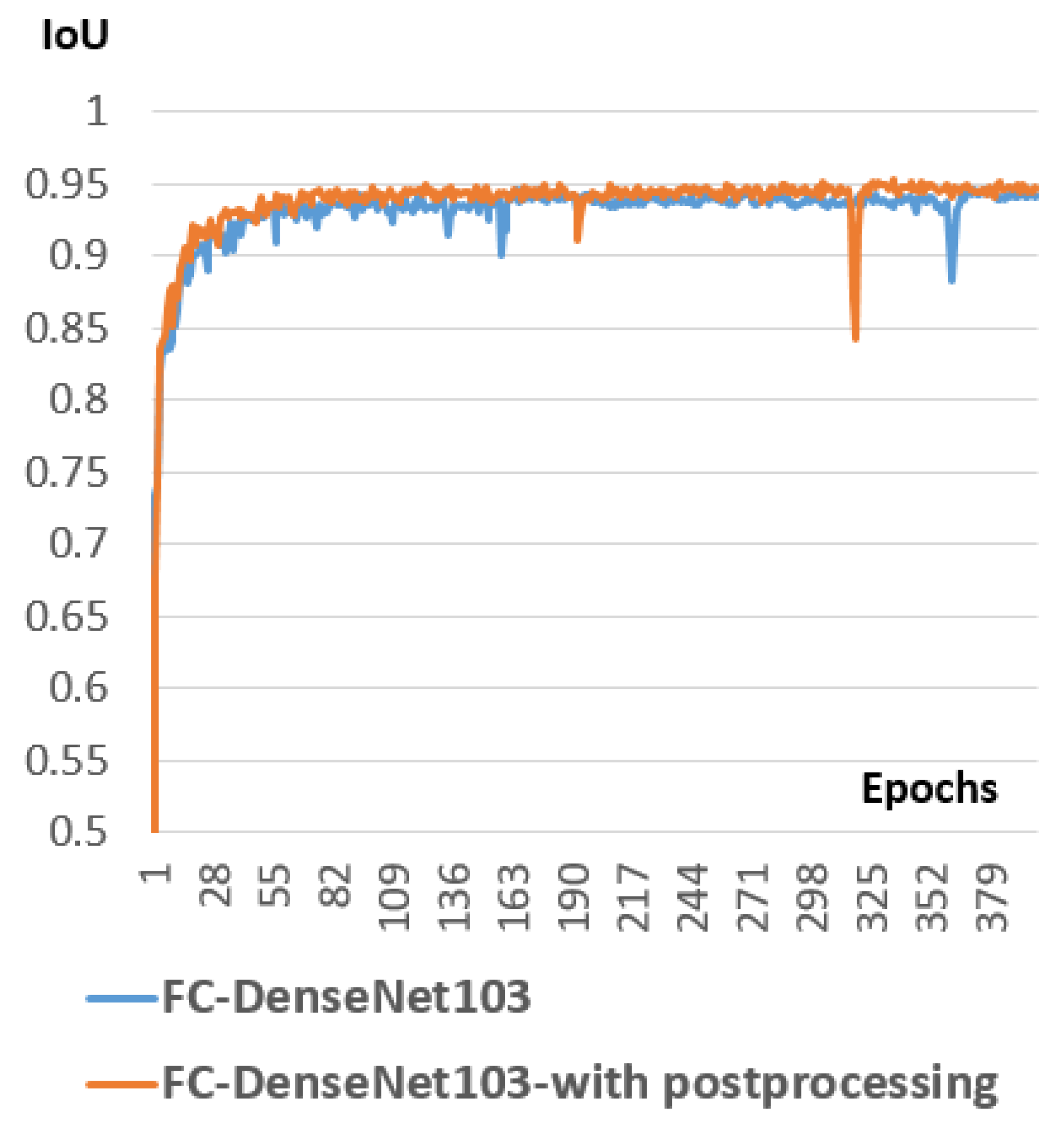
Improving the performance of the chosen model by adding a post-processing step and selecting the right configuration setting for both models.
The rest of the paper is organized as follows:
Section 2 was dedicated for a review about some related works that studied the spine anatomy in ultrasound imagery.
Section 3 introduced the architecture of the selected models (FC-DenseNets and U-Net) and the deep learning components (ASPP and DSC) that we used for spinal cord ultrasound image segmentation.
Section 4 discussed the experiments we made and the best approach to adopt for the different targeted scenarios.
2. Related Works
The analysis of ultrasound medical images of the spine was the subject of many studies in recent years. Ultrasound is a safe, radiation-free imaging modality that is easy to use by neurosurgeons compared to other medical imaging modalities. Pinter et al. [
5] introduced a real-time method to automatically delineate the transverse processes parts of the vertebrae using ultrasound medical imagery. The method creates from the shadows cast by each transverse process a three-dimensional volume of the surface of the spine. Baum et al. [
6] used the results of this study to build a step-wise method for identification of the visible landmarks in ultrasound to visualize a 3D model of the spine. Sass et al. [
7] developed a method to make a navigated three-dimensional intraoperative ultrasound for spine surgery. They registered the patient automatically by using an intraoperative computed tomography before mapping it to a preoperative image data to get visualized navigation of the common parts of the spine. Hetherington et al. [
15] developed a deep learning model named SLIDE to discriminate many parts of the spinal column using only 2d ultrasound with 88% cross-validation accuracy. The model acts in real-time (40 frames per second). Conversano et al. [
16] developed a method for estimation of spine mineral density from ultrasound images. This method is useful for the diagnosis of osteoporosis. Zhou et al. [
1] developed an automated measurement of the spine curvature from 3D Ultrasound Imaging. The method is useful for the diagnosis of scoliosis. Inklebarger et al. [
17] developed a method to visualize transabdominal lumbar spine image using portable ultrasound. They specified a settings of machine configurations and probe selection to follow in order to obtain a profitable image. Karnik et al. [
18] made a review of applications of the ultrasound imaging for the analysis of neonatal spine with emphasis on cases where it is the primary imaging modality to use. Di Pietro et al. [
19] made a similar study on the examination of the neonatal and infant spine from the ultrasound. Ungi et al. [
20] also made a review about applications of tracked ultrasound modality in navigation during spine Interventions. Chen et al. [
21] made a pairwise registration of 2D ultrasound (US) and 3D computed tomography of the spine. The model is done using Convolutional Neural Network (CNN) before the refinement of the registration using an orientation code mutual information metric. Shajudeen et al. [
22] developed a method for automatic segmentation of the spine bones surface in ultrasound images. The method can be extended to any bone surface presented in ultrasound images. Hurdle et al. [
23] made a review about the use of ultrasound imagery for guidance in the diagnosis of many spinal pains.
From the above, we observed that a great number of studies had treated the use of ultrasound imagery in the diagnosis of clinical symptoms associated with the spine. Nevertheless, they are limited to the identification and segmentation of the bones of the spine. Up to our knowledge, no one treated the identification and segmentation of the spinal cord from ultrasound imaging. During laminectomy surgery, ultrasound images of the spinal cord could be visualized and analyzed. Surgeons usually refer to these images to study the effect of the laminectomy surgery and the spinal cord pulsation to confirm the adequacy of spinal cord decompression. Hence, we targeted this problem in this study and aim to provide an automatic solution for the segmentation of spinal cord using the recent advances in deep learning algorithms.
3. Proposed Method
This section gives an in-depth representation of the architecture of the models that have proven efficiency in our task of segmentation of the spinal cord inside ultrasound imagery. As U-Net and FC-DenseNets have proven efficiency in this mission, we will explain in detail the related concepts such as Convolutional Neural Network (CNN), U-net, DenseNets, and Fully Convolutional DenseNets. We will introduce later the Atrous Spatial Pyramid Pooling (ASPP) and the Depthwise Separable Convolution (DSC) and emphasize their usefulness in improving the performance of the chosen models.
3.1. CNN: Convolutional Neural Network
If we have data that contains N training samples
, where
x represents the annotated input,
y represents the label, Convolutional neural network (CNN) is able to construct a model
F that maps the relationship between the input data
x and the output data
y. The CNN is built by stacking a series of layers that perform operations like convolution using a kernel function, non-linear activation, and max pooling. A training process of the model is used to give this built CNN model the set of parameters that best fits this mapping relationship with the minimal error. The training process includes five steps. The first step makes the initialization of the parameters and weights of CNN with random values. The second step is the forward propagation phase. During this step, a training sample
is passed to the network,
is transferred from the input layer to the output layer. Finally, we get the output
, which is formulated as:
L is the number of layers, is the weight vector of the Lth layer .
The third step consists of estimating the loss function or the cost function, which is the function that calculates the error margin between the resulting output
and the correct output value
. The fourth step will make the correction of the weight vector
,
,
,
to minimize this loss function following this optimization problem:
where
l is the loss function. Usually, like here in our work, the cross-entropy loss function is used, as we made in the training phase of the model we adopted in this paper. In fact, we used a weighted version of cross-entropy to alleviate the imbalance between the class Spinal Cord and the class Other. To solve the numerical optimization problem, we use back-propagation and stochastic gradient descent methods. After this step, a more adequate set of parameters and weights is given to our model. In the fifth step, we will repeat the second, third, and fourth steps through all of the training data:
. Usually, this training will end by converging the loss function into a small value. This convergence is assured especially when we use a state of the art architecture like the architectures that we tested in this paper. Passing the full training data one time through the CNN network is named one epoch. Training CNNs usually involves running multiple epochs. Many techniques are used to make the cost function decrease faster. First, the Batch normalization technique [
24] introduced in 2015 is becoming a state of the art method used, like we used here in our work, for normalizing the output of convolutional layers and fully connected layer before applying the non-linear activation function. This has a significant effect on making the loss function converge faster. Also, optimizers are also used to make the loss function converge faster. We used in our experiments the ADAM (Adaptive Moment Estimation) [
25] introduced in 2015 as it is becoming a state of the art gradient descent optimizer.
3.2. Semantic Segmentation and U-Net Architecture
The semantic segmentation is the task of classifying every pixel inside an image into a meaningful category. In medical image analysis, semantic segmentation is a highly pertinent task. Computer-aided diagnosis relies heavily on the accurate segmentation of the organs and structures of interest in the captured medical image modalities (MRI, CT, Fluoroscopy, Ultrasound…). The success made by Convolutional Neural Networks had profoundly influenced the area of semantic segmentation. Many architectures had been proposed. Among the most used architectures in medical image segmentation, U-Net [
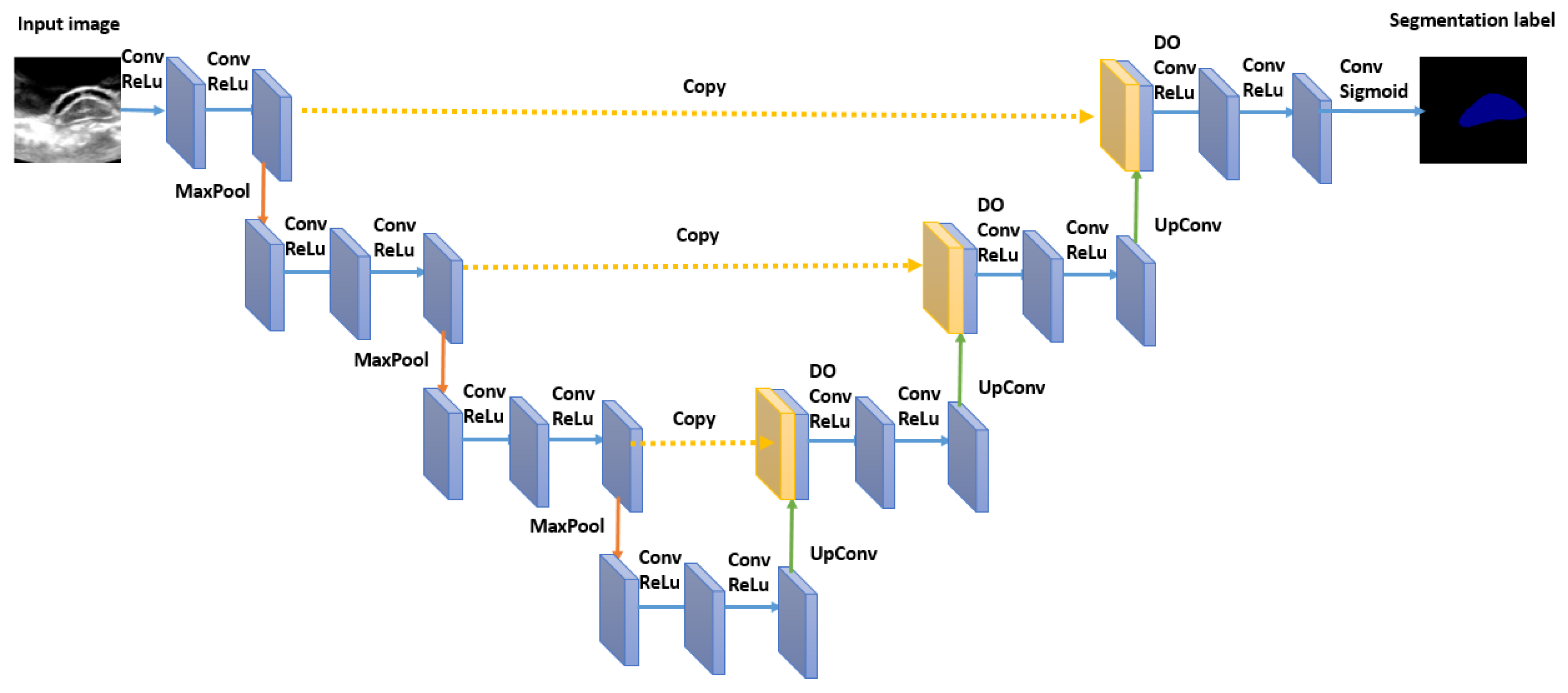
26] is a state of the art model,
Figure 5 represents the architecture of U-Net. U-Net model is an encoder-decoder architecture. The encoder is the contracting part on the left, and the decoder is the expansive path on the right side. The encoder part contains a series of 3 × 3 convolutions followed by a ReLU (Rectified Linear Unit) activation function. A 2 × 2 max-pooling operation is done after a series of consecutive convolutions for downsampling. The decoder part consists of upsampling the feature vector obtained at the end of the encoder part by a series of 2 × 2 up-convolutions to reconstruct the segmentation map at the size of the input image at the end. Between every up-convolutions operations, there are a series of 3 × 3 convolutions followed by ReLu similarly to the decoder part. Skip connections are added by copying the feature map of the encoder part with the correspondingly feature map of the decoder part. In the end, a sigmoid activation function is used to generate for each feature vector the desired class category. The network is trained end to end using back-propagation and stochastic gradient descent. As being discussed in the experimental part of this study, U-Net has proven efficiency where the spinal cord pattern is not previously learned by in the train set.
3.3. DenseNets and Fully Convolutional DenseNets
In 2017, Huang et al. [
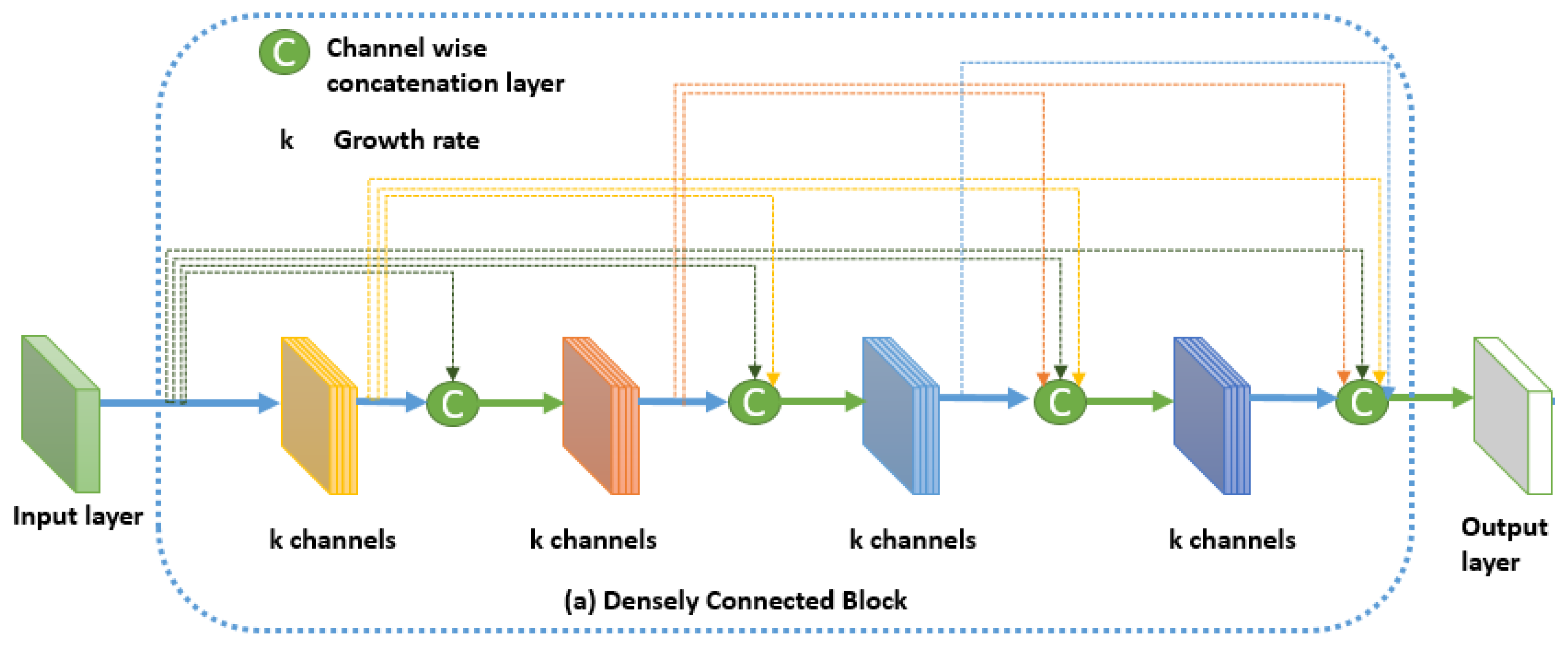
27] proposed the architecture of Densely Connected Convolutional Networks (or DenseNet) for image classification tasks. In this network, every layer is connected to every other layer in a feed-forward manner. Consequently, the input of every layer contains a concatenation of the feature maps of all preceding layers. Also, its own feature map is used as input in all subsequent layers. The network outperforms the state of the art networks on image classification problem, making it a strong feature extractor that can be used as a building block for other tasks like semantic segmentation. The network presents many advantages over its competitors. It mitigates the vanishing-gradient problem and reinforces feature propagation and reuse with smaller number of parameters. The architecture of the Dense block in Densenet is illustrated in
Figure 6.
Formally, if we consider an image
that passes through the DenseNet network that contains
L layers. Each layer applies a transformation
with
l refers to the layer index.
is composed of Convolution, Batch Normalization [
24], ReLU [
28] and Pooling [
29]. If we set
as the output of the
lth layer,
for the traditional convolutional network. But in the dense block of the Densenet architecture, the
lth layer of receives as input the feature map of all the preceding layers as expressed in Equation (
3):
where
is the concatenation of the feature maps generated in the preceding layers.
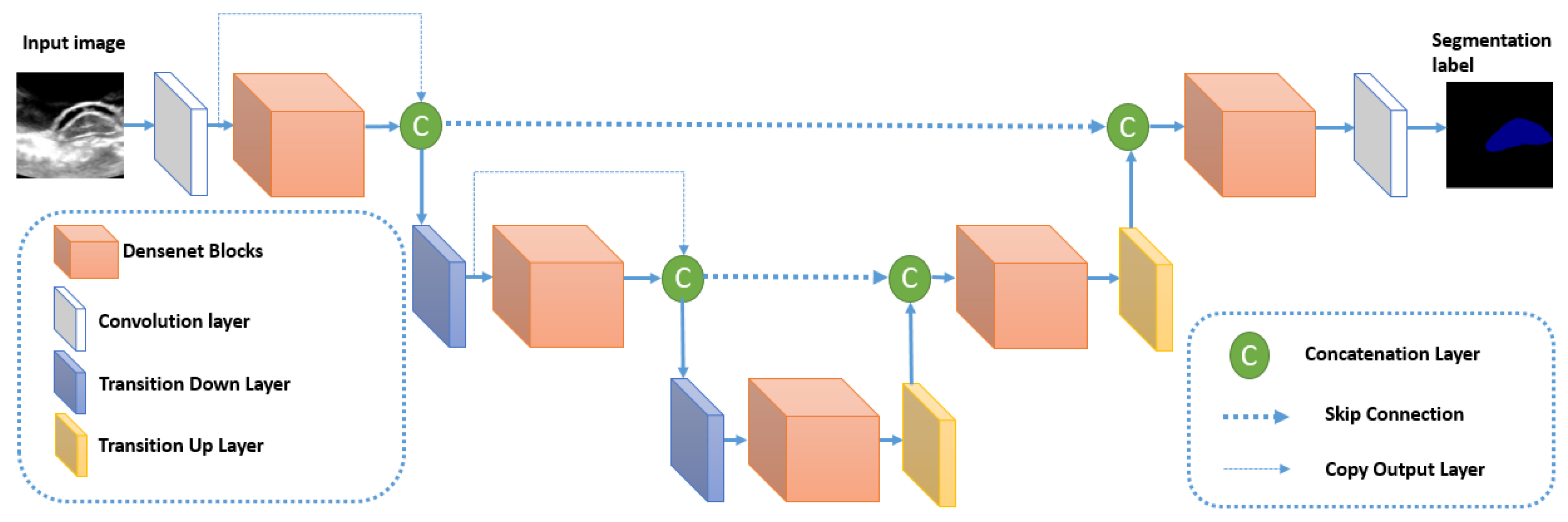
The architecture of Densenet is reused within the semantic segmentation context in the algorithm Fully Convolutional DenseNet (or FC-DenseNet) [
30] by merging it inside an U-Net like model. The architecture of FC-Densenet is illustrated in
Figure 7. The encoder path of FC-DenseNet corresponds to a DenseNet network that contains dense blocks separated by normal layers (Convolutions, Batch normalization, ReLu, and pooling). These normal layers form the transition down block that reduces the spatial resolution of each feature map using the pooling operation. The last layer of the encoder path is denoted as the bottleneck of the network. FC-Densenet adds a decoder path that aims to recover the original spatial resolution of the image. This decoder part contains Dense Blocks separated by Transition up blocks. The Dense blocks are similar to their corresponding Dense blocks in the encoder. The Transition Up block contains the up-sampling operations (transposed convolutions) necessary to compensate the down-sampling operations (pooling) in the encoder path. Similarly to U-Net, the Dense block of the two paths are connected by skip connections to guide the reconstruction of the input spatial resolution through the up-sampling part of the network. We can note that in the down-sampling path, the output of the dense block is concatenated to the output of the previous transition block to form the input for the next Transition down block. This operation is not used in the up-sampling path to reduce computations because already we have skip connection concatenated to every Dense Block input. The last layer in FC-Densenet is a 1 × 1 convolution followed by a softmax layer to generate for every pixel the per class distribution. The network is trained using cross-entropy loss calculated in a pixel-wise manner. The network can be trained from scratch without the need to train a feature extractor on external data as done in many state of the art segmentation algorithms. In our study, FC-DenseNet is proven to be the best tested algorithm in segmenting spinal cord when the pattern is already learned inside the training set. In this cases study, it outperforms U-Net.
3.4. Atrous Spatial Pyramid Pooling
As the shape and the size of objects inside the image may differ, the concept of image pyramid [
31] had been introduced to improve segmentation precision. It consists on extracting features from different scales in a pyramid like approach before interpolating and merging them. But calculating the feature maps for every scale separately increases the size of the network and leads to heavy computations with risk of over-fitting. This is why a relevant method that combines the multiscale information in an efficient way is needed. Spatial pyramid pooling (SPP) [
32] is proposed to treat this problem. SPP was first proposed to solve the issue of random input size of proposals in object detection [
32]. SPP divides the randomly sized images into spatial bins before applying the pooling operation on every bin and concatenating them to obtain the fixed feature map size associated with this input image. Although its efficiency in capturing multi-scale features from the image, SPP is not well adapted to image segmentation because the pooling operations lose the pixel details needed for this task. Hence, we substituted the normal pooling layers in SPP by atrous convolutions with different sampling rates. Then, the extracted features from every sampling rate are merged to obtain the final feature vector. This method is called Atrous Spatial Pyramid Pooling (ASPP). The atrous convolution helps to have different receptive field for a convolution kernel by merely changing the sampling rate. This approach is the base of the state of the art model DeepLabv 3 plus [
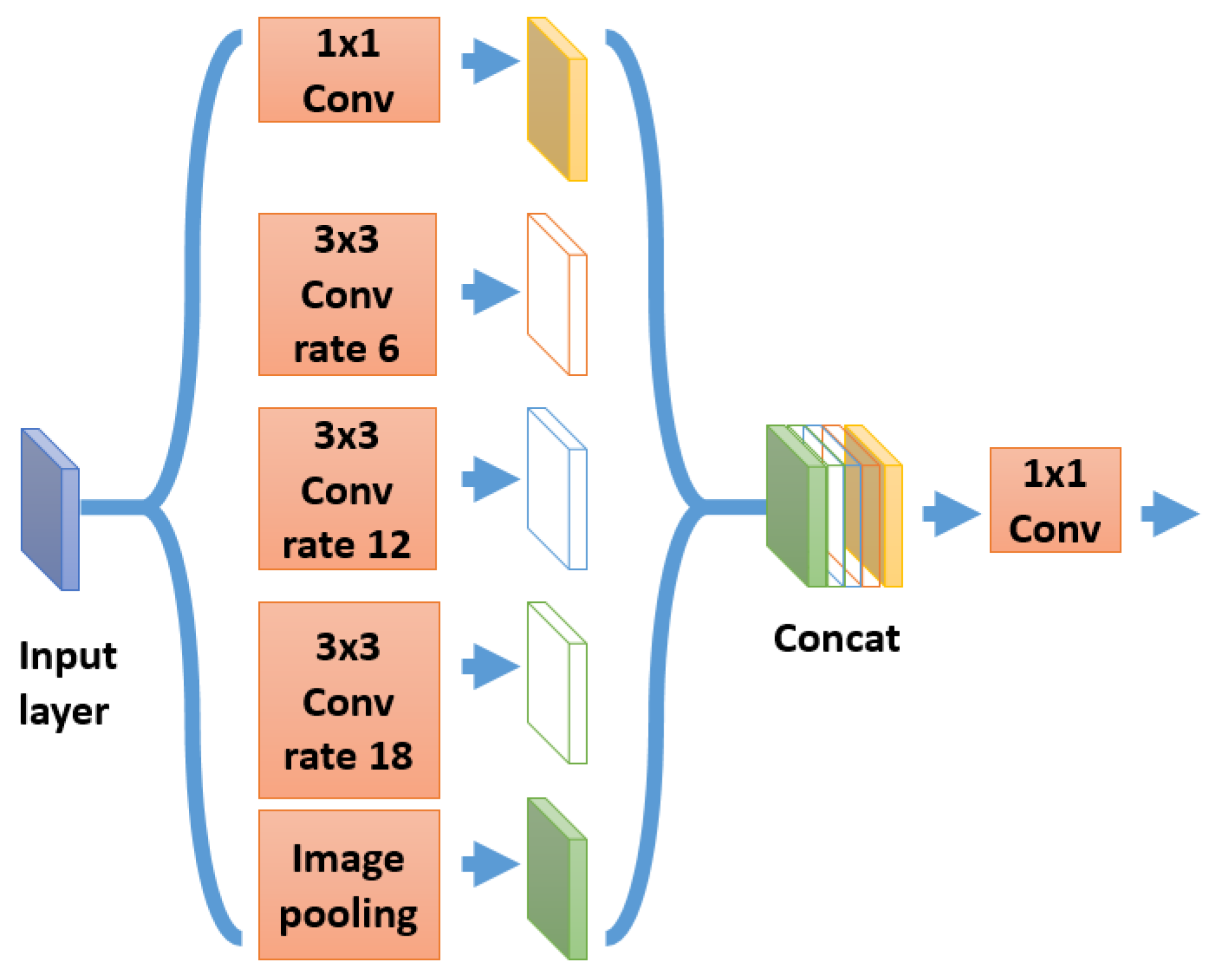
33], which is a segmentation model different in architecture from U-Net and FC-DenseNet. It is currently the best algorithm tested on PASCAL VOC dataset [
34]. This is why we decided to study the effect of inserting an ASPP module inside the FC-DenseNet.
Figure 8 illustrates the version of ASPP used in DeepLab v3 plus and in our experiments.
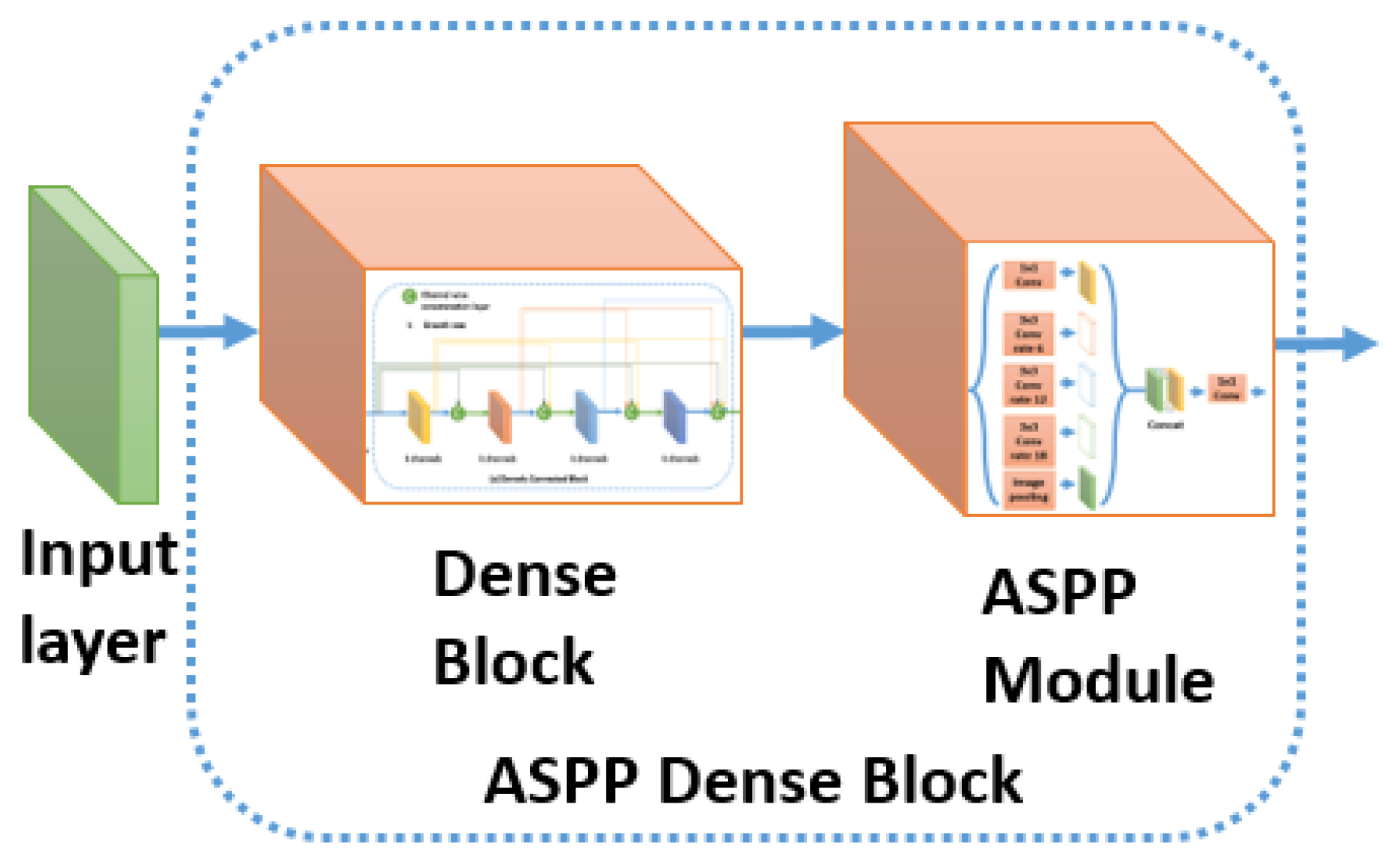
Figure 9 shows the insertion of the ASPP module inside the DenseNet block to form an ASPP Dense Block. We will study the effect of this insertion inside the experimental part.
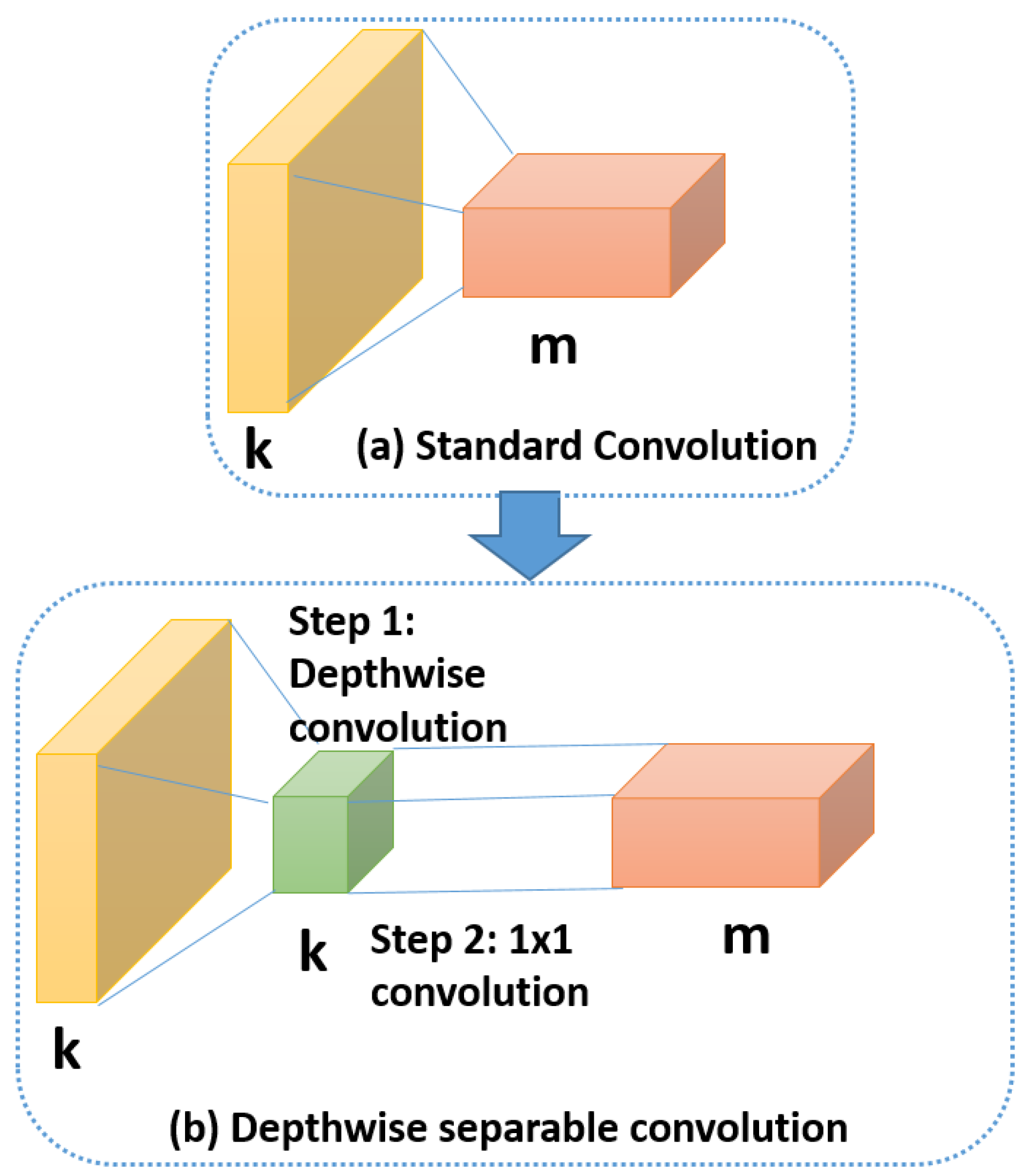
3.5. Depthwise Separable Convolution
Recently, MobileNet [
35] had been introduced for efficient memory use and firstly designed for mobile and embedded devices. MobileNet is based on using Depth-wise Separable Convolutions (DSC), which have two benefits over traditional convolutions. First, they have a lower number of parameters to train as compared to standard convolutions. This makes the model more generalized and reduces overfitting. Second, they need a lesser number of computations for the training. DSC comprises of separating the standard convolution into two successive convolutions. The first convolution is performed separately over each channel of the input layer. Then, we apply a
convolution to the output feature maps from the previous step to get the final output layer.
Figure 10 illustrates the difference between standard convolution and depthwise separable convolution.
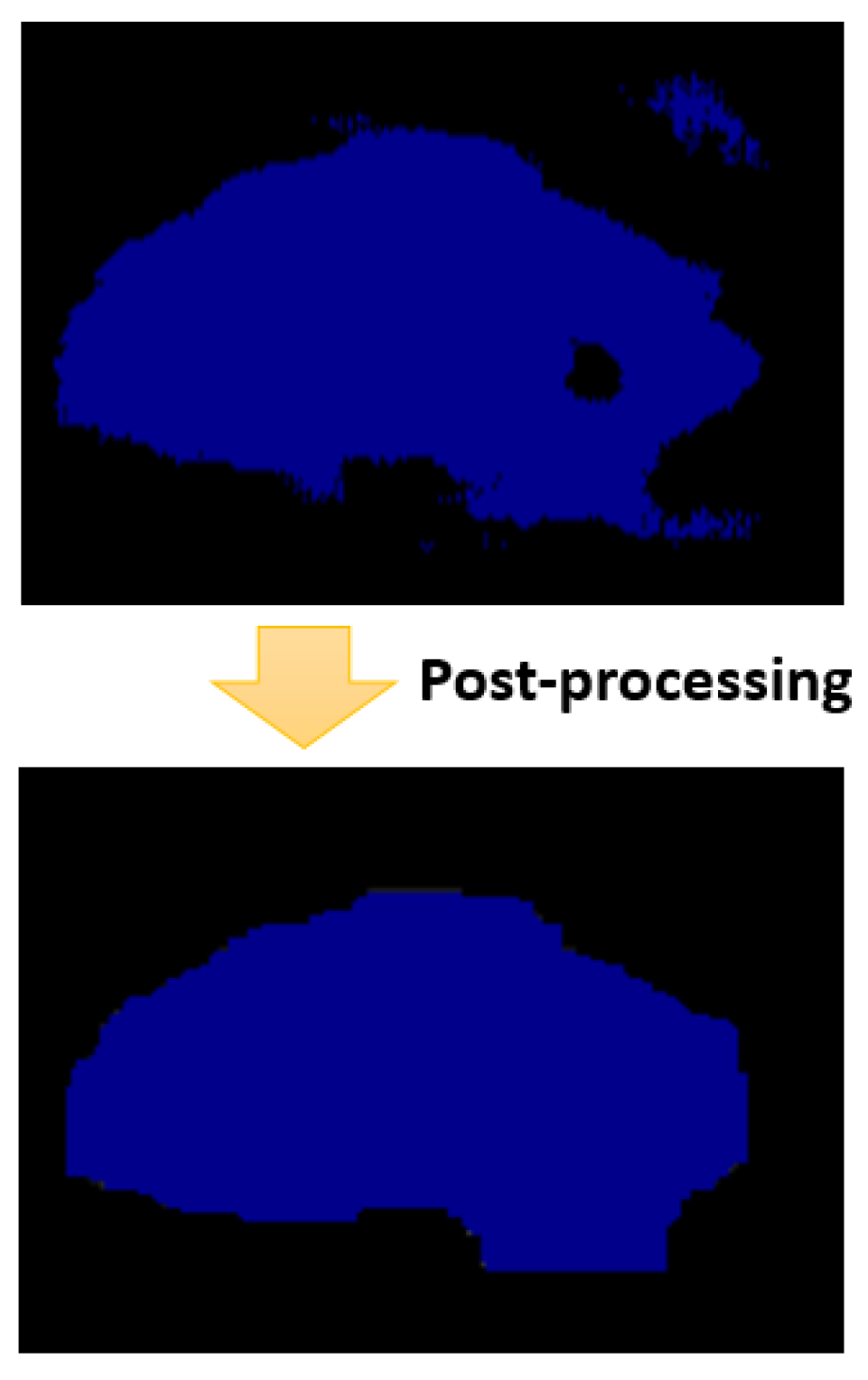
3.6. Postprocessing
To raffinate the segmentation map generated by the semantic segmentation model, we applied a list of post-processing operations to improve the accuracy of segmentation.
Figure 11 represents the status of a segmentation map of the spinal cord before and after the post-processing step.
The post-processing step is divided into four sub-steps. In every sub-step, we apply a different morphological operation on the segmentation map generated from the previous sub-step. The first sub-step is to remove small objects that the number of pixels is less than 1000. In fact, we always have in the ground truth one connected blob that corresponds to the spinal cord, with a number of pixels that is surely bigger than 1000 pixels. So, we removed small objects that contains a number of connected pixels less than 1000, and the degree of connectivity is set to 1. In fact, these removed objects correspond evidently for false-positive pixels. After that, we apply the second sub-step to the segmentation map generated from the first sub-step. This sub-step corresponds to removing small holes inside the spinal cord boundary. In fact, we are sure that the connected pixels corresponding to the spinal cord contain no holes. Hence, we removed these holes based on this property. Then, the third sub-step corresponds to apply a morphological closing using a square filter of size to make the boundary smooth and comparable to the boundary in real images. Then, we apply the last sub-step, which corresponds to a morphological opening using a square filter of size . This intended to recover effects of the closing operation by filtering out the components that probably exceed the real spinal cord boundary. We will study in detail the effect of the post-processing step in the experimental part.