Automated Detection of Sleep Stages Using Deep Learning Techniques: A Systematic Review of the Last Decade (2010–2020)
Abstract
1. Introduction
2. Medical Background
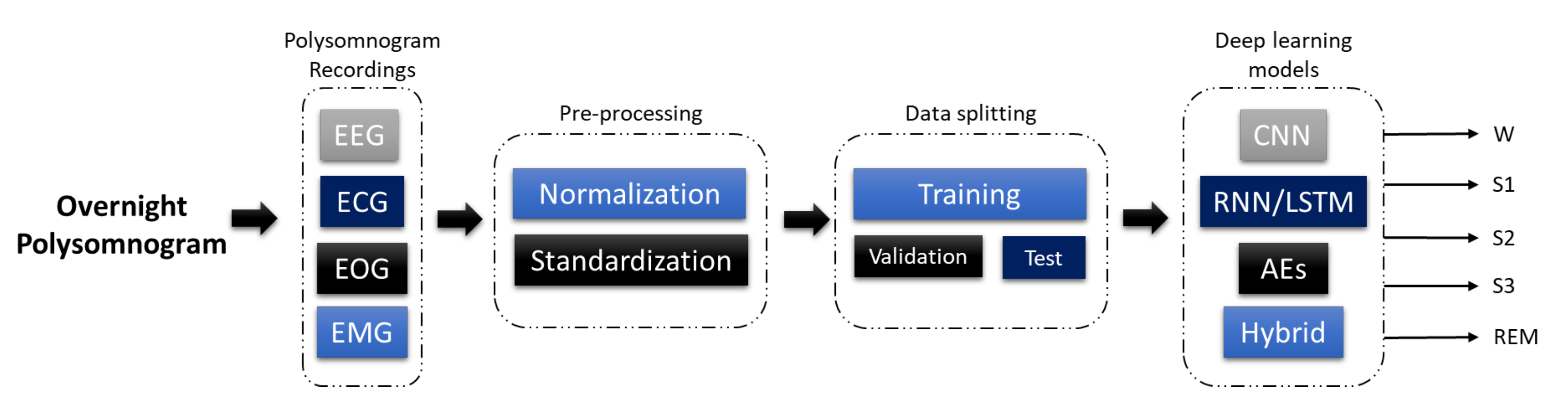
3. Programmed Diagnostic Tools (PDTs) for Polysomnogram (PSG) Analysis
4. DL Models
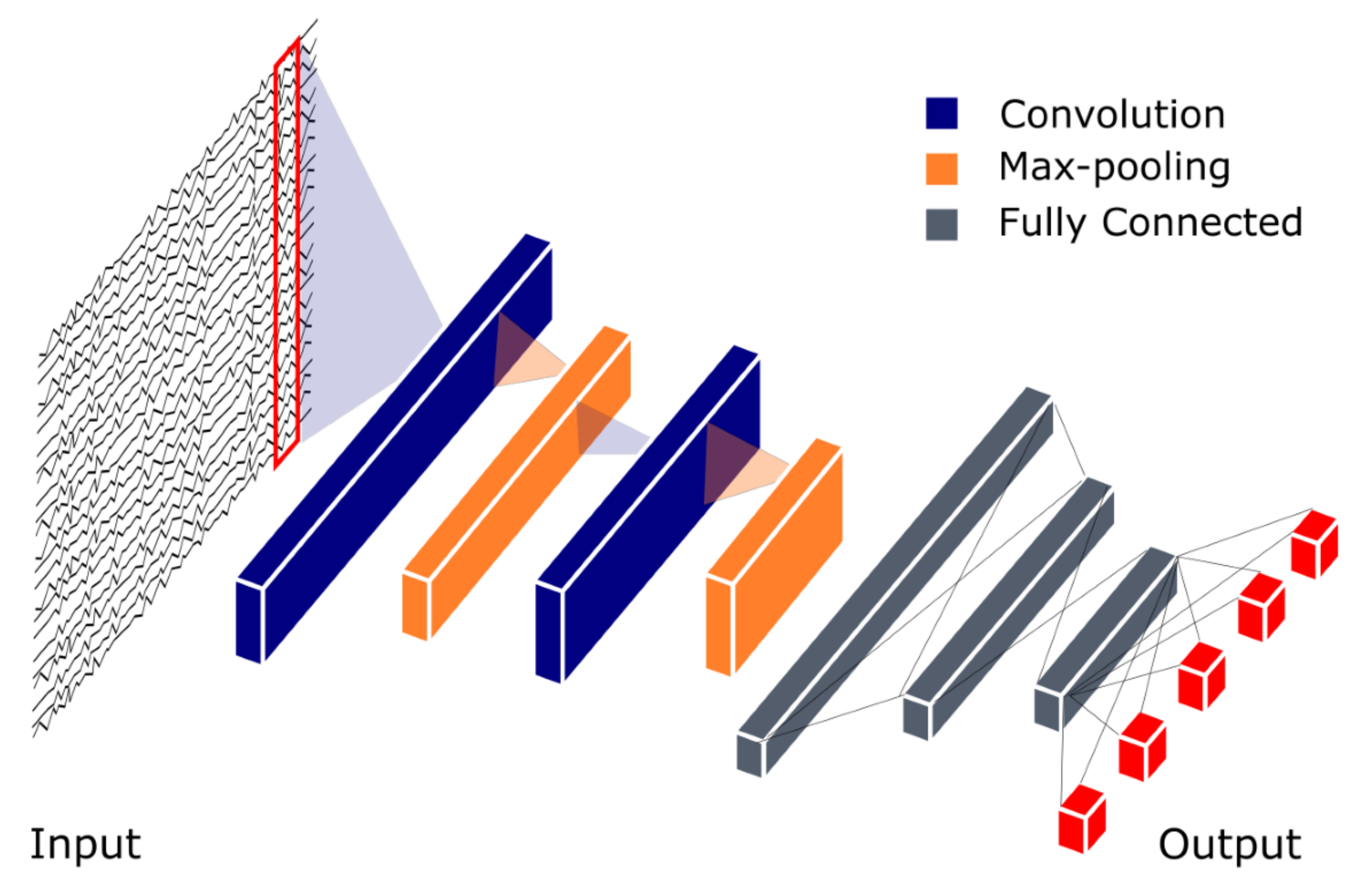
4.1. Convolutional Neural Network (CNN)
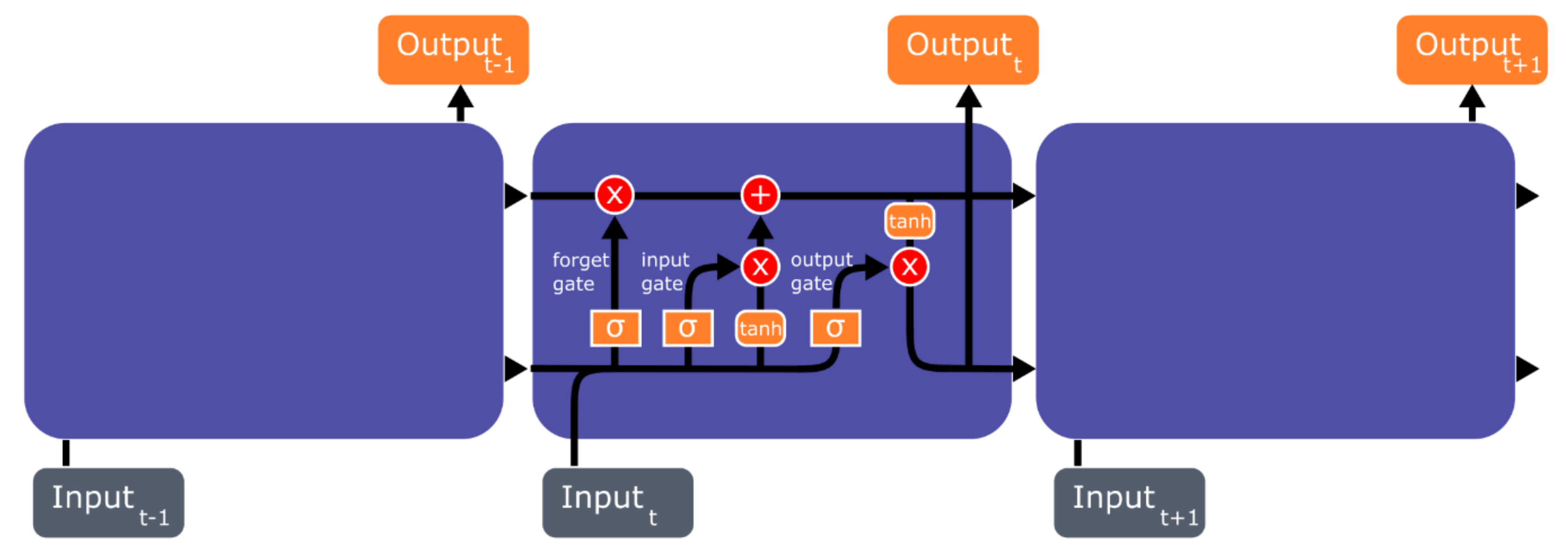
4.2. Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM)
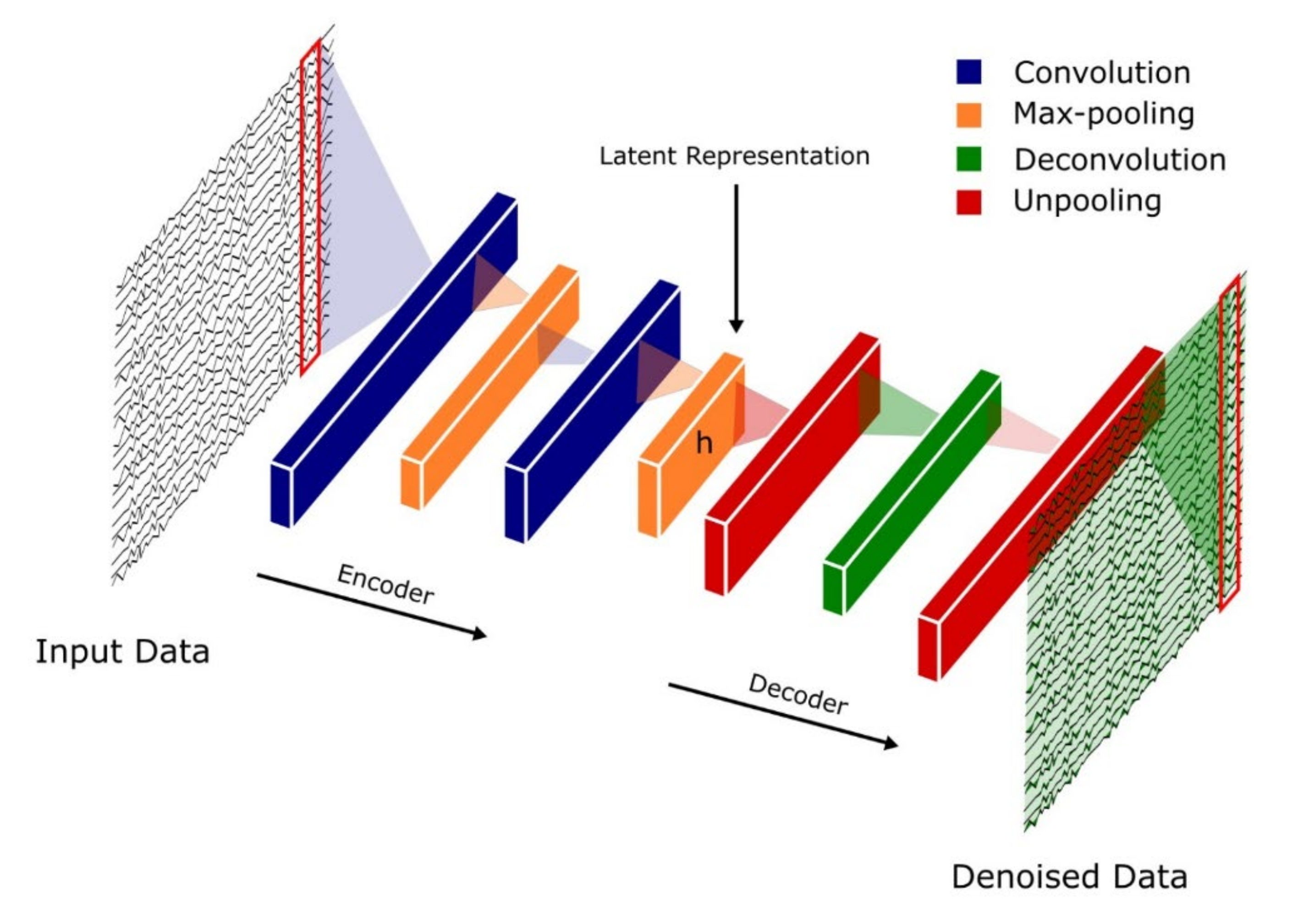
4.3. Autoencoders (AEs)
4.4. Hybrid Models
5. Sleep Stages Classification Using DL Models
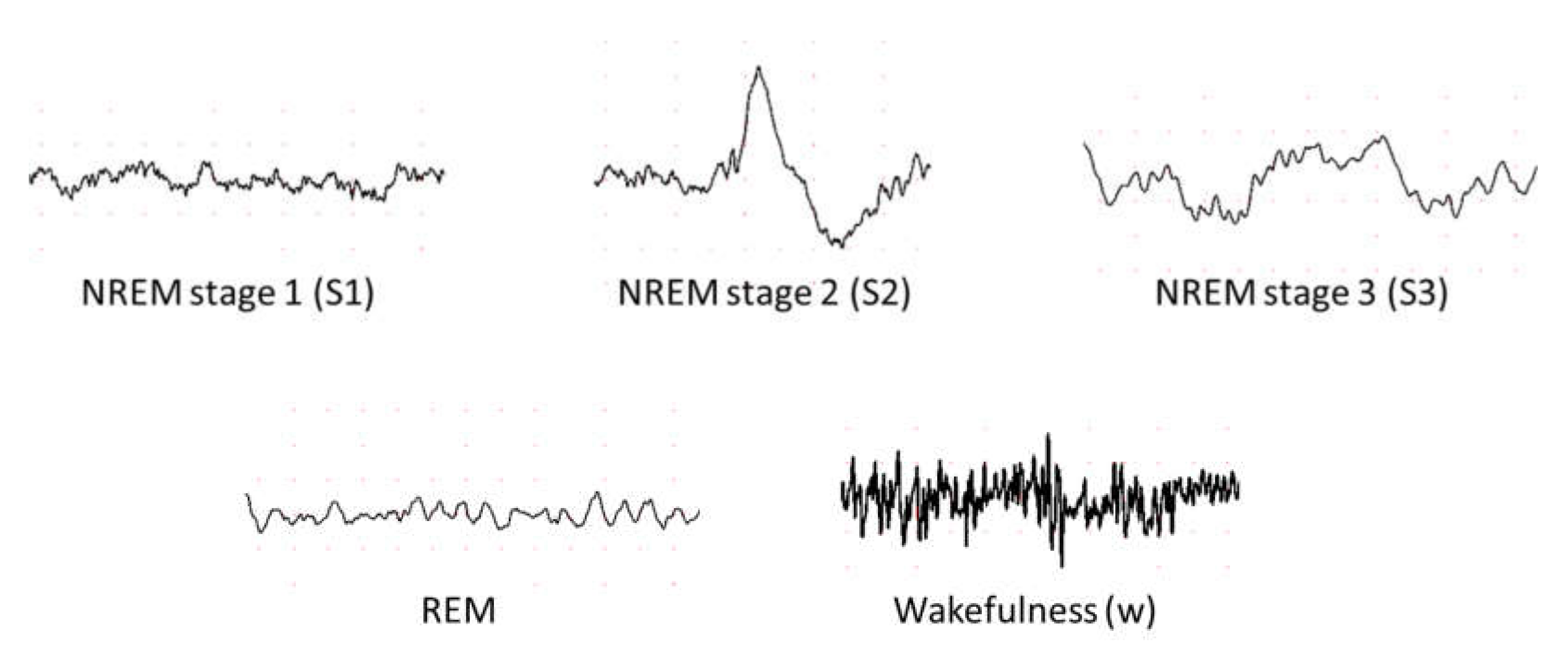
5.1. Different Stages of Sleep
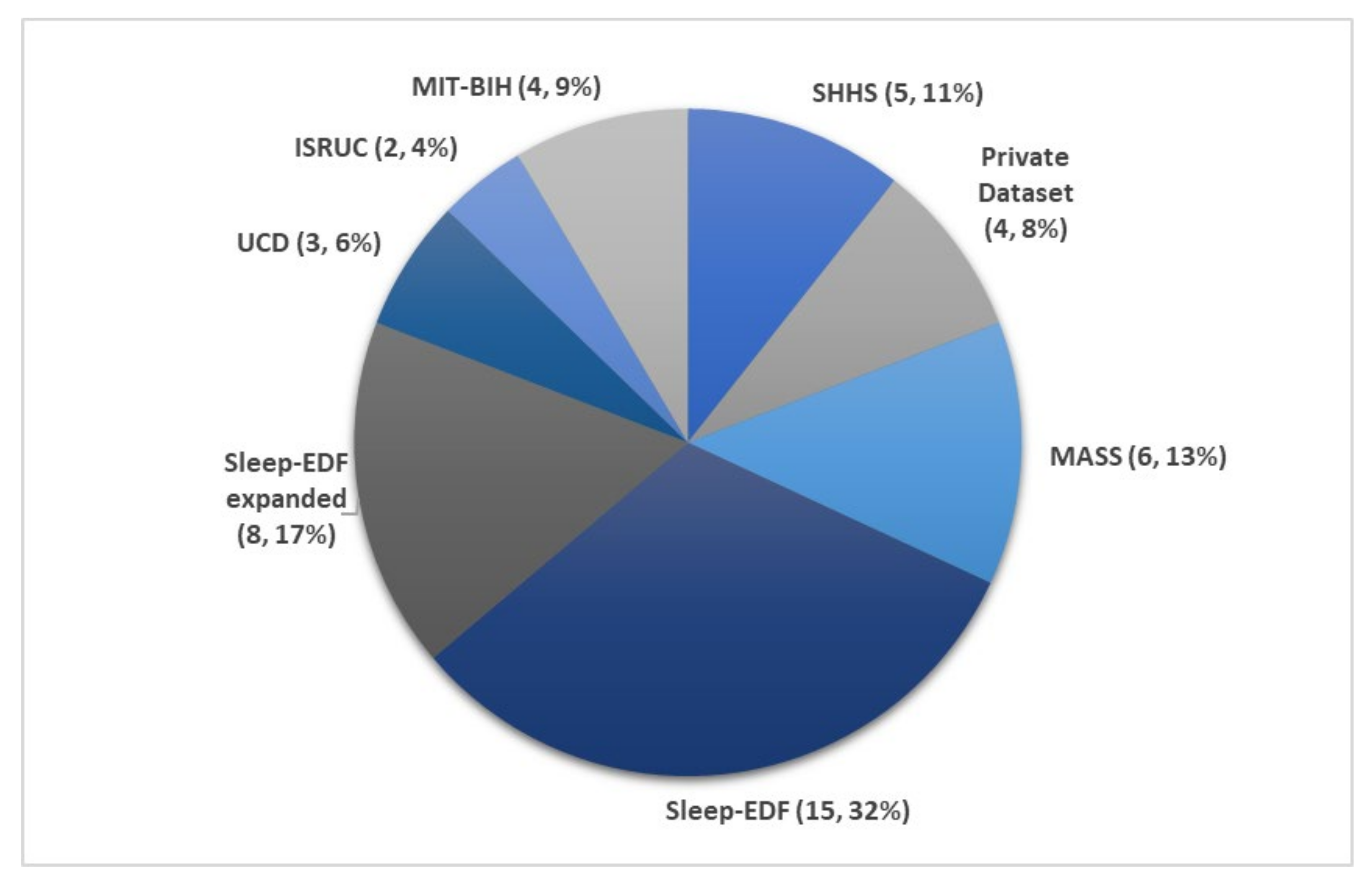
5.2. Sleep Databases
5.3. DL Techniques Used in Automatic Sleep Stage Classification
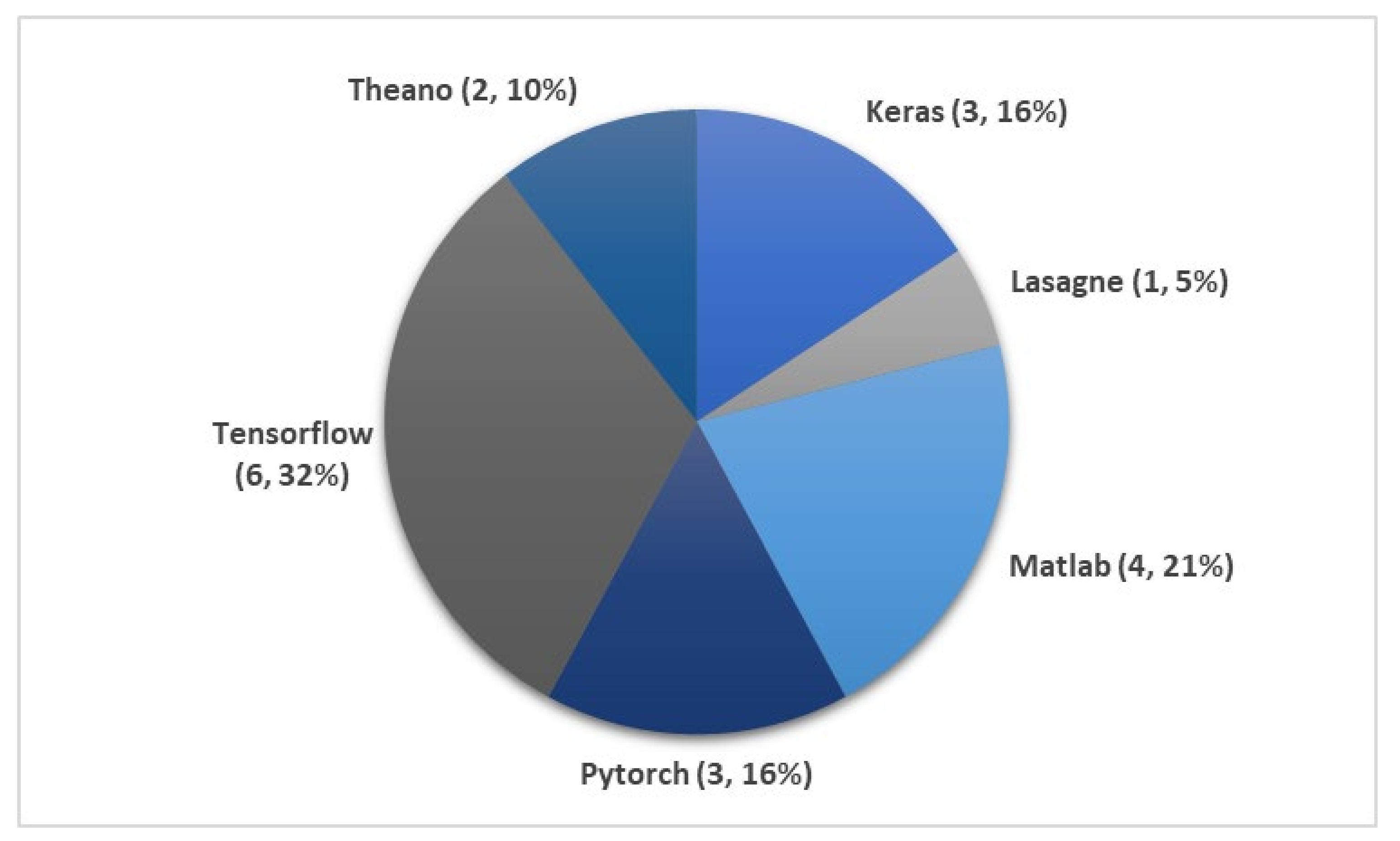
| Author | Signals | Samples | Approach | Tools/Programming Languages | Accuracy (%) |
|---|---|---|---|---|---|
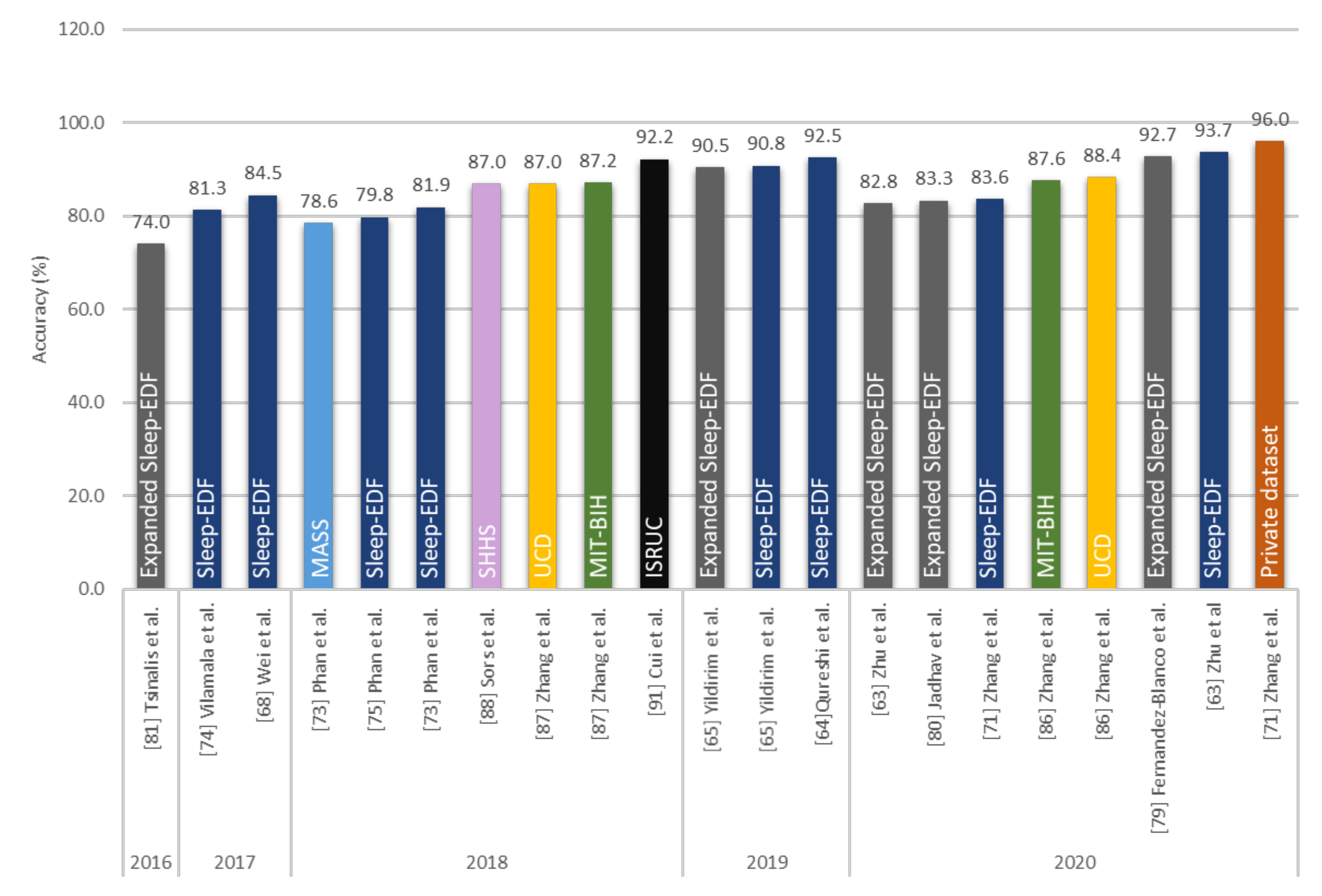
| Zhu et al. [63] 2020 | EEG | 15,188 | attention CNN | − | 93.7 |
| Qureshi et al. [64] 2019 | EEG | 41,900 | CNN | − | 92.5 |
| Yildirim et al. [65] 2019 | EEG | 15,188 | 1D-CNN | Keras | 90.8 |
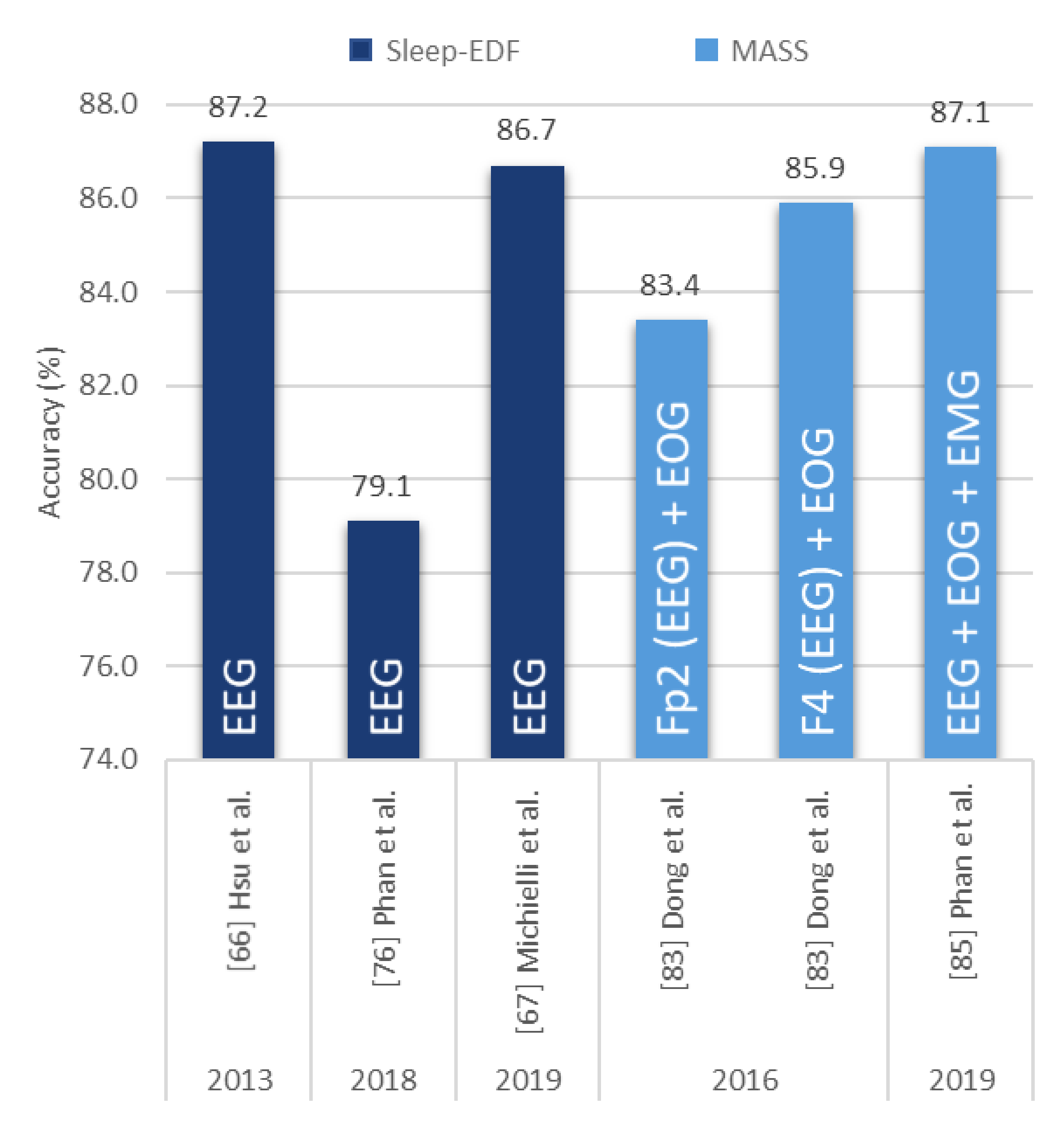
| Hsu et al. [66] 2013 | EEG | 2880 | Elman RNN | − | 87.2 |
| Michielli et al. [67] 2019 | EEG | 10,280 | RNN-LSTM | MATLAB | 86.7 |
| Wei et al. [68] 2017 | EEG | − | CNN | − | 84.5 |
| Mousavi et al. [69] 2019 | EEG | 42,308 | CNN-BiRNN | TensorFlow | 84.3 |
| Seo et al. [70] 2020 | EEG | 42,308 | CRNN | PyTorch | 83.9 |
| Zhang et al. [71] 2020 | EEG | − | CNN | − | 83.6 |
| Supratak et al. [72] 2017 | EEG | 41,950 | CNN-BiLSTM | TensorFlow | 82.0 |
| Phan et al. [73] 2019 | EEG | − | Multi-task CNN | TensorFlow | 81.9 |
| Vilamala et al. [74] 2017 | EEG | − | CNN | − | 81.3 |
| Phan et al. [75] 2018 | EEG | − | 1-max CNN | − | 79.8 |
| Phan et al. [76] 2018 | EEG | − | Attentional RNN | − | 79.1 |
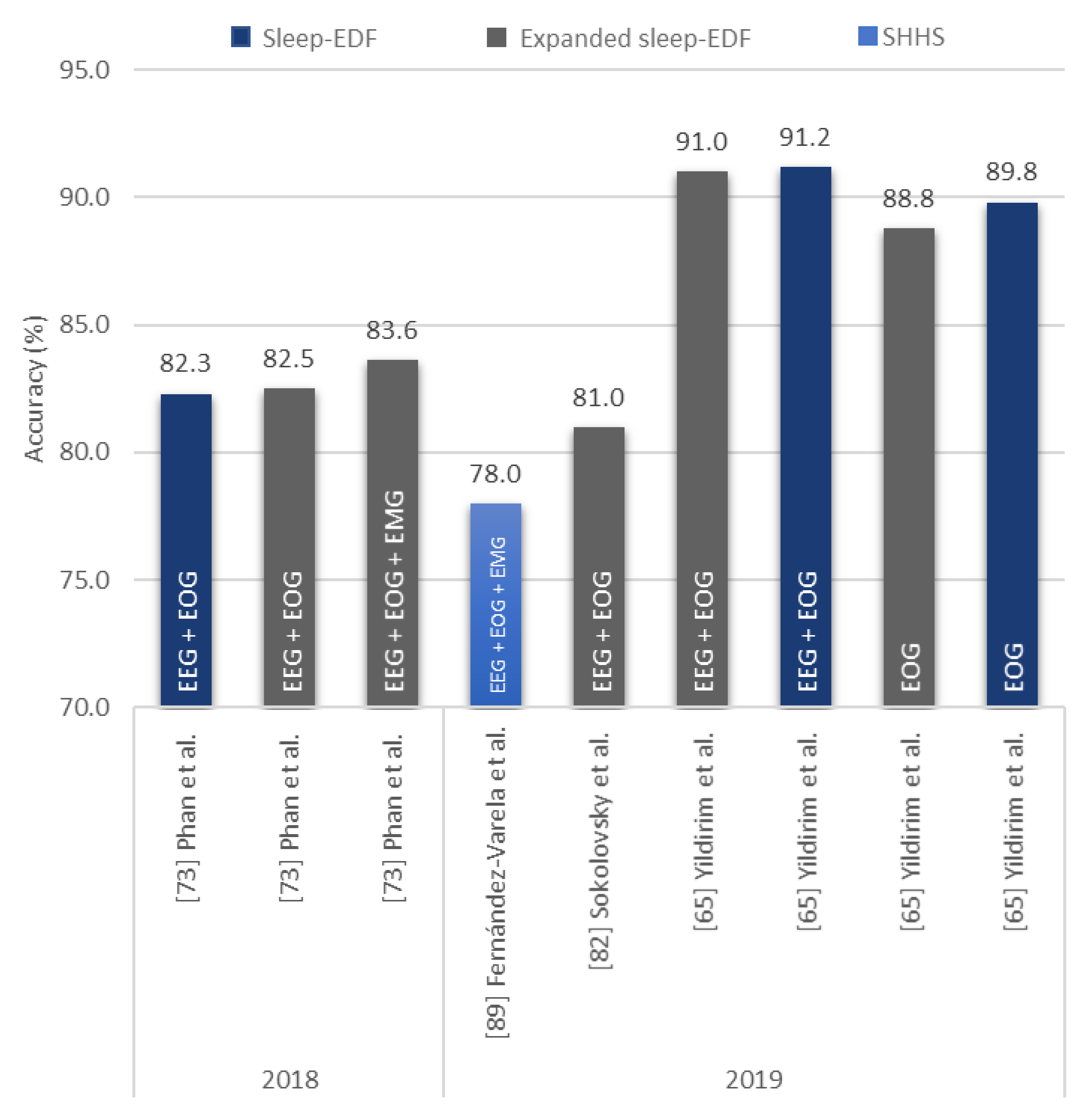
| Yildirim et al. [65] 2019 | EOG | 15,188 | 1D-CNN | Keras | 89.8 |
| Yildirim et al. [65] 2019 | EEG + EOG | 15,188 | 1D-CNN | Keras | 91.2 |
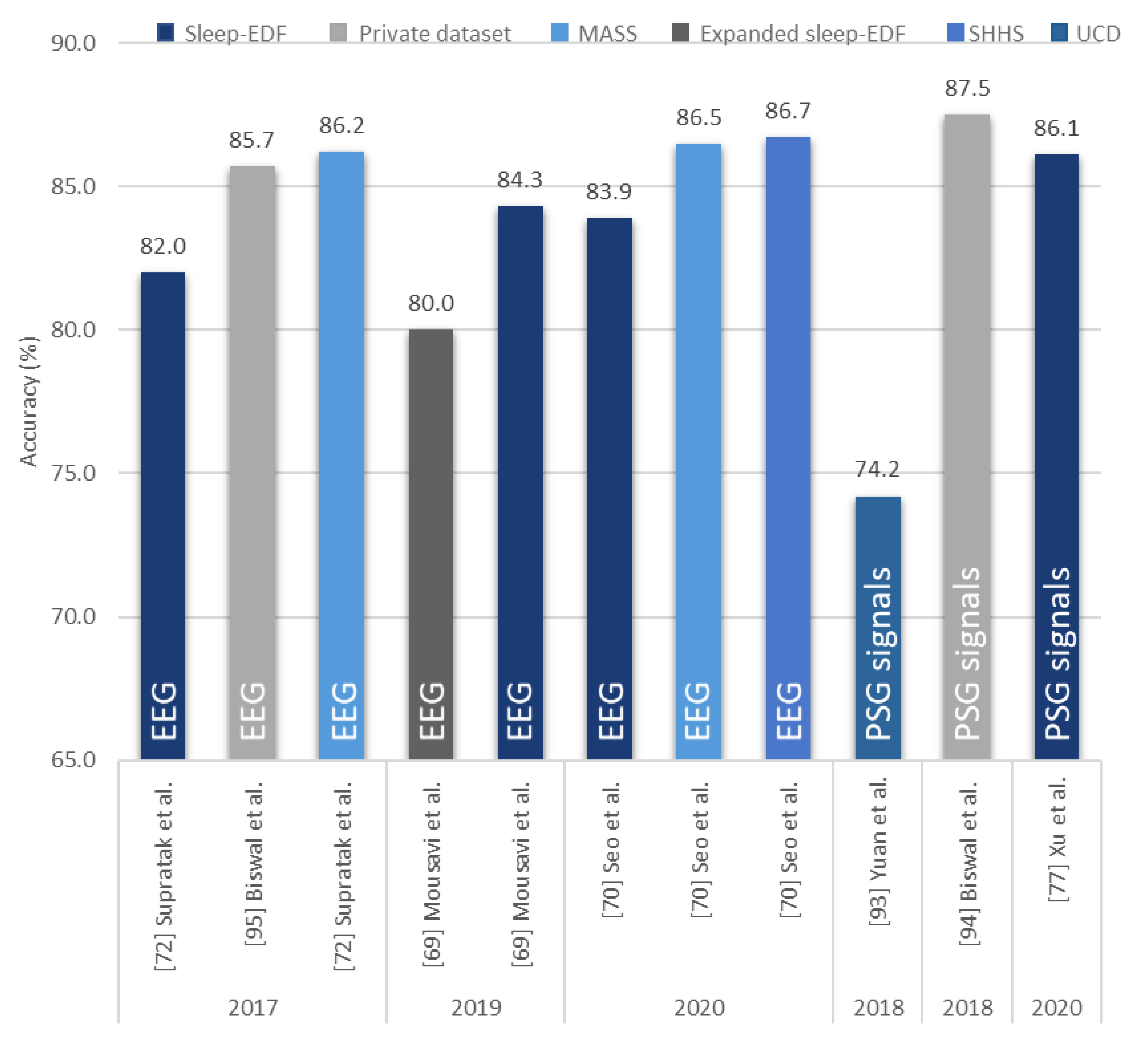
| Xu et al. [77] 2020 | PSG signals | − | DNN | − | 86.1 |
| Phan et al. [73] 2019 | EEG + EOG | − | Multi-task CNN | TensorFlow | 82.3 |
| Author | Signals | Samples | Approach | Tools/Programming Languages | Accuracy (%) |
|---|---|---|---|---|---|
| Wang et al. [78] 2018 | EEG | − | C-CNN | − | − |
| Wang et al. [78] 2018 | EEG | − | RNN-biLSTM | − | − |
| Fernandez-Blanco et al. [79] 2020 | EEG | − | CNN | − | 92.7 |
| Yildirim et al. [65] 2019 | EEG | 127,512 | 1D-CNN | Keras | 90.5 |
| Jadhav et al. [80] 2020 | EEG | 62,177 | CNN | − | 83.3 |
| Zhu et al. [63] 2020 | EEG | 42,269 | attention CNN | − | 82.8 |
| Mousavi et al. [69] 2019 | EEG | 222,479 | 1D-CNN | TensorFlow | 80.0 |
| Tsinalis et al. [81] 2016 | EEG | − | 2D-CNN | Lasagne + Theano | 74.0 |
| Yildirim et al. [65] 2019 | EOG | 127,512 | 1D-CNN | Keras | 88.8 |
| Yildirim et al. [65] 2019 | EEG + EOG | 127,512 | 1D-CNN | Keras | 91.0 |
| Sokolovsky et al. [82] 2019 | EEG + EOG | − | CNN | TensorFlow + Keras | 81.0 |
| Author | Signals | Samples | Approach | Tools/Programming Languages | Accuracy (%) |
|---|---|---|---|---|---|
| Seo et al. [70] 2020 | EEG | 57,395 | CRNN | PyTorch | 86.5 |
| Supratak et al. [72] 2017 | EEG | 58,600 | CNN-BiLSTM | TensorFlow | 86.2 |
| Phan et al. [73] 2019 | EEG | − | Multi-task CNN | TensorFlow | 78.6 |
| Dong et al. [83] 2018 | EOG F4 | − | MNN RNN-LSTM | Theano | 85.9 |
| Dong et al. [83] 2018 | EOG Fp2 | − | MNN RNN-LSTM | Theano | 83.4 |
| Chambon et al. [84] 2018 | EEG/EOG + EMG | − | 2D-CNN | Keras | − |
| Phan et al. [85] 2019 | EEG + EOG + EMG | − | Hierarchical RNN | TensorFlow | 87.1 |
| Phan et al. [73] 2019 | EEG + EOG + EMG | − | Multi-task CNN | TensorFlow | 83.6 |
| Phan et al. [73] 2019 | EEG + EOG | − | Multi-task CNN | TensorFlow | 82.5 |
| Database | Author | Signals | Samples | Approach | Tools/Programming Languages | Accuracy (%) |
|---|---|---|---|---|---|---|
| MIT-BIH | Zhang et al. [86] 2020 | EEG | − | Orthogonal CNN | − | 87.6 |
| Zhang et al. [87] 2018 | EEG | − | CUCNN | MATLAB | 87.2 | |
| SHHS | Sors et al. [88] 2018 | EEG | 5793 | CNN | − | 87.0 |
| Seo et al. [70] 2020 | EEG | 5,421,338 | CRNN | PyTorch | 86.7 | |
| Fernández-Varela et al. [89] 2019 | EEG + EOG + EMG | 1,209,971 | 1D-CNN | − | 78.0 | |
| Zhang et al. [90] 2019 | EEG + EOG + EMG | 5793 | CNN-LSTM | − | − | |
| SHHS | Li et al. [60] 2018 | ECG HRV | 400,547 | CNN | MATLAB | 65.9 |
| MIT-BIH | Li et al. [60] 2018 | ECG HRV | 2829 | CNN | MATLAB | 75.4 |
| Tripathy et al. [61] 2018 | EEG + HRV | 7500 | DNN Autoencoder | MATLAB | 73.7 |
| Database | Author | Signals | Samples | Approach | Tools/Programming Languages | Accuracy (%) |
|---|---|---|---|---|---|---|
| ISRUC | Cui et al. [91] 2018 | EEG | − | CNN | − | 92.2 |
| Yang et al. [92] 2018 | EEG | − | CNN-LSTM | − | − | |
| UCD | Zhang et al. [86] 2020 | EEG | − | Orthogonal CNN | − | 88.4 |
| Zhang et al. [87] 2018 | EEG | − | CUCNN | MATLAB | 87.0 | |
| Yuan et al. [93] 2019 | Multivariate PSG signals | 287,840 | Hybrid CNN | PyTorch | 74.2 | |
| Private datasets | Zhang et al. [71] 2020 | EEG | 264,736 | CNN | − | 96.0 |
| Biswal et al. [94] 2018 | PSG signals | 10,000 | RCNN | PyTorch | 87.5 | |
| Biswal et al. [95] 2017 | EEG | 10,000 | RCNN | TensorFlow | 85.7 | |
| Class = 4 | ||||||
| Radha et al. [62] 2019 | ECG HRV | 541,214 | LSTM | − | 77.0 |
6. Discussion
6.1. Proposed CNN-Based Models
6.2. Proposed RNN/LSTM-Based Models
6.3. Proposed Hybrid Models
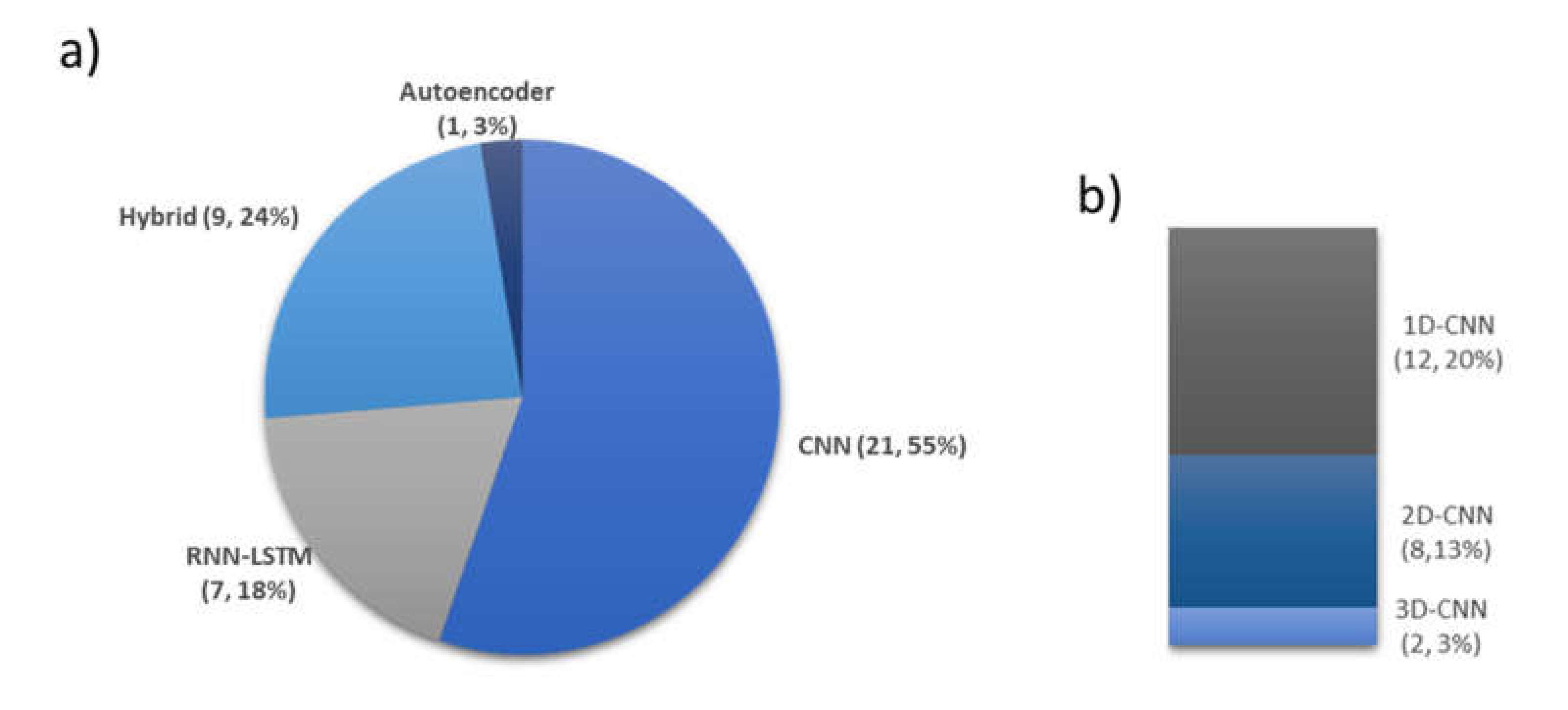
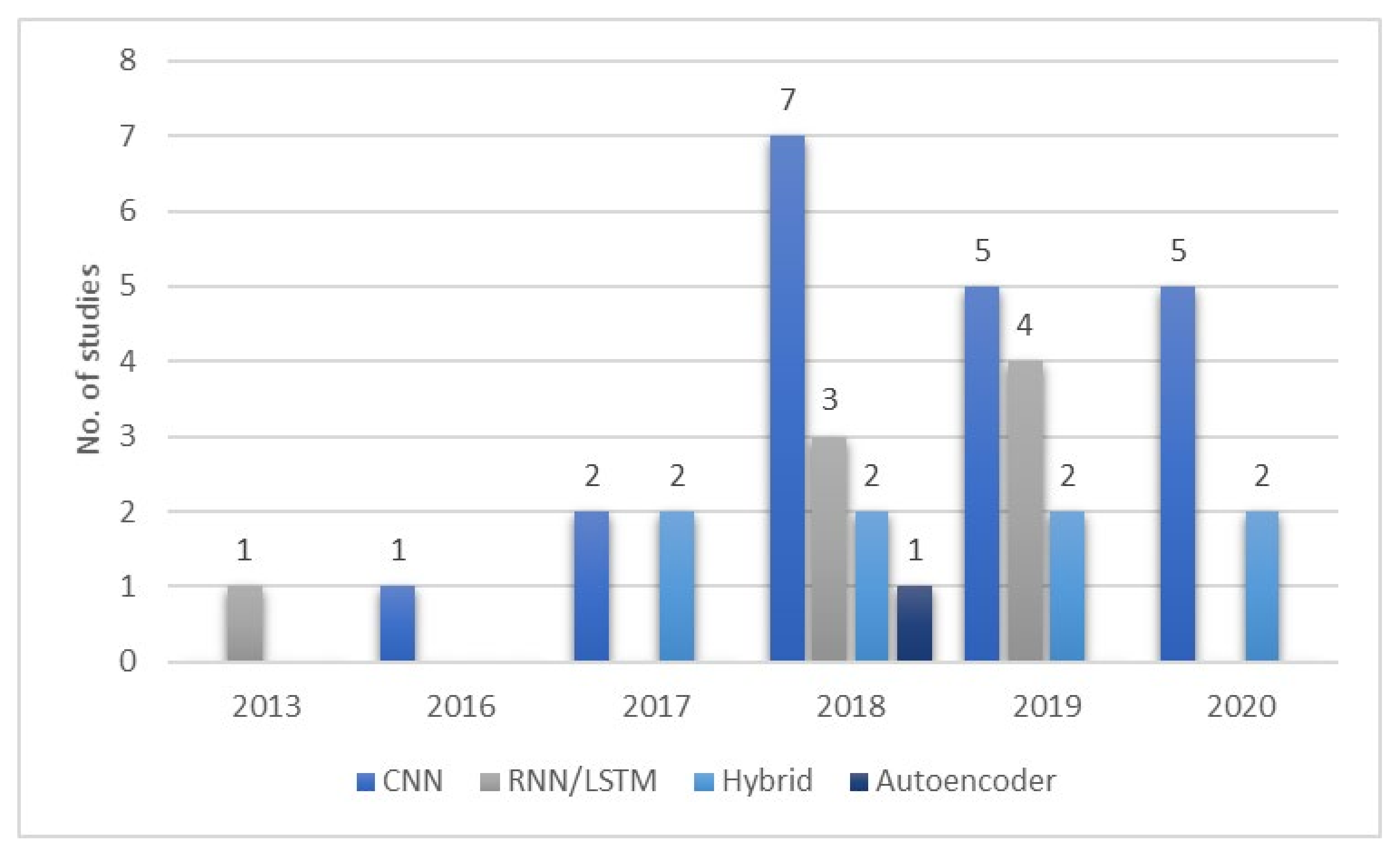
- Numerous studies (15 from Figure 10) employed CNN models with EEG signals, and that CNN models are effective in recognizing characteristic features of sleep EEG.
- One-dimensional (1D)-CNN models were used more often than 2D- and 3D-CNN models. From Figure 9b, 12, 8, and 2 1D, 2D, and 3D models were used, respectively.
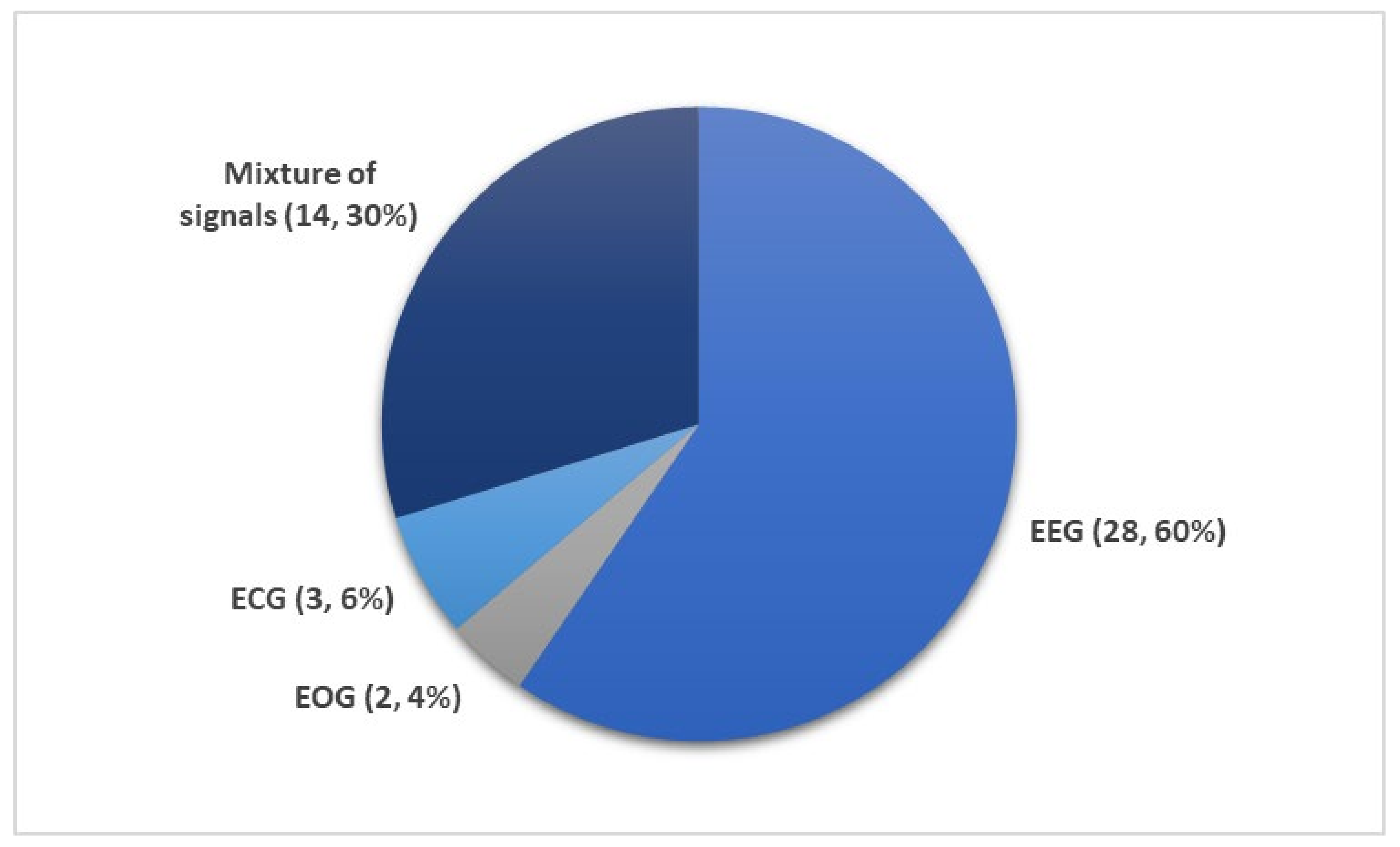
- Most studies (60% from Figure 8) used EEG signals and achieved high classification accuracy.
- EEG signals were mainly used in studies that explored a mixture of PSG signals. In other words, EEG could be a reference signal when considering mixture of PSG signals to train and evaluate newly proposed models.
- It is difficult to compare various models and identify the best performing approach, because the majority of studies used data from only one sleep database to train and test the model.
- There is a lack of studies that utilized other PSG recordings, such as EOG, EMG, or ECG signals. Studies that used these PSG recordings also did not perform equally well as those that used only EEG signals. Hence, this limits the implementation of these PSG recordings in real world applications for automated sleep stages classification.
7. Future Work
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Laposky, A.; Bass, J.; Kohsaka, A.; Turek, F.W. Sleep and circadian rhythms: Key components in the regulation of energy metabolism. FEBS Lett. 2007, 582, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.W.; Duffy, J.F. Sleep, sleep disorders, and sexual dysfunction. World J. Men’s Health 2019, 37, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Institute of Medicine (US), Committee on Sleep Medicine and Research. Sleep Disorders and Sleep Deprivation: An Unmet Public Health Problem; Colten, H.R., Altevogt, B.M., Eds.; National Academies Press: Washington, DC, USA, 2006. [Google Scholar]
- Stranges, S.; Tigbe, W.; Gómez-Olivé, F.X.; Thorogood, M.; Kandala, N.-B. Sleep problems: An emerging global epidemic? Findings from the INDEPTH WHO-SAGE study among more than 40,000 older adults from 8 countries across Africa and Asia. Sleep 2012, 35, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Schulz, H. Rethinking sleep analysis. J. Clin. Sleep Med. 2008, 4, 99–103. [Google Scholar] [CrossRef]
- Spriggs, W.H. Essentials of Polysomnography; Jones & Bartlett Learning: Burlington, MA, USA, 2014. [Google Scholar]
- Silber, M.H.; Ancoli-Israel, S.; Bonnet, M.H.; Chokroverty, S.; Grigg-Damberger, M.M.; Hirshkowitz, M.; Kapen, S.; A Keenan, S.; Kryger, M.H.; Penzel, T.; et al. The visual scoring of sleep in adults. J. Clin. Sleep Med. 2007, 3, 121–131. [Google Scholar] [CrossRef]
- Corral, J.; Pepin, J.-L.; Barbé, F. Ambulatory monitoring in the diagnosis and management of obstructive sleep apnoea syndrome. Eur. Respir. Rev. 2013, 22, 312–324. [Google Scholar] [CrossRef]
- Jung, R.; Kuhlo, W. Neurophysiological studies of abnormal night sleep and the Pickwickian syndrome. Prog. Brain Res. 1965, 18, 140–159. [Google Scholar] [CrossRef]
- Bahammam, A. Obstructive sleep apnea: From simple upper airway obstruction to systemic inflammation. Ann. Saudi Med. 2011, 31, 1–2. [Google Scholar] [CrossRef][Green Version]
- Marshall, N.S.; Wong, K.K.H.; Liu, P.Y.; Cullen, S.R.J.; Knuiman, M.; Grunstein, R.R. Sleep apnea as an independent risk factor for all-cause mortality: The Busselton health study. Sleep 2008, 31, 1079–1085. [Google Scholar] [CrossRef]
- Hirotsu, C.; Tufik, S.; Andersen, M.L. Interactions between sleep, stress, and metabolism: From physiological to pathological conditions. Sleep Sci. 2015, 8, 143–152. [Google Scholar] [CrossRef]
- Schilling, C.; Schredl, M.; Strobl, P.; Deuschle, M. Restless legs syndrome: Evidence for nocturnal hypothalamic-pituitary-adrenal system activation. Mov. Disord. 2010, 25, 1047–1052. [Google Scholar] [CrossRef] [PubMed]
- Hungin, A.P.S.; Close, H. Sleep disturbances and health problems: Sleep matters. Br. J. Gen. Pract. 2010, 60, 319–320. [Google Scholar] [CrossRef]
- Hudgel, D.W. The role of upper airway anatomy and physiology in obstructive sleep. Clin. Chest Med. 1992, 13, 383–398. [Google Scholar] [PubMed]
- Shahar, E.; Whitney, C.W.; Redline, S.; Lee, E.T.; Newman, A.B.; Nieto, F.J.; O’Connor, G.; Boland, L.L.; Schwartz, J.E.; Samet, J.M. Sleep-disordered Breathing and Cardiovascular Disease. Am. J. Respir. Crit. Care Med. 2001, 163, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Mohsenin, V. Obstructive sleep apnea and hypertension: A critical review. Curr. Hypertens. Rep. 2014, 16, 482. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, J.S.; Patel, S.R. Obstructive sleep apnea. Ann. Intern. Med. 2014, 161. [Google Scholar] [CrossRef]
- Williamson, A.; Lombardi, D.A.; Folkard, S.; Stutts, J.; Courtney, T.K.; Connor, J.L. The link between fatigue and safety. Accid. Anal. Prev. 2011, 43, 498–515. [Google Scholar] [CrossRef]
- Léger, D.; Guilleminault, C.; Bader, G.; Lévy, E.; Paillard, M. Medical and socio-professional impact of insomnia. Sleep 2002, 25, 625–629. [Google Scholar] [CrossRef]
- Iber, C.; Ancoli-Israel, S.; Chesson, A.L.; Quan, S.F. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specification; American Academy of Sleep Medicine: Darien, IL, USA, 2007. [Google Scholar]
- Svetnik, V.; Ma, J.; Soper, K.A.; Doran, S.; Renger, J.J.; Deacon, S.; Koblan, K.S. Evaluation of automated and semi-automated scoring of polysomnographic recordings from a clinical trial using zolpidem in the treatment of insomnia. Sleep 2007, 30, 1562–1574. [Google Scholar] [CrossRef][Green Version]
- Pittman, M.S.D.; Macdonald, R.M.M.; Fogel, R.B.; Malhotra, A.; Todros, K.; Levy, B.; Geva, D.A.B.; White, D.P. Assessment of automated scoring of polysomnographic recordings in a population with suspected sleep-disordered breathing. Sleep 2004, 27, 1394–1403. [Google Scholar] [CrossRef]
- Anderer, P.; Gruber, G.; Parapatics, S.; Woertz, M.; Miazhynskaia, T.; Klösch, G.; Saletu, B.; Zeitlhofer, J.; Barbanoj, M.J.; Danker-Hopfe, H.; et al. An E-health solution for automatic sleep classification according to Rechtschaffen and Kales: Validation study of the Somnolyzer 24 × 7 utilizing the siesta database. Neuropsychobiology 2005, 51, 115–133. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Bhat, S.; Faust, O.; Adeli, H.; Chua, E.C.-P.; Lim, W.J.E.; Koh, J.E.W. Nonlinear dynamics measures for automated EEG-based sleep stage detection. Eur. Neurol. 2015, 74, 268–287. [Google Scholar] [CrossRef] [PubMed]
- Mirza, B.; Wang, W.; Wang, J.; Choi, H.; Chung, N.C.; Ping, P. Machine learning and integrative analysis of biomedical big data. Genes 2019, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Razaghi, H.; Barika, R.; Ciaccio, E.J.; Acharya, U.R. A review of automated sleep stage scoring based on physiological signals for the new millennia. Comput. Methods Programs Biomed. 2019, 176, 81–91. [Google Scholar] [CrossRef]
- Shoeibi, A.; Ghassemi, N.; Khodatars, M.; Jafari, M.; Hussain, S.; Alizadehsani, R.; Moridian, P.; Khosravi, A.; Hosseini-Nejad, H.; Rouhani, M.; et al. Epileptic Seizure Detection Using Deep Learning Techniques: A Review. arXiv 2020, arXiv:2007.01276. [Google Scholar]
- Faust, O.; Hagiwara, Y.; Hong, T.J.; Lih, O.S.; Acharya, U.R. Deep learning for healthcare applications based on physiological signals: A review. Comput. Methods Programs Biomed. 2018, 161, 1–13. [Google Scholar] [CrossRef]
- Silva, D.B.; Cruz, P.P.; Molina, A.; Molina, A.M. Are the long–short term memory and convolution neural networks really based on biological systems? ICT Express 2018, 4, 100–106. [Google Scholar] [CrossRef]
- Hubel, D.H.; Wiesel, T.N. Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J. Physiol. 1962, 160, 106–154. [Google Scholar] [CrossRef]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef]
- Tabian, I.; Fu, H.; Khodaei, Z.S. A convolutional neural network for impact detection and characterization of complex composite structures. Sensors 2019, 19, 4933. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Goehring, T.; Keshavarzi, M.; Carlyon, R.P.; Moore, B.C.J. Using recurrent neural networks to improve the perception of speech in non-stationary noise by people with cochlear implants. J. Acoust. Soc. Am. 2019, 146, 705. [Google Scholar] [CrossRef] [PubMed]
- Coto-Jiménez, M. Improving post-filtering of artificial speech using pre-trained LSTM neural networks. Biomimetics 2019, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Lyu, C.; Chen, B.; Ren, Y.; Ji, D. Long short-term memory RNN for biomedical named entity recognition. BMC Bioinform. 2017, 18, 462. [Google Scholar] [CrossRef] [PubMed]
- Graves, A.; Liwicki, M.; Fernández, S.; Bertolami, R.; Bunke, H.; Schmidhuber, J. A Novel connectionist system for unconstrained handwriting recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 31, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharma, A.; Tsunoda, T. Brain wave classification using long short-term memory network based OPTICAL predictor. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Kim, B.-H.; Pyun, J.-Y. ECG identification for personal authentication using LSTM-based deep recurrent neural networks. Sensors 2020, 20, 3069. [Google Scholar] [CrossRef]
- Yu, Y.; Si, X.; Hu, C.; Zhang, J.-X. A Review of recurrent neural networks: LSTM cells and network architectures. Neural Comput. 2019, 31, 1235–1270. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Gers, F.A.; Schmidhuber, J.; Cummins, F. Learning to forget: Continual prediction with LSTM. In Proceedings of the 9th International Conference on Artificial Neural Networks—ICANN’99, Edinburgh, UK, 7–10 September 1999. [Google Scholar]
- Masuko, T. Computational cost reduction of long short-term memory based on simultaneous compression of input and hidden state. In Proceedings of the 2017 IEEE Automatic Speech Recognition and Understanding Workshop, Okinawa, Japan, 16–20 December 2017; pp. 126–133. [Google Scholar] [CrossRef]
- Dash, S.; Acharya, B.R.; Mittal, M.; Abraham, A.; Kelemen, A. (Eds.) Deep Learning Techniques for Biomedical and Health Informatics; Springer International Publishing: Cham, Switzerland, 2020. [Google Scholar]
- Rumelhart, D.E.; McClelland, J.L. Learning internal representations by error propagation. In Parallel Distributed Processing: Explorations in the Microstructure of Cognition: Foundations; MIT Press: Cambridge, MA, USA, 1987; pp. 318–362. [Google Scholar]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep learning for computer vision: A brief review. Comput. Intell. Neurosci. 2018, 2018, 1–13. [Google Scholar] [CrossRef]
- Testolin, A.; Diamant, R. Combining denoising autoencoders and dynamic programming for acoustic detection and tracking of underwater moving targets. Sensors 2020, 20, 2945. [Google Scholar] [CrossRef] [PubMed]
- Trabelsi, A.; Chaabane, M.; Ben-Hur, A. Comprehensive evaluation of deep learning architectures for prediction of DNA/RNA sequence binding specificities. Bioinformatics 2019, 35, i269–i277. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Liao, B.; Xu, X.; Yang, J. A hybrid deep learning model for predicting protein hydroxylation sites. Int. J. Mol. Sci. 2018, 19, 2817. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Sugita, Y.; Koga, E.; Shirakawa, S.; Inoue, K.; Uchida, S.; Kuwahara, H.; Kousaka, M.; Kobayashi, T.; Tsuji, Y.; et al. Proposed sments and amendments to ‘A Manual of Standardized Terminology, Techniques and Scoring System for Sleep Stages of Human Subjects’, the Rechtschaffen & Kales (1968) standard. Psychiatry Clin. Neurosci. 2001, 55, 305–310. [Google Scholar] [CrossRef]
- Carley, D.W.; Farabi, S.S. Physiology of sleep. Diabetes Spectr. 2016, 29, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.-K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef]
- Kemp, B.; Zwinderman, A.; Tuk, B.; Kamphuisen, H.; Oberye, J. Analysis of a sleep-dependent neuronal feedback loop: The slow-wave microcontinuity of the EEG. IEEE Trans. Biomed. Eng. 2000, 47, 1185–1194. [Google Scholar] [CrossRef]
- Zhang, G.-Q.; Cui, L.; Mueller, R.; Tao, S.; Kim, M.; Rueschman, M.; Mariani, S.; Mobley, D.R.; Redline, S. The national sleep research resource: Towards a sleep data commons. J. Am. Med. Inform. Assoc. 2018, 25, 1351–1358. [Google Scholar] [CrossRef]
- Quan, S.F.; Howard, B.V.; Iber, C.; Kiley, J.P.; Nieto, F.J.; O’Connor, G.; Rapoport, D.M.; Redline, S.; Robbins, J.; Samet, J.M.; et al. The sleep heart health study: Design, rationale, and methods. Sleep 1997, 20, 1077–1085. [Google Scholar] [CrossRef]
- Ichimaru, Y.; Moody, G. Development of the polysomnographic database on CD-ROM. Psychiatry Clin. Neurosci. 1999, 53, 175–177. [Google Scholar] [CrossRef]
- Khalighi, S.; Sousa, T.; Santos, J.M.; Nunes, U. ISRUC-Sleep: A comprehensive public dataset for sleep researchers. Comput. Methods Programs Biomed. 2016, 124, 180–192. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, C.; Gosselin, N.; Carrier, J.; Nielsen, T. Montreal archive of sleep studies: An open-access resource for instrument benchmarking and exploratory research. J. Sleep Res. 2014, 23, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, Q.C.; Liu, C.; Shashikumar, S.P.; Nemati, S.; Clifford, G.D. Deep learning in the cross-time frequency domain for sleep staging from a single-lead electrocardiogram. Physiol. Meas. 2018, 39, 124005. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, R.; Acharya, U.R. Use of features from RR-time series and EEG signals for automated classification of sleep stages in deep neural network framework. Biocybern. Biomed. Eng. 2018, 38, 890–902. [Google Scholar] [CrossRef]
- Radha, M.; Fonseca, P.; Moreau, A.; Ross, M.; Cerny, A.; Anderer, P.; Long, X.; Aarts, R.M. Sleep stage classification from heart-rate variability using long short-term memory neural networks. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Zhu, T.; Luo, W.; Yu, F. Convolution-and attention-based neural network for automated sleep stage classification. Int. J. Environ. Res. Public Health 2020, 17, 4152. [Google Scholar] [CrossRef]
- Qureshi, S.; Karrila, S.; Vanichayobon, S. GACNN SleepTuneNet: A genetic algorithm designing the convolutional neuralnetwork architecture for optimal classification of sleep stages from a single EEG channel. Turk. J. Electr. Eng. Comput. Sci. 2019, 27, 4203–4219. [Google Scholar] [CrossRef]
- Yıldırım, Ö.; Baloglu, U.B.; Acharya, U.R. A deep learning model for automated sleep stages classification using PSG signals. Int. J. Environ. Res. Public Health 2019, 16, 599. [Google Scholar] [CrossRef]
- Hsu, Y.-L.; Yang, Y.-T.; Wang, J.-S.; Hsu, C.-Y. Automatic sleep stage recurrent neural classifier using energy features of EEG signals. Neurocomputing 2013, 104, 105–114. [Google Scholar] [CrossRef]
- Michielli, N.; Acharya, U.R.; Molinari, F. Cascaded LSTM recurrent neural network for automated sleep stage classification using single-channel EEG signals. Comput. Biol. Med. 2019, 106, 71–81. [Google Scholar] [CrossRef]
- Wei, L.; Lin, Y.; Wang, J.; Ma, Y. Time-Frequency Convolutional Neural Network for Automatic Sleep Stage Classification Based on Single-Channel EEG. In Proceedings of the 2017 IEEE 29th International Conference on Tools with Artificial Intelligence, Boston, MA, USA, 6–8 November 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 88–95. [Google Scholar]
- Mousavi, S.; Afghah, F.; Acharya, U.R. SleepEEGNet: Automated sleep stage scoring with sequence to sequence deep learning approach. PLoS ONE 2019, 14, e0216456. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Back, S.; Lee, S.; Park, D.; Kim, T.; Lee, K. Intra- and inter-epoch temporal context network (IITNet) using sub-epoch features for automatic sleep scoring on raw single-channel EEG. Biomed. Signal Process. Control 2020, 61, 102037. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, M.; Li, Y.; Su, M.; Xu, Z.; Wang, C.; Kang, D.; Li, H.; Mu, X.; Ding, X.; et al. Automated multi-model deep neural network for sleep stage scoring with unfiltered clinical data. Sleep Breath. 2020, 24, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Supratak, A.; Dong, H.; Wu, C.; Guo, Y. DeepSleepNet: A model for automatic sleep stage scoring based on raw single-channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1998–2008. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.; Andreotti, F.; Cooray, N.; Chén, O.Y.; de Vos, M. Joint classification and prediction CNN framework for automatic sleep stage classification. IEEE Trans. Biomed. Eng. 2019, 66, 1285–1296. [Google Scholar] [CrossRef]
- Vilamala, A.; Madsen, K.H.; Hansen, L.K. Deep convolutional neural networks for interpretable analysis of EEG sleep stage scoring. In Proceedings of the 2017 IEEE 27th International Workshop on Machine Learning for Signal Processing (MLSP), Tokyo, Japan, 25–28 September 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Phan, H.; Andreotti, F.; Cooray, N.; Chen, O.Y.; de Vos, M. DNN filter bank improves 1-max pooling CNN for single-channel EEG automatic sleep stage classification. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 453–456. [Google Scholar] [CrossRef]
- Phan, H.; Andreotti, F.; Cooray, N.; Chen, O.Y.; De Vos, M. Automatic Sleep Stage Classification Using Single-Channel EEG: Learning Sequential Features with Attention-Based Recurrent Neural Networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018. [Google Scholar]
- Xu, M.; Wang, X.; Zhangt, X.; Bin, G.; Jia, Z.; Chen, K. Computation-Efficient Multi-Model Deep Neural Network for Sleep Stage Classification. In Proceedings of the ASSE ’20: 2020 Asia Service Sciences and Software Engineering Conference, Nagoya, Japan, 13–15 May 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 1–8. [Google Scholar]
- Wang, Y.; Wu, D. Deep Learning for Sleep Stage Classification. In Proceedings of the 2018 Chinese Automation Congress (CAC), Xi’an, China, 30 November–2 December 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 3833–3838. [Google Scholar]
- Fernandez-Blanco, E.; Rivero, D.; Pazos, A. Convolutional neural networks for sleep stage scoring on a two-channel EEG signal. Soft Comput. 2019, 24, 4067–4079. [Google Scholar] [CrossRef]
- Jadhav, P.; Rajguru, G.; Datta, D.; Mukhopadhyay, S. Automatic sleep stage classification using time-frequency images of CWT and transfer learning using convolution neural network. Biocybern. Biomed. Eng. 2020, 40, 494–504. [Google Scholar] [CrossRef]
- Tsinalis, O.; Matthews, P.M.; Guo, Y.; Zafeiriou, S. Automatic Sleep Stage Scoring with Single-Channel EEG Using Convolutional Neural Networks; Imperial College London: London, UK, 2016. [Google Scholar]
- Sokolovsky, M.; Guerrero, F.; Paisarnsrisomsuk, S.; Ruiz, C.; Alvarez, S.A. Deep learning for automated feature discovery and classification of sleep stages. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 17. [Google Scholar] [CrossRef]
- Dong, H.; Supratak, A.; Pan, W.; Wu, C.; Matthews, P.M.; Guo, Y. Mixed neural network approach for temporal sleep stage classification. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 324–333. [Google Scholar] [CrossRef]
- Chambon, S.; Galtier, M.N.; Arnal, P.J.; Wainrib, G.; Gramfort, A. A deep learning architecture for temporal sleep stage classification using multivariate and multimodal time series. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 758–769. [Google Scholar] [CrossRef]
- Phan, H.; Andreotti, F.; Cooray, N.; Chén, O.Y.; de Vos, M. SeqSleepNet: End-to-end hierarchical recurrent neural network for sequence-to-sequence automatic sleep staging. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yao, R.; Ge, W.; Gao, J. Orthogonal convolutional neural networks for automatic sleep stage classification based on single-channel EEG. Comput. Methods Programs Biomed. 2020, 183, 105089. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, Y. Complex-valued unsupervised convolutional neural networks for sleep stage classification. Comput. Methods Programs Biomed. 2018, 164, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Sors, A.; Bonnet, S.; Mirek, S.; Vercueil, L.; Payen, J.-F. A convolutional neural network for sleep stage scoring from raw single-channel EEG. Biomed. Signal Process. Control 2018, 42, 107–114. [Google Scholar] [CrossRef]
- Fernández-Varela, I.; Hernández-Pereira, E.; Alvarez-Estevez, D.; Moret-Bonillo, V. A Convolutional Network for Sleep Stages Classification. arXiv 2019, arXiv:1902.05748v1. [Google Scholar] [CrossRef]
- Zhang, L.; Fabbri, D.; Upender, R.; Kent, D.T. Automated sleep stage scoring of the Sleep Heart Health Study using deep neural networks. Sleep 2019, 42. [Google Scholar] [CrossRef]
- Cui, Z.; Zheng, X.; Shao, X.; Cui, L. Automatic sleep stage classification based on convolutional neural network and fine-grained segments. Complexity 2018, 2018, 1–13. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, X.; Yuan, F. A Study on Automatic Sleep Stage Classification Based on CNN-LSTM. In Proceedings of the ICCSE’18: The 3rd International Conference on Crowd Science and Engineering, Singapore, 28–31 July 2018; Association for Computing Machinery: New York, NY, USA, 2018; pp. 1–5. [Google Scholar]
- Yuan, Y.; Jia, K.; Ma, F.; Xun, G.; Wang, Y.; Su, L.; Zhang, A. A hybrid self-attention deep learning framework for multivariate sleep stage classification. BMC Bioinform. 2019, 20, 1–10. [Google Scholar] [CrossRef]
- Biswal, S.; Sun, H.; Goparaju, B.; Westover, M.B.; Sun, J.; Bianchi, M.T. Expert-level sleep scoring with deep neural networks. J. Am. Med. Inform. Assoc. 2018, 25, 1643–1650. [Google Scholar] [CrossRef]
- Biswal, S.; Kulas, J.; Sun, H.; Goparaju, B.; Westover, M.B.; Bianchi, M.T.; Sun, J. SLEEPNET: Automated Sleep Staging System via Deep Learning. arXiv 2017, arXiv:1707.08262. [Google Scholar]
- Hoshide, S.; Kario, K. Sleep Duration as a risk factor for cardiovascular disease—A review of the recent literature. Curr. Cardiol. Rev. 2010, 6, 54–61. [Google Scholar] [CrossRef]
- Woods, S.L.; Froelicher, E.S.S.; Motzer, S.U.; Bridges, S.J. Cardiac Nursing, 5th ed.; Lippincott Williams and Wilkins: London, UK, 2005. [Google Scholar]
- Krieger, J. Breathing during sleep in normal subjects. Clin. Chest Med. 1985, 6, 577–594. [Google Scholar] [PubMed]
- Madsen, P.L.; Schmidt, J.F.; Wildschiodtz, G.; Friberg, L.; Holm, S.; Vorstrup, S.; Lassen, N.A. Cerebral O2 metabolism and cerebral blood flow in humans during deep and rapid-eye-movement sleep. J. Appl. Physiol. 1991, 70, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Klosh, G.; Kemp, B.; Penzel, T.; Schlogl, A.; Rappelsberger, P.; Trenker, E.; Gruber, G.; Zeithofer, J.; Saletu, B.; Herrmann, W.; et al. The SIESTA project polygraphic and clinical database. IEEE Eng. Med. Boil. Mag. 2001, 20, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Yıldırım, Ö.; Talo, M.; Ay, B.; Baloglu, U.B.; Aydin, G.; Acharya, U.R. Automated detection of diabetic subject using pre-trained 2D-CNN models with frequency spectrum images extracted from heart rate signals. Comput. Biol. Med. 2019, 113, 103387. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.-H.; Vicnesh, J.; Koh, J.E.; Oh, S.L.; Arunkumar, N.; Abdulhay, E.; Ciaccio, E.J.; Acharya, U.R. Autism spectrum disorder diagnostic system using HOS bispectrum with EEG Signals. Int. J. Environ. Res. Public Health 2020, 17, 971. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Kim, J.-M. Automated bearing fault diagnosis using 2D analysis of vibration acceleration signals under variable speed conditions. Shock. Vib. 2016, 2016, 1–11. [Google Scholar] [CrossRef]
- Kiranyaz, S.; Avci, O.; Abdeljaber, O.; Ince, T.; Gabbouj, M.; Inman, D.J. 1D convolutional neural networks and applications: A survey. Mech. Syst. Signal Process. 2021, 151. [Google Scholar] [CrossRef]
- Patanaik, A.; Ong, J.L.; Gooley, J.J.; Ancoli-Israel, S.; Chee, M.W.L. An end-to-end framework for real-time automatic sleep stage classification. Sleep 2018, 41. [Google Scholar] [CrossRef]
- Terzano, M.G.; Parrino, L.; Sherieri, A.; Chervin, R.; Chokroverty, S.; Guilleminault, C.; Hirshkowitz, M.; Mahowald, M.; Moldofsky, H.; Rosa, A.; et al. Atlas, rules, and recording techniques for the scoring of cyclic alternating pattern (CAP) in human sleep. Sleep Med. 2001, 2, 537–553. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loh, H.W.; Ooi, C.P.; Vicnesh, J.; Oh, S.L.; Faust, O.; Gertych, A.; Acharya, U.R. Automated Detection of Sleep Stages Using Deep Learning Techniques: A Systematic Review of the Last Decade (2010–2020). Appl. Sci. 2020, 10, 8963. https://doi.org/10.3390/app10248963
Loh HW, Ooi CP, Vicnesh J, Oh SL, Faust O, Gertych A, Acharya UR. Automated Detection of Sleep Stages Using Deep Learning Techniques: A Systematic Review of the Last Decade (2010–2020). Applied Sciences. 2020; 10(24):8963. https://doi.org/10.3390/app10248963
Chicago/Turabian StyleLoh, Hui Wen, Chui Ping Ooi, Jahmunah Vicnesh, Shu Lih Oh, Oliver Faust, Arkadiusz Gertych, and U. Rajendra Acharya. 2020. "Automated Detection of Sleep Stages Using Deep Learning Techniques: A Systematic Review of the Last Decade (2010–2020)" Applied Sciences 10, no. 24: 8963. https://doi.org/10.3390/app10248963
APA StyleLoh, H. W., Ooi, C. P., Vicnesh, J., Oh, S. L., Faust, O., Gertych, A., & Acharya, U. R. (2020). Automated Detection of Sleep Stages Using Deep Learning Techniques: A Systematic Review of the Last Decade (2010–2020). Applied Sciences, 10(24), 8963. https://doi.org/10.3390/app10248963







