Pretreatment Method for DNA Barcoding to Analyze Gut Contents of Rotifers
Abstract
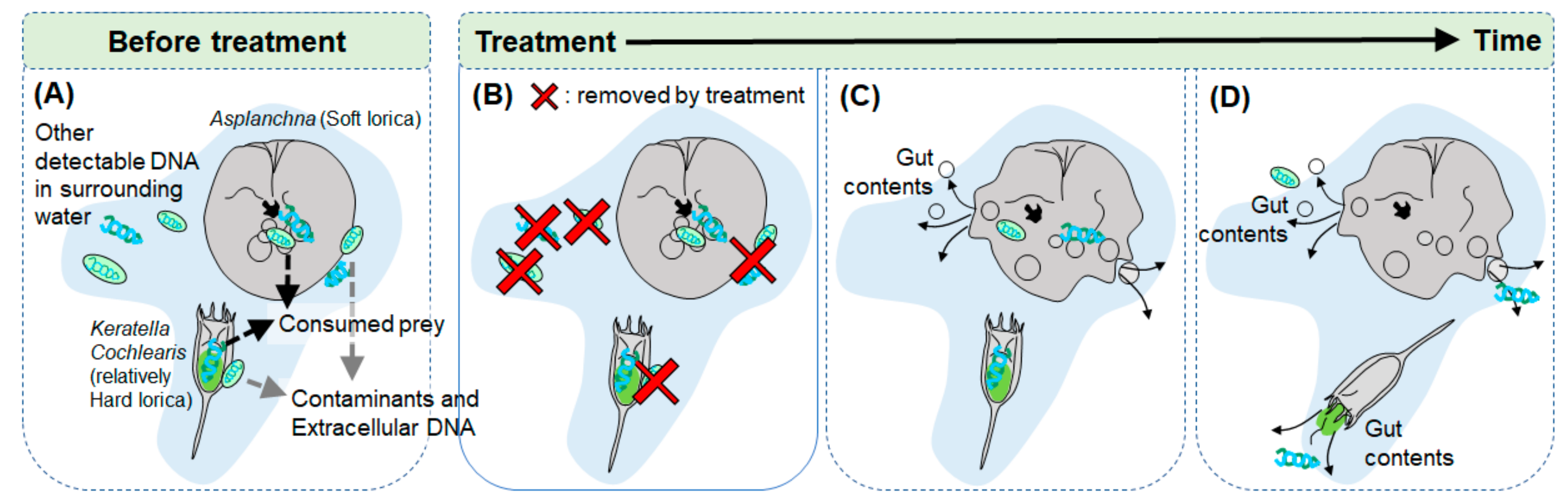
1. Introduction
2. Materials and Methods
2.1. Selection of Chemicals for Eliminating Extracellular DNAs
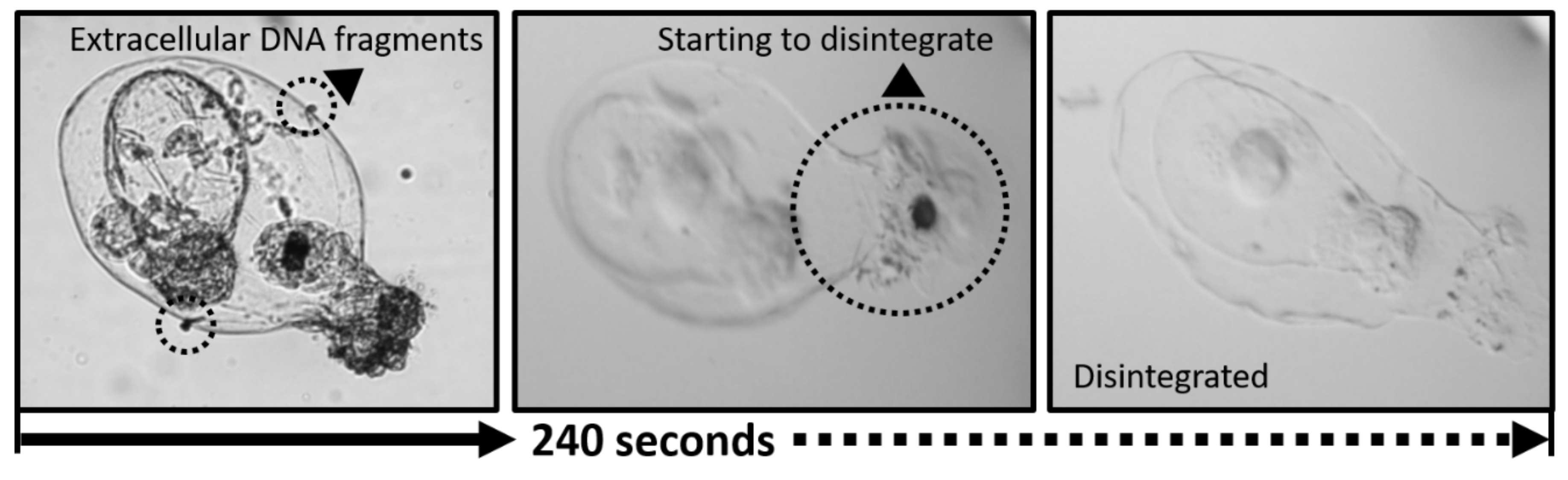
2.2. Responses of Rotifers Lorica to Bleach
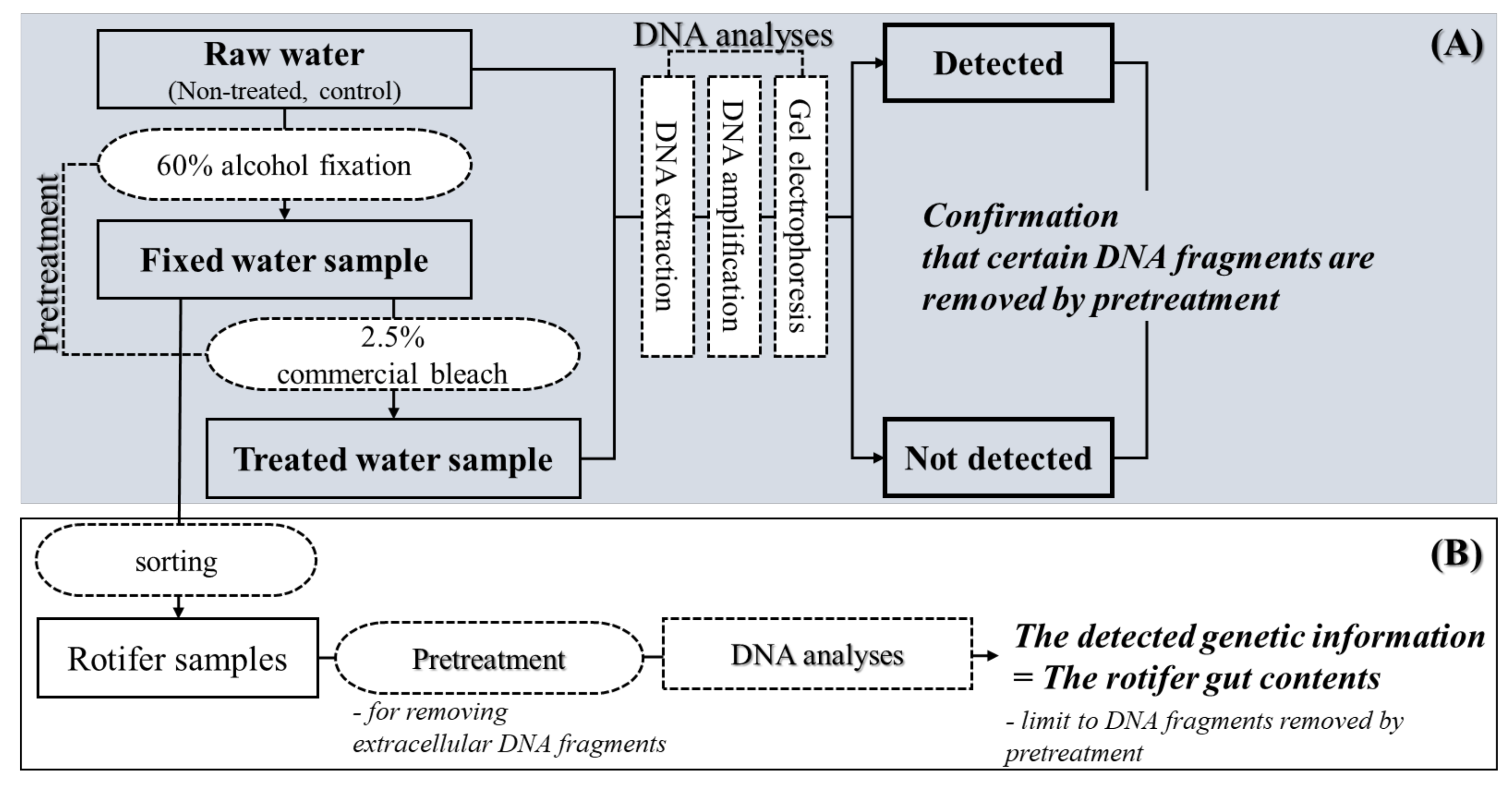
2.3. Application and Effectiveness Verification of Set Treatment Concentration and Time
2.4. DNA Analysis Precedure
| Food source | Primer | Sequence (5’-3’) | Base Pair | Ref. | |
|---|---|---|---|---|---|
| Chlorophyceae (18s rRNA) | ChloroF | TGG CCT ATC TTG TTG GTC TGT | 473 bp | [42] | |
| ChloroR | GAA TCA ACC TGA CAA GGC AAC | ||||
| 94 °C, 3 min -> 35cycles [94 °C, 1 min -> 55 °C **, 1 min -> 72 °C, 1 min] -> 72 °C, 10 min | |||||
| Diatomea (18s rRNA; V4) | M13F- D512for | TGT AAA ACG GCC AGT ATT CCA GCT CCA ATA GCG | 390-410 bp | [43] | |
| M13R-D978rev | CAG GAA ACA GCT ATG ACG ACT ACG ATG GTA TCT AAT C | ||||
| 94 °C, 2 min ->5cycles [94 °C, 45s -> 53 °C **, 45s -> 72 °C, 1 min] ->35cycles [94 °C, 45s -> 51 °C **, 45s -> 72 °C, 1 min] -> 72 °C, 10 min | |||||
| Cyanobacteria (16S rRNA; ITS *) | 16S27F | AGA GTT TGA TCC TGG CTC AG | 422 bp | [44] | |
| 23S30R | CTT CGC CTC TGT GTG CCT AGG T | ||||
| 94 °C, 5 min -> 10cycles [94 °C, 45s -> 57 °C **, 45 s -> 68 °C, 2 min] -> 25cycles [92 °C, 45s -> 54 °C **, 45 s -> 68 °C, 2 min] -> 68 °C, 7 min | |||||
| Bacteria (16S rDNA; nearly full-length) | Forward | GAG TTG GAT CCT GGC TCA G | About 2000 bp | [45,46] | |
| Reverse | AAG GAG GGG ATC CAG CC | ||||
| 95 °C, 3 min -> 35cycles [94 °C, 1min -> 60 °C **, 1 min -> 72 °C, 2 min] -> 72 °C, 3 min | |||||
| Ciliophora (18S rRNA) | Cil F | TGG TAG TGT ATT GGA CWA CCA | 600-670 bp | [47] | |
| Cil R- | 1 | TCT GAT CGT CTT TGA TCC CTT | |||
| 2 | TCT RAT CGT CTT TGA TCC CCT A | ||||
| 3 | TCT GAT TGT CTT TGA TCC CCT | ||||
| 95 °C, 5 min -> 35cylces [94 °C, 45 s -> 55 °C **, 1 min -> 72 °C, 1 min] -> 72 °C, 10 min | |||||
| Heterotrophic nanoflagellates (18s rRNA) | EukA *** | AAC CTG GTT GAT CCT GCC AGT | 800–900 bp | [38,39] | |
| EukB *** | TGA TCC TTC TGC AGG TTC ACC TAC | ||||
| 95 °C, 2 min -> 35cycles [95 °C, 30 s -> 55 °C **, 30 s -> 72 °C, 2 min] -> 72 °C, 7 min | |||||
3. Results
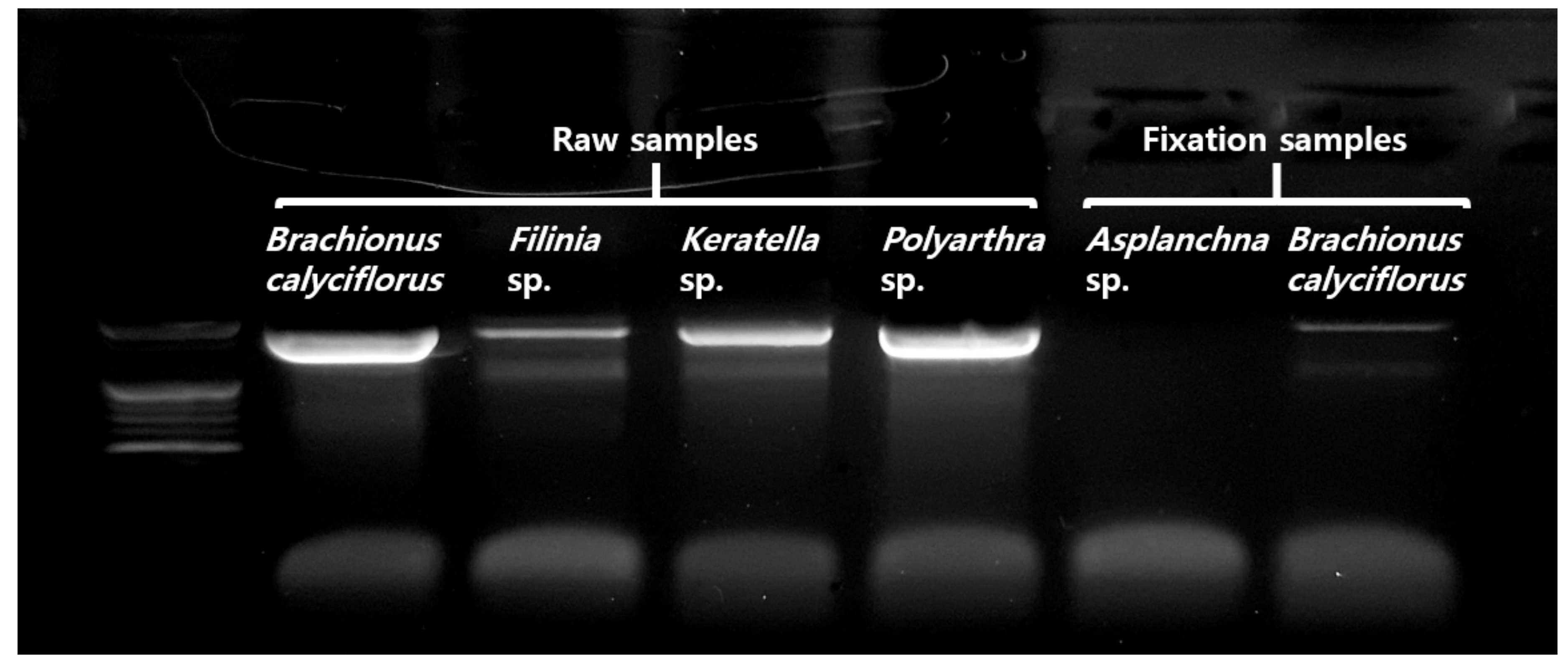
3.1. Responses of Rotifers Lorica to Commercial Bleach Treatment
3.2. Application and Effectiveness Verification of Set Pretreatment Concentration and Time
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Treatments | Procedure | Ref. |
|---|---|---|
| Psoralen + UV irradiation |
1. 8-methoxypsoralen of 100 µg∙mLᅳ1 2. Irradiation with long-wave (365nm) UV light for 1 h | [25] |
| Hydrogen peroxide + Bleach * + UV irradiation | 1. Soaked in hydrogen peroxide (3–30%) for 10–30 min 2. Rinsed with distilled water 3 *. Rinsed thoroughly with 10% bleach 4. Rinsed with distilled water 5. UV irradiated for 10 min | [26] |
| Acid wash + Ethanol + UV irradiation | 1. Soaked in 15% HCl for 10 min 2. Rinsed with 70% ethanol for 10 min 3. Rinsed in sterile double-distilled water for 30 min 4. UV irradiation (254 nm) for 15 min | [27] |
| Bleach * + Ethanol | 1 *. Soaked in 10% bleach for ~10 min 2. Rinsed with 70% ethanol | [28] |
| Bleach * + EDTA | 1 *. Immersed in 20% bleach for 2 min 2. Rinsed with distilled water | [29] |
| 1. 0.5M EDTA at 55°C in a 2-day | ||
| UV irradiation + Bleach * | 1. UV irradiation (254 nm) for 10 min 2 *. Soaked for approximately 5 min in a 5% bleach solution | [30] |
| Acid wash + Bleach * + UV irradiation | 1. 30% acetic acid 2. Rinsed with ultrapure water 3 *. Immersed for 10 min in 10% sodium hypochlorite with sporadic shaking 4. Exposed to UV irradiation (254 nm) for at least 10 min | [31] |
| Ethidium Monoazied (EMA) or Propidium Monoazied (PMA) | 1. EMA or PMA added following a conventional procedure in accordance with the manufacturer’s protocol (PowerSoil DNA extraction kit (Mo Bio Lab, Inc.) | [32] |
| Bleach * | 1 *. Exposure to 2.5% bleach for 40 min or overnight | [33] |
References
- Carrillo, P.; Medina-Sánchez, J.M.; Villar-Argaiz, M.; Delgado-Molina, J.A.; Bullejos, F.J. Complex interactions in microbial food webs: stoichiometric and functional approaches. Limnetica 2006, 25, 189–204. [Google Scholar]
- Wallace, R.L.; Snell, T.W.; Claudia, R.; Thomas, N. Rotifera vol. 1: Biology, Ecology and Systematics, 2nd ed.; Backhuys: Leiden, The Nederlands, 2006; p. 94. [Google Scholar]
- Pree, B.; Larsen, A.; Egge, J.K.; Simonelli, P.; Madhusoodhanan, R.; Tsagaraki, T.M.; Vage, S.; Erga, S.R.; Bratbak, G.; Thingstad, T.F. Dampened copepod-mediated trophic cascades in a microzooplakton-dominated microbial food web: a mesocosm Study. Limnol. Oceanogr. 2017, 62, 1031–1044. [Google Scholar] [CrossRef]
- Gurav, M.N.; Pejaver, M.K. Survey of rotifers to evaluate the water quality of the river Gadhi and its reservoir. Ecol. Environ. Conserv. 2013, 19, 417–423. [Google Scholar]
- Neto, G.; José, A.; Silva, L.C.D.; Saggio, A.A.; Rocha, O. Zooplankton communities as eutrophication bioindicators in tropical reservoirs. Biota Neotropica 2014, 14, e20140018. [Google Scholar]
- Moore, M.V.; De Stasio, B.T., Jr.; Huizenga, K.N.; Silow, E.A. Trophic coupling of the microbial and the classical food web in Lake Baikal, Siberia. Freshwater Biol. 2019, 64, 138–151. [Google Scholar]
- Oh, H.J.; Jeong, H.G.; Nam, G.S.; Oda, Y.; Dai, W.; Lee, E.H.; Kong, D.; Hwang, S.J.; Chang, K.H. Comparison of taxon-based and trophi-based response patterns of rotifers community to water quality: applicability of the rotifer functional group as an indicator of water quality. Anim. Cells Syst. 2017, 21, 133–140. [Google Scholar] [CrossRef]
- Rothhaupt, K. Differences in particle size-dependent feeding efficiencies of closely related rotifer species. Limnol. Oceanogr. 1990, 35, 16–23. [Google Scholar] [CrossRef]
- Arndt, H. Rotifers as predators on components of the microbial web (bacteria, heterotrophic flagellates, ciliates)—a review. In Rotifer Symposium VI.; Gilbert, J.J., Lubzens, E., Miracle, M.R., Eds.; Springer: Dordrecht, The Nederlands, 1993; pp. 231–246. [Google Scholar]
- Thouvenot, A.; Debroas, D.; Richardot, M.; Devaux, J. Impact of natural metazooplankton assemblage on planktonic microbial communities in a newly flooded reservoir. J. Plankton Res. 1999, 21, 179–199. [Google Scholar] [CrossRef][Green Version]
- Mohr, S.; Adrian, R. Reproductive success of the rotifer Brachionus calyciflorus feeding on ciliates and flagellates of different trophic modes. Freshwater Biol. 2002, 47, 1832–1839. [Google Scholar] [CrossRef]
- Devetter, M.; Sed’a, J. Rotifer fecundity in relation to components of microbial food web in a eutrophic reservoir. Hydrobiologia 2003, 504, 167–175. [Google Scholar] [CrossRef]
- Pourriot, R. Food and feeding habits of Rotifera. Arch. Hydrobiol. Beihefte 1977, 8, 243–260. [Google Scholar]
- Hebert, P.D.; Cywinska, A.; Ball, S.L.; Dewaard, J.R. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. Ser. B Biol. Sci. 2003, 270, 313–321. [Google Scholar] [CrossRef]
- Barnes, M.A.; Turner, C.R. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 2016, 17, 1–17. [Google Scholar] [CrossRef]
- Kress, W.J.; García-Robledo, C.; Uriarte, M.; Erickson, D.L. DNA barcodes for ecology, evolution, and conservation. Trends Ecol. Evol. 2015, 30, 25–35. [Google Scholar] [CrossRef]
- Symondson, W.O.C. Molecular identification of prey in predator diets. Mol. Ecol. 2002, 11, 627–641. [Google Scholar] [CrossRef]
- Carreon-Martinez, L.; Johnson, T.B.; Ludsin, S.A.; Heath, D.D. Utilization of stomach content DNA to determine diet diversity in piscivorous fishes. J. Fish Biol. 2011, 78, 1170–1182. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.; Gim, J.A.; Jeong, K.S.; Kim, H.S.; Joo, G.J. Application of DNA barcoding for identification of freshwater carnivorous fish diets: Is number of prey items dependent on size class for Micropterus salmoides? Ecol. Evol. 2014, 4, 219–229. [Google Scholar] [CrossRef]
- Jo, H.; Ventura, M.; Vidal, N.; Gim, J.S.; Buchaca, T.; Barmuta, L.A.; Jeppesen, E.; Joo, G.J. Discovering hidden biodiversity: the use of complementary monitoring of fish diet based on DNA barcoding in freshwater ecosystems. Ecol. Evol. 2016, 6, 219–232. [Google Scholar] [CrossRef]
- Craig, C.; Kimmerer, W.J.; Cohen, C.S. A DNA-based method for investigating feeding by copepod nauplii. J. Plankton Res. 2013, 36, 271–275. [Google Scholar] [CrossRef]
- Ho, T.W.; Hwang, J.S.; Cheung, M.K.; Kwan, H.S.; Wong, C.K. DNA-based study of the diet of the marine calanoid copepod Calanus sinicus. J. Exp. Mar. Biol. Ecol. 2017, 494, 1–9. [Google Scholar] [CrossRef]
- Hochberg, R.; Wallace, R.L.; Walsh, E.J. Soft bodies, hard jaws: an introduction to the symposium, with rotifers as models of jaw diversity. Integr. Comp. Biol. 2015, 55, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Jin, W.; Zhou, Y.; Wang, P.; Zhao, W. Hidden defensive morphology in rotifers, benefits, costs, and fitness consequences. Sci. Rep. 2017, 7, 4488. [Google Scholar]
- Jinno, Y.; Yoshiura, K.; Niikawa, N. Use of psoralen as extinguisher of contaminated DNA in PCR. Nucleic Acids Res. 1990, 18, 6739. [Google Scholar] [CrossRef]
- Merriwether, D.A.; Rothhammer, F.; Ferrell, R.E. Genetic variation in the New World: ancient teeth, bone, and tissue as sources of DNA. Experientia 1994, 50, 592–601. [Google Scholar] [CrossRef] [PubMed]
- Lalueza, C.; Perez-Perez, A.; Prats, E.; Cornudella, L.; Turbon, D. Lack of founding Amerindian mitochondrial DNA lineages in extinct aborigines from Tierra del Fuego-Patagonia. Hum. Mol. Gen. 1997, 6, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Stone, A.C.; Stoneking, M. mtDNA analysis of a prehistoric Oneota population: implications for the peopling of the New World. Am. J. Hum. Genet. 1998, 62, 1153–1170. [Google Scholar] [CrossRef] [PubMed]
- Kolman, C.J.; Tuross, N. Ancient DNA analysis of human populations. Am. J. Phys. Anthropol. 2000, 111, 5–23. [Google Scholar] [CrossRef]
- Kaestle, F.A.; Smith, D.G. Ancient mitochondrial DNA evidence for prehistoric population movement: The Numic expansion. Am. J. Phys. Anthropol. 2001, 115, 1–12. [Google Scholar] [CrossRef]
- Montiel, R.; Malgosa, A.; Francalacci, P. Authenticating ancient human mitochondrial DNA. Hum. Biol. 2001, 73, 689–713. [Google Scholar] [CrossRef]
- Wagner, A.O.; Malin, C.; Knapp, B.A.; Illmer, P. Removal of free extracellular DNA from environmental samples by ethidium monoazide and propidium monoazide. Appl. Environ. Microbial. 2008, 74, 2537–2539. [Google Scholar] [CrossRef]
- Greenstone, M.H.; Weber, D.C.; Coudron, T.A.; Payton, M.E.; Hu, J.S. Removing external DNA contamination from arthropod predators destined for molecular gut-content analysis. Mol. Ecol. Resour. 2012, 12, 464–469. [Google Scholar] [CrossRef]
- Rutala, W.A.; Weber, D.J. Disinfection, sterilization, and antisepsis: An overview. Am. J. Infect. Control. 2016, 44, e1–e6. [Google Scholar] [CrossRef] [PubMed]
- Laspoumaderes, C.; Modenutti, B.; Balseiro, E. Herbivory versus omnivory: linking homeostasis and elemental imbalance in copepod development. J. Plankton Res. 2010, 32, 1573–1582. [Google Scholar] [CrossRef]
- Gilbert, J.J.; Williamson, C.E. Predator-prey behavior and its effect on rotifer survival in associations of Mesocyclops edax, Asplanchna girodi, Polyarthra vulgaris, and Keratella cochlearis. Oecologia 1978, 37, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Pilliod, D.S.; Goldberg, C.S.; Arkle, R.S.; Waits, L.P. Estimating occupancy and abundance of stream amphibians using environmental DNA from filtered water samples. Can. J. Fish. Aquat. Sci. 2013, 70, 1123–1130. [Google Scholar] [CrossRef]
- Mukherjee, I.; Hodoki, Y.; Nakano, S.I. Seasonal dynamics of heterotrophic and plastidic protists in the water column of Lake Biwa, Japan. Aquat. Microb. Ecol. 2017, 80, 123–137. [Google Scholar] [CrossRef]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Moro, C.V.; Crouzet, O.; Rasconi, S.; Thouvenot, A.; Coffe, G.; Batisson, I.; Bohatier, J. New design strategy for development of specific primer sets for PCR-based detection of Chlorophyceae and Bacillariophyceae in environmental samples. Appl. Environ. Microbiol. 2009, 75, 5729–5733. [Google Scholar] [CrossRef]
- Zimmermann, J.; Jahn, R.; Gemeinholzer, B. Barcoding diatoms: evaluation of the V4 subregion on the 18S rRNA gene including new primers and protocols. Org. Divers. Evol. 2011, 11, 173–192. [Google Scholar] [CrossRef]
- Taton, A.; Grubisic, S.; Brambilla, E.; De Wit, R.; Wilmotte, A. Cyanobacterial diversity in natural and artificial microbial mats of Lake Fryxell (McMurdo Dry Valleys, Antarctica): a morphological and molecular approach. Appl. Environ. Microbiol. 2003, 69, 5157–5169. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.W.; Min, B.R.; Choi, Y.K. The genetic diversity of bacterial communities in the groundwater. Korean J. Environ. Biol. 2000, 18, 53–61. [Google Scholar]
- Lara, E.; Berney, C.; Harms, H.; Chatzinotas, A. Cultivation-independent analysis reveals a shift in ciliate 18S rRNA gene diversity in a polycyclic aromatic hydrocarbon-polluted soil. FEMS Microbiol. Ecol. 2007, 62, 365–373. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.H.; Doi, H.; Nishibe, Y.; Nakano, S.I. Feeding habits of omnivorous Asplanchna: comparison of diet composition among Asplanchna herricki, A. priodonta and A. girodi in pond ecosystems. J. Limnol. 2010, 69, 209–216. [Google Scholar] [CrossRef]
- Kleinow, W. Biochemical studies on Brachionus plicatilis: hydrolytic enzymes, integument proteins and composition of trophi. In Rotifer Symposium VI.; Gilbert, J.J., Lubzens, E., Miracle, M.R., Eds.; Springer: Dordrecht, The Nederlands, 1993; pp. 231–246. [Google Scholar]
- Kim, K.H.; Park, D.E.; Oh, S. Effects of heat treatment on the nutritional quality of milk: II. destruction of microorganisms in milk by heat treatment. J. Milk Sci. Biotechnol. 2017, 35, 55–72. [Google Scholar] [CrossRef]
- Choi, H.J.; Kim, B.H.; Kim, J.D.; Han, M.S. Streptomyces neyagawaensis as a control for the hazardous biomass of Microcystis aeruginosa (Cyanobacteria) in eutrophic freshwaters. Biol. Control 2005, 33, 335–343. [Google Scholar] [CrossRef]
- Shunyu, S.; Yongding, L.; Yinwu, S.; Genbao, L.; Dunhai, L. Lysis of Aphanizomenon flos-aquae (Cyanobacterium) by a bacterium Bacillus cereus. Biol. Control 2006, 39, 345–351. [Google Scholar] [CrossRef]
- Ooms-Wilms, A.L. Are bacteria an important food source for rotifers in eutrophic lakes? J. Plankton Res. 1997, 19, 1125–1141. [Google Scholar] [CrossRef]
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Cárdenas, P. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar]
- Simek, K.; Jezbera, J.; Horňak, K.; Vrba, J.; Seda, J. Role of diatom-attached choanoflagellates of the genus Salpingoeca as pelagic bacterivores. Aquat. Microb. Ecol. 2004, 36, 257–269. [Google Scholar] [CrossRef]
- Frenken, T.; Wierenga, J.; van Donk, E.; Declerck, S.A.; de Senerpont Domis, L.N.; Rohrlack, T.; Van de Waal, D.B. Fungal parasites of a toxic inedible cyanobacterium provide food to zooplankton. Limnol. Oceanogr. 2018, 63, 2384–2393. [Google Scholar] [CrossRef]
| Target | Treatments and Conditions | Ref. |
|---|---|---|
| Contaminated DNA for extinguishing the template activity | Incubation of DNA with a psoralen, 8-methoxypsoralen in the dark for 30 min to overnight and subsequent irradiation of UV (365 nm) for 1 hr. | [25] |
| Bone for removal of contamination | Washed in sterile distilled water, followed by 10% bleach * | [26] |
| Teeth for destroying any contaminating DNA on the surface | Soaked in hydrogen peroxide (3–30%) for 10–30 min, rinsed with distilled water, rinsed thoroughly with 10% bleach *, rinsed again with distilled water and UV irradiated for 10 min | |
| Teeth and cortical bone pieces to prevent extraneous contaminations (dirt, carbonate deposits, acid residues) | Soaked for 10 min in 15 % HCL, for 10 min in 70 % ethanol and rinsed in sterile double-distilled water for 30 min | [27] |
| Tooth for prevention of contamination | Soaked for ~ 10 min in 10% bleach * and then rinsed with 70 % ethanol | [28] |
| Skeletal material (e.g., powdered bone) for reducing DNA contamination | - Immersing in 20 % bleach * for 2 min followed by extensive ddH2O washing - 2 days treatment with 0.5 M EDTA at 55 | [29] |
| Bone fragments for elimination of any minor surface contamination | 10 min on each side with UV light (254 nm) and soaked for approximately 5 min in a 5 % bleach solution* (in some cases) | [30] |
| Teeth for removal of dirt and other contaminants | Treated with 30 % acetic acid, rinsed with ultrapure water, immersed for 10 min in 10 % sodium hypochlorite* and exposed to UV light (254 nm) for at least 10 min on each side | [31] |
| Environmental samples for removal of free extracellular DNA | Added Ethidium or Propidium Monoazied (EMA or PMA) following a conventional procedure in accordance with the manufacturer’s protocol (PowerSoil DNA extraction kit (MoBio Lab, Inc.) | [32] |
| External DNA contamination of arthropod gut-content | 40 min of end-over-end rotation in 2.5 % commercial bleach * | [33] |
| Concentration (Final) and Duration Time | Rotifer Species | ||||
|---|---|---|---|---|---|
| Brachionus Forficula | Keratella sp. | Trichocerca sp. | Polyarthra sp. | Asplanchna sp. | |
| 20% | 60 s | 90 s | 120 s | 60 s | 35 s |
| 10% | 210 s | 180 s | 150 s | 90 s | 45 s |
| 5% | 300 s | 240 s | 300 s | 210 s | 120 s |
| 2.5% | 450 s | 300 s | 450 s | 300 s | 240 s |
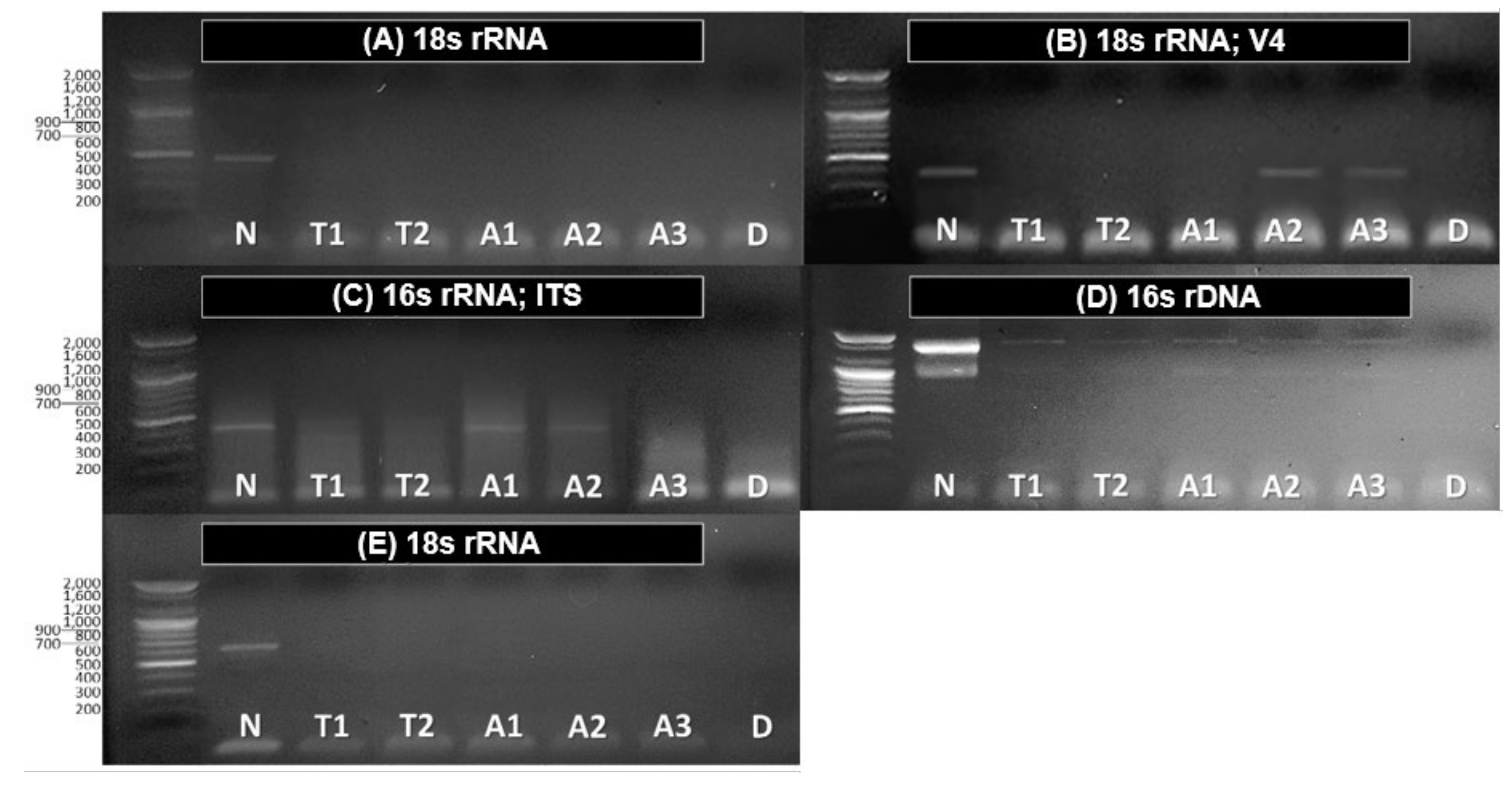
| Samples. | 18s rRNA | 18s rRNA; V4 | 16S rRNA; ITS | 16S rDNA | 18S rRNA |
|---|---|---|---|---|---|
| Non-treated | Chlamydomonas nivalis (99%) Vitreochlamys nekrassovii (99%) | Aulacoseira granulate (100%) Aulacoseira ambigua(99%) | Chlorophyta sp. (98%) | Bacillus cereus (81%) | Tintinnidium fluviatile (90%) |
| Treated | Not-detected | Choanoflagellate (97%) Meira nashicola (99%) | Not-detected | Bacillus thuringiensis (85%) | Not-detected |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, H.-J.; Krogh, P.H.; Jeong, H.-G.; Joo, G.-J.; Kwak, I.-S.; Hwang, S.-J.; Gim, J.-S.; Chang, K.-H.; Jo, H. Pretreatment Method for DNA Barcoding to Analyze Gut Contents of Rotifers. Appl. Sci. 2020, 10, 1064. https://doi.org/10.3390/app10031064
Oh H-J, Krogh PH, Jeong H-G, Joo G-J, Kwak I-S, Hwang S-J, Gim J-S, Chang K-H, Jo H. Pretreatment Method for DNA Barcoding to Analyze Gut Contents of Rotifers. Applied Sciences. 2020; 10(3):1064. https://doi.org/10.3390/app10031064
Chicago/Turabian StyleOh, Hye-Ji, Paul Henning Krogh, Hyun-Gi Jeong, Gea-Jae Joo, Ihn-Sil Kwak, Sun-Jin Hwang, Jeong-Soo Gim, Kwang-Hyeon Chang, and Hyunbin Jo. 2020. "Pretreatment Method for DNA Barcoding to Analyze Gut Contents of Rotifers" Applied Sciences 10, no. 3: 1064. https://doi.org/10.3390/app10031064
APA StyleOh, H.-J., Krogh, P. H., Jeong, H.-G., Joo, G.-J., Kwak, I.-S., Hwang, S.-J., Gim, J.-S., Chang, K.-H., & Jo, H. (2020). Pretreatment Method for DNA Barcoding to Analyze Gut Contents of Rotifers. Applied Sciences, 10(3), 1064. https://doi.org/10.3390/app10031064






