Abstract
Sleep is vital for one’s general well-being, but it is often neglected, which has led to an increase in sleep disorders worldwide. Indicators of sleep disorders, such as sleep interruptions, extreme daytime drowsiness, or snoring, can be detected with sleep analysis. However, sleep analysis relies on visuals conducted by experts, and is susceptible to inter- and intra-observer variabilities. One way to overcome these limitations is to support experts with a programmed diagnostic tool (PDT) based on artificial intelligence for timely detection of sleep disturbances. Artificial intelligence technology, such as deep learning (DL), ensures that data are fully utilized with low to no information loss during training. This paper provides a comprehensive review of 36 studies, published between March 2013 and August 2020, which employed DL models to analyze overnight polysomnogram (PSG) recordings for the classification of sleep stages. Our analysis shows that more than half of the studies employed convolutional neural networks (CNNs) on electroencephalography (EEG) recordings for sleep stage classification and achieved high performance. Our study also underscores that CNN models, particularly one-dimensional CNN models, are advantageous in yielding higher accuracies for classification. More importantly, we noticed that EEG alone is not sufficient to achieve robust classification results. Future automated detection systems should consider other PSG recordings, such as electroencephalogram (EEG), electrooculogram (EOG), and electromyogram (EMG) signals, along with input from human experts, to achieve the required sleep stage classification robustness. Hence, for DL methods to be fully realized as a practical PDT for sleep stage scoring in clinical applications, inclusion of other PSG recordings, besides EEG recordings, is necessary. In this respect, our report includes methods published in the last decade, underscoring the use of DL models with other PSG recordings, for scoring of sleep stages.
Keywords:
sleep disorder; obstructive sleep disorder; overnight polysomnogram; EEG; EMG; ECG; HRV signals; deep learning 1. Introduction
Sleep is crucial for the maintenance and regulation of various biological functions at a molecular level [1], which helps humans to restore physical and mental wellbeing and proper brain function during the day [2]. There are two primary types of sleep: non-rapid eye movement (NREM) and rapid eye movement (REM) sleep. NREM sleep comprises four stages, after which, it continues into the REM sleep stage. NREM and REM sleep stages are connected and cyclically alternated through the sleep process wherein unbalanced cycling or the absence of sleep stages give rise to sleep disorders [3]. Unfortunately, sleep disorders, which lead to poor sleep quality, is often neglected [4]. Stranges et al. [4] highlighted that sleep-related problems is a looming global health issue. In their study, datasets from the World Health Organization (WHO) and International Network for the Demographic Evaluation of Populations and Their Health (INDEPTH) were used to investigate the prevalence of sleep problems in low-income countries. It was reported that 16.6% of the adult population, which amounts to approximately 150 million, have sleep problems and current trends indicate that this figure will increase to 260 million by 2030.
To date, it is mandatory that sleep stage scoring is done manually by human experts [5,6]. However, human experts have limited capacity to handle slow changes in background electroencephalography (EEG) and learn the different rules to score sleep stages for various polysomnogram (PSG) recordings [6]. Furthermore, evaluations by human experts are prone to inter- and intra-observer variabilities that can negatively affect the quality of sleep stage scoring [7]. Another important factor affecting sleep stage scoring is the patient convenience and the cost of diagnosis. As such, a sleep lab is a highly controlled environment that requires dedicated facilities and highly trained personnel. Hence, sleep labs tend to be in urban centers and patients must travel there to spend one or multiple nights in the facility. These factors make sleep labs inconvenient for patients and the cost per diagnosis is high. Other diagnostic methods, such as portable monitoring devices for sleep stages, exhibit some advantages, such as enhancing access to patients, low cost, and user-friendliness. However, these advantages are outweighed by several disadvantages, such as having diagnostic limitations, failure of device, reliability concerns, and underestimating the apnea/hypopnea index, amongst others [8]. To improve the situation requires a fundamental change in the sleep stage scoring process. We need machines to replace the labor carried out by human experts. This can only be done with systems that understand sleep stages in much of the same way as human experts do. Deep learning (DL) is hailed as a method to mechanize knowledge work, such as sleep stage scoring. However, before we join and adopt this technology, it is prudent to investigate both capabilities and limitations of current DL methods.
This paper aims to capture both capabilities and limitations of current DL methods in sleep stage classification. It is intended to provide deep cohesive information for experts to consolidate and extend their knowledge in the field. This knowledge might also be of interest to policy makers and healthcare administrators because DL technologies are going to shape future sleep stage scoring systems. This review paper summarizes the various DL models employed in the last 10 years and their performances as sleep stage classification systems. This information is valuable for those who plan to use established techniques to address a related problem. This review will also help establish the distinctiveness of a study, because any claim for novelty requires an overview of established methods. In this paper, we focused the review process on DL techniques, because during our practice in the field we found that, among the various artificial intelligence technologies, DL is the most suitable to be developed into a decision support tool for sleep stage scoring. In the foreseeable future, all studies on the topic would either include DL or mention DL-based techniques as a reference point.
To support our claim that DL technology will benefit sleep stage scoring, we have structured the remainder of the manuscript as follows. Section 3 and Section 4 describe programmed diagnostic tools (PDTs) and various DL tools, respectively. Section 5 describes the guidelines for sleep stage classification and the publicly available databases with sleep recordings to train and evaluate DL models. Section 6 discusses the key findings of automated sleep stage classification studies based on different DL models. In Section 7, we elaborate on the potential future direction of sleep analysis. Section 8 concludes the paper by highlighting our review findings, which includes a discussion of the best DL models and PSG signals employed for automated sleep stage classification.
2. Medical Background
The discovery of obstructive sleep apnea (OSA) in 1965 is lauded as the greatest progress in the history of sleep medicine [9]. For many years, OSA was regarded as the occasional closure of the upper airway, thus, early treatments, such as tracheostomy, prioritized primarily on reducing the obstruction to airway [10]. However, recent studies show that OSA is linked to the risk of cardiovascular disease and death [11], emphasizing the need to consider other factors for treatment options.
The emergence of sleep disorders, such as insomnia, OSA, and various other sleep-related disorders further contribute to poor sleep quality [12,13]. Some sleep disorders manifest themselves in sleep interruptions, such as early morning awaking, and the lack or absence of restful sleep [14]. In OSA, this is more severe with symptoms such as extreme daytime drowsiness, snoring and repeated occurrences of interruptions to the respiratory airflow during sleep, which stems from the collapse of the upper airway in the throat [15]. These in turn affect the cardiovascular physiology, causing cardiovascular diseases, such as stroke, angina, and heart failure [16]. OSA has also been linked to higher morbidity and mortality rates [17] and low quality of vital scores [18]. Some studies had also showed that a lack of sleep increases fatigue during the day, which decreases the performance of individuals at work and threatens their occupational safety [19,20].
Overnight polysomnogram (PSG) is currently the “gold standard” to measure multiple physiologic parameters of sleep, and it is used to score sleep stages [6]. These recordings include electroencephalograms (EEGs), electrooculograms (EOGs), electromyograms (EMGs), electrocardiogram (ECGs), respiratory efforts, airflow, and blood oxygenation [5]. According to the American Academy of Sleep Medicine guidelines [21], sleep should be scored by segmenting these PSG recordings into 30-s fragments, also known as epochs. Each epoch is then scored and categorized based on the sleep stages that appear most often. For example, an epoch with one characteristic of Sleep Stage 1 and two characteristics of Sleep Stage 2 will be classified as Sleep Stage 2.
Technology has been shown to help improve convenience and bring down healthcare costs. In this case, such technology could take the form of a cost-effective programmed diagnostic tool (PDT) for automated detection of sleep disorders. Some studies had demonstrated the ability of PDTs to perform as well as experts in sleep stage scoring [22,23,24], and even outperforming the experts in the detection of microstructures of sleep, such as arousal and cyclic alternating pattern. These highlight PDT versatility and the potential to first augment (and perhaps replace) human experts in the analysis of sleep recordings [6,25].
3. Programmed Diagnostic Tools (PDTs) for Polysomnogram (PSG) Analysis
PDTs are based on artificial intelligence, namely conventional machine learning and DL techniques.
Unfortunately, machine learning is often subjected to the curse of dimensionality or p(features) >> n(samples), a common problem that surfaces when training machine learning models with high dimensional data, such as PSG recordings, which cause the model to overfit [26]. Hence, data reduction step such as feature extraction is helpful for traditional machine learning algorithms to prevent overfitting problem when handling high dimensional PSG recordings [27]. Often, the feature extraction from PSG recordings is done manually through experience and acquired skills of human observers [27,28]. The extracted features are fed to standard machine learning models, such as the support vector machine or random forest classifiers, to classify PSG recordings into respective sleep stages. However, this explicit feature engineering involves converting PSG recordings to a low dimensional vector which can result in information loss [27]. Furthermore, the aforementioned machine learning techniques are not ideally suited to handle high dimensional data because of the lack of depth to capture relevant relationships between the covariates in large volumes of data. As a result, standard machine learning is limited in its ability to reliably classify PSG recordings with high precision and accuracy.
On the other hand, DL models can be trained with raw PSG recordings without the need for information reduction [27]. DL techniques attempt to train the model, such that they learn how to make sense of data on their own by extracting features automatically from PSG signals. The extracted knowledge determines the inference. When a large data volume is available, DL models often outperform machine learning models because they can utilize all the available information and make accurate predictions [29]. In addition, DL models learn features that are useful to make inference and neglect those that are not. Hence, DL techniques are more suitable than traditional machine learning techniques when dealing with high-dimensional PSG signals and are thus considered state-of-art methods for automated sleep stage classification.
4. DL Models
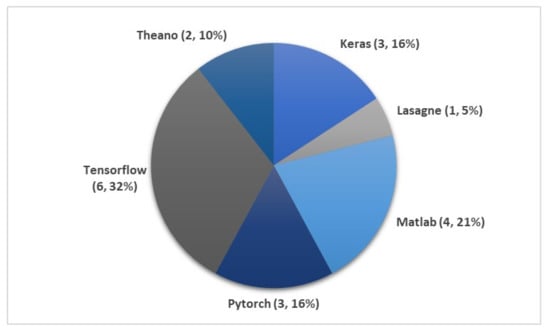
In contrast to other programming languages (such as MATLAB), the Python programming language includes a plethora of libraries that can facilitate the development of DL models with greater ease. Figure 1 shows that over 75% of the automated sleep stages classification studies employed DL tools from Python (TensorFlow, Theano, Keras, PyTorch, Lasagne). Keras is a high-level Application Programming Interface (API) for Python that can use either TensorFlow or Theano as its backend. The programming process in TensorFlow and Theano is also simplified, such that the cognitive load for human to build a DL model is significantly reduced.
4.1. Convolutional Neural Network (CNN)
The first CNN was created to mimic the human visual system. In the visual cortex, simple and complex cortical cells break down visual information into simpler representations, so that it is easier for the brain to perceive and classify the image [30,31]. In an analysis of multiple light recordings, a typical CNN model comprises three main layers: convolutional, pooling, and fully connected, as depicted in Figure 2. In this model, an input data is first broken down by learned filters at the convolutional layer to extract important features, and feature maps are created as outputs. These feature maps contain different kinds of information about the data characteristics [32]. The pooling layer follows immediately after the convolutional layer, and it is responsible for reducing the feature map dimension. By doing so, the feature map complexity is reduced, and the visual information is broken down further. Another desired effect of this architecture is the reduction of overfitting [33]. Multiple convolutional and pooling layers can be incorporated in a model to make it “deeper” and increase its recognition ability to classify complex images. After a series of convolutional and pooling layers, the resulting feature map is flattened into a single list of vectors before it is fed to the fully connected layers, which establishes connections between output and input via learnable weights [32]. The superiority of this architecture was first demonstrated in image recognition and classification by Krizhevsky et al. [34], where their proposed CNN model was awarded the top five award in an image classification competition, known as ImageNet Large Scale Visual Recognition Competition (ILSVRC-2012).
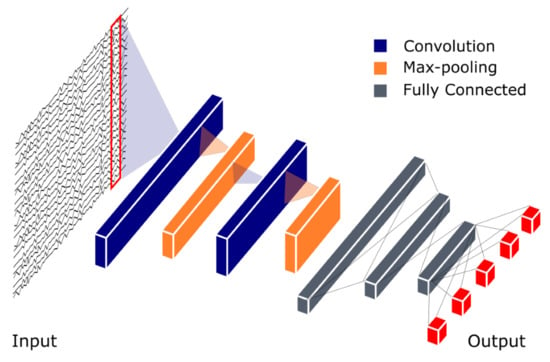
Figure 2.
Basic Convolutional Neural Network (CNN) model architecture for polysomnogram (PSG) recording analysis. The CNN output model is represented by five red boxes indicating five prediction classes for example W (Wakefulness), S1–S3 (Non-REM sleep Stages 1–3) and REM (rapid eye movement) sleep.
4.2. Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM)
Both RNN and LSTM models are designed for sequential data processing, such as speech [35], [36], text [37], and handwriting recognition [38]. These models attempt to recognize a pattern in the sequence. Previously, Kumar et al. [39] proposed LSTM model to analyze brain EEG for brain-computer interface (BCI) systems. Their model achieved a low misclassification rate of 3.09% and 2.07% for two publicly available BCI datasets. A recent study by Kim et al. [40] demonstrated the effectiveness of the LSTM and RNN in analyzing bio signals. The LSTM-based deep RNN model, proposed in their study, achieved exemplary performance: 100% accuracy, precision, and sensitivity in personal authentication based on ECG signals. This makes them suitable for classifying bio signals like PSG recordings, which have distinct patterns in different sleep stages.
It needs to be noted that very early versions of RNNs were incapable of learning long-term dependencies, because these RNNs were unable to form connections between old and new data when a large information gap existed between them [41]. This resulted in a phenomenon known as vanishing gradient problem, where the error signals vanished during backpropagation, which eventually led to a model breakdown. Hochreiter and Schmidhuber [42] developed LSTM to solve the vanishing gradient problem, but Ger et al. [43] showed that LSTM was still not able to efficiently learn sequences that were very long or continuous. The reason for this failure was that the internal values of the memory cell in LSTM models had grown out of bound from the continuous input stream, despite the fact that the LSTM model was programmed to reset itself when faced with this kind of problem. As a remedy, “forget gates” were introduced to LSTMs, which remove data that were no longer relevant in memory cells, thereby forgetting and resetting information in memory cells at appropriate times [43]. Useful information, on the other hand, was continuously backpropagated, thus, allowing these models to memorize relevant information and recognize patterns in long-term dependencies. The architecture of the LSTM model is shown in Figure 3.
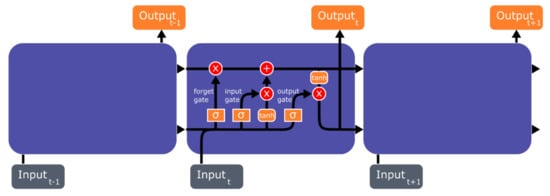
Figure 3.
Basic Long Short-Term Memory (LSTM) model architecture.
However, high computational complexity is the downside of RNN and LSTM, and they need a large memory bandwidth to train [44,45]. As such, hardware designers often experience difficulty when dealing with RNN or LSTM models, because these models occupy a large amount of cloud space, which is not scalable. Therefore, reducing the computational complexity of RNN and LSTM models must be considered for real-time or mobile applications.
4.3. Autoencoders (AEs)
Rumelhart et al. [46] were the first to propose Autoencoders (AEs), a DL technique that is specialized in dimensionality reduction and denoising of data. The key player in an autoencoder operation is the latent (hidden) representation, h, as shown in Figure 4. It plays the role of a bottleneck, which retains only those features that are necessary to reconstruct the input data on the AE output [47]. This latent representation (features) is often used in data classification tasks. This implies that AEs and CNNs have common characteristics and attempt to extract and learn only important features. While CNNs make use of convolutional layers to concentrate the extracted features and improve recognition ability, AEs make use of latent representation (h) to compress the data received from the encoder unit. This step retains salient features and removes irrelevant data. Thus, the operation of AEs includes denoising and reducing the data dimension while extracting features simultaneously. This reduces the computational complexity which in certain classification tasks makes AE models easier to train [48]. However, AEs are also associated with disadvantages, such as poor data compressibility and the inability to train a model effectively for certain tasks. For instance, the latent representation (h) will fail to capture salient features if errors are present in the encoder unit [47]. This is because h cannot get rid of errors; instead, it will compute the average of the input data rather than retain salient features for the decoder unit. The goal of AEs is to reconstruct the input data as shown in Figure 4, hence, the encoder units of AEs have to ensure that there are minimum errors before feeding the input data into the latent representation, h.
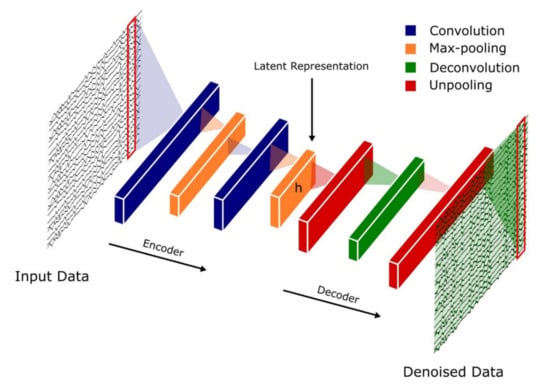
Figure 4.
Basic autoencoder architecture.
4.4. Hybrid Models
Hybrid models are based on either CNN–RNN or CNN–LSTM models. The idea of creating such hybrids is to combine the advantages associated with both CNNs and RNN/LSTMs, in terms of feature extraction and pattern recognition ability in sequential data [49,50]. In these hybrid models, the convolutional layers are at the frontline of the model to extract important features from PSG signals, while RNN or LSTM layers would attempt to recognize patterns in feature maps received from the convolutional layers.
5. Sleep Stages Classification Using DL Models
5.1. Different Stages of Sleep
According to Rechtschaffen and Kales (R and K) [51], humans can experience six discrete stages during sleep: (1) wakefulness (W), (2) rapid eye movement (REM) sleep, and (3) four stages of non-REM (NREM) sleep (S1 to S4) [52]. Based on the sleep electroencephalogram (EEG) characteristics, W occurs when the brain is most active, which is represented by high frequency of alpha rhythms. In the NREM sleep, these alpha rhythms eventually diminish when entering the S1 wherein theta rhythm dominates instead. In the S2, sleep spindles and occasional K-complex waveform will appear. The K-complex waveform usually lasts for approximately 1 to 2 s. The S3 sleep occurs when low frequency delta rhythms appear intermittently and eventually, they dominate in the S4 sleep. Finally, REM sleep usually follows after the S4 sleep. In the REM sleep, theta rhythms resurface again, but unlike in the S1 sleep, theta rhythms are accompanied with EEG flattening [52]. Following the guidelines from American Academy of Sleep Medicine (AASM), the S3 and S4 sleep stages can be merged into one sleep stage S3, because of the similarity in their characteristics [21]. Since the delta rhythms are the slowest EEG waves, S3 and S4 sleep stages are known as Slow Wave Sleep (SWS) or the deep sleep. Thus, most sleep classification studies are based on five: W, S1, S2, S3, and REM sleep stages, instead of six (Figure 5).
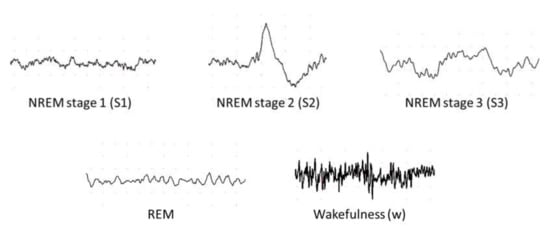
Figure 5.
Examples of electroencephalography (EEG) signals in different sleep stages.
5.2. Sleep Databases
Eight main sleep databases have been used for automated sleep stage classifications. Five of the databases are free to download from PhysioNet [53], namely the Sleep-EDF [54], the expanded Sleep-EDF [54], the St. Vincent’s University Hospital/University College Dublin Sleep Apnea Database (UCD) [53], the Sleep Heart Health Study (SHHS) [55,56], and the Massachusetts Institute of Technology-Beth Israel Hospital (MIT-BIH) [57] database. The ISRUC-Sleep datasets [58] can be downloaded from the official websites. Permission is required to obtain the sleep datasets from the Montreal Archive of Sleep Studies (MASS) [59].
The PSG recordings, in most of the sleep databases, are scored according to R and K rules [51], wherein scoring is done based on wakefulness, NREM sleep and REM sleep. NREM sleep is then subdivided into four stages (S1 to S4). Exceptions are ISRUC and MASS which follow the AASM guideline and partition the recordings into five sleep stages instead of six [21].
5.3. DL Techniques Used in Automatic Sleep Stage Classification
The development of a program diagnostic tool (PDT) for automatic sleep stage classification using DL techniques is shown in Figure 6. First, PSG recordings have to be pre-processed to achieve standardization or normalization. Depending on the requirement and architecture of the proposed DL model, additional steps to convert the PSG recordings into the right input format is required; for example, converting one-dimensional (1D) signals into a two-dimensional (2D) format to train 2D-CNN models. Subsequently, the pre-processed signals are split into training, validation, and testing sets. The training set is used to train the model, the validation set is to fine-tune the model, and the testing set is used to evaluate the model’s performance. A well-trained model can accurately classify PSG recordings into the five sleep stages.
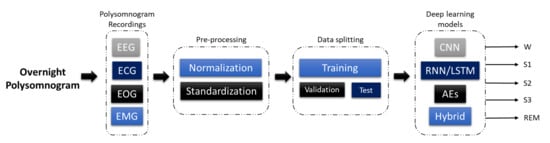
Figure 6.
Programmed diagnostic tool (PDT) block diagram with DL for automated sleep stage classification.
Figure 7 illustrates the number of times each sleep database had been used by studies for automated sleep stage classification using DL techniques, from 2010 to 2020. The DL methods and accuracy obtained from the respective sleep databases are summarized as follows: Sleep-EDF (Table 1), expanded Sleep-EDF (Table 2), MASS (Table 3), MIT-BIH, and SHHS (Table 4), and studies that used the remaining two sleep databases (ISRUC and UCD) and private datasets are listed in Table 5. With the exception of three studies [60,61,62], which classified sleep into four stages, all automated sleep stage classification studies, in Table 1, Table 2, Table 3, Table 4 and Table 5, followed the AASM guidelines [21] and classified sleep into five stages. In studies with sleep databases following the R and K rules [51], (i.e., Sleep-EDF, expanded Sleep-EDF, UCD, SHHS, and MIT-BIH), the S3 and S4 stages were often combined manually before pre-processing the PSG signals.
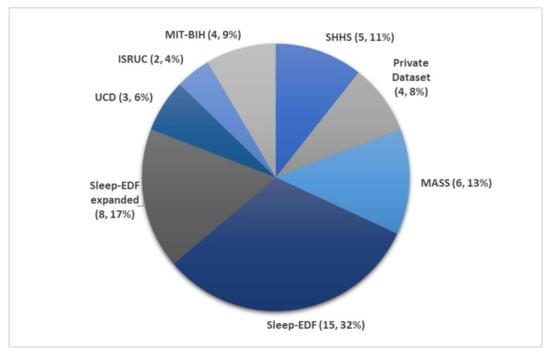
Figure 7.
Pie chart representation of the frequency in which each sleep database was used in automated sleep stage classification studies. The total number of studies was 47, as listed in Table 1, Table 2, Table 3, Table 4 and Table 5. * Summary statistics: using various databases for sleep stage classification.

Table 1.
Summary of automated sleep stage classification approaches with DL applied to PSG recordings in Sleep-EDF dataset.
Table 1.
Summary of automated sleep stage classification approaches with DL applied to PSG recordings in Sleep-EDF dataset.
| Author | Signals | Samples | Approach | Tools/Programming Languages | Accuracy (%) |
|---|---|---|---|---|---|
| Zhu et al. [63] 2020 | EEG | 15,188 | attention CNN | − | 93.7 |
| Qureshi et al. [64] 2019 | EEG | 41,900 | CNN | − | 92.5 |
| Yildirim et al. [65] 2019 | EEG | 15,188 | 1D-CNN | Keras | 90.8 |
| Hsu et al. [66] 2013 | EEG | 2880 | Elman RNN | − | 87.2 |
| Michielli et al. [67] 2019 | EEG | 10,280 | RNN-LSTM | MATLAB | 86.7 |
| Wei et al. [68] 2017 | EEG | − | CNN | − | 84.5 |
| Mousavi et al. [69] 2019 | EEG | 42,308 | CNN-BiRNN | TensorFlow | 84.3 |
| Seo et al. [70] 2020 | EEG | 42,308 | CRNN | PyTorch | 83.9 |
| Zhang et al. [71] 2020 | EEG | − | CNN | − | 83.6 |
| Supratak et al. [72] 2017 | EEG | 41,950 | CNN-BiLSTM | TensorFlow | 82.0 |
| Phan et al. [73] 2019 | EEG | − | Multi-task CNN | TensorFlow | 81.9 |
| Vilamala et al. [74] 2017 | EEG | − | CNN | − | 81.3 |
| Phan et al. [75] 2018 | EEG | − | 1-max CNN | − | 79.8 |
| Phan et al. [76] 2018 | EEG | − | Attentional RNN | − | 79.1 |
| Yildirim et al. [65] 2019 | EOG | 15,188 | 1D-CNN | Keras | 89.8 |
| Yildirim et al. [65] 2019 | EEG + EOG | 15,188 | 1D-CNN | Keras | 91.2 |
| Xu et al. [77] 2020 | PSG signals | − | DNN | − | 86.1 |
| Phan et al. [73] 2019 | EEG + EOG | − | Multi-task CNN | TensorFlow | 82.3 |

Table 2.
Summary of automated sleep stage classification approaches with DL applied to PSG recordings in Expanded Sleep-EDF dataset.
Table 2.
Summary of automated sleep stage classification approaches with DL applied to PSG recordings in Expanded Sleep-EDF dataset.
| Author | Signals | Samples | Approach | Tools/Programming Languages | Accuracy (%) |
|---|---|---|---|---|---|
| Wang et al. [78] 2018 | EEG | − | C-CNN | − | − |
| Wang et al. [78] 2018 | EEG | − | RNN-biLSTM | − | − |
| Fernandez-Blanco et al. [79] 2020 | EEG | − | CNN | − | 92.7 |
| Yildirim et al. [65] 2019 | EEG | 127,512 | 1D-CNN | Keras | 90.5 |
| Jadhav et al. [80] 2020 | EEG | 62,177 | CNN | − | 83.3 |
| Zhu et al. [63] 2020 | EEG | 42,269 | attention CNN | − | 82.8 |
| Mousavi et al. [69] 2019 | EEG | 222,479 | 1D-CNN | TensorFlow | 80.0 |
| Tsinalis et al. [81] 2016 | EEG | − | 2D-CNN | Lasagne + Theano | 74.0 |
| Yildirim et al. [65] 2019 | EOG | 127,512 | 1D-CNN | Keras | 88.8 |
| Yildirim et al. [65] 2019 | EEG + EOG | 127,512 | 1D-CNN | Keras | 91.0 |
| Sokolovsky et al. [82] 2019 | EEG + EOG | − | CNN | TensorFlow + Keras | 81.0 |

Table 3.
Summary of automated sleep stages classification approaches with DL applied to PSG recordings in Montreal Archive of Sleep Studies (MASS) dataset.
Table 3.
Summary of automated sleep stages classification approaches with DL applied to PSG recordings in Montreal Archive of Sleep Studies (MASS) dataset.
| Author | Signals | Samples | Approach | Tools/Programming Languages | Accuracy (%) |
|---|---|---|---|---|---|
| Seo et al. [70] 2020 | EEG | 57,395 | CRNN | PyTorch | 86.5 |
| Supratak et al. [72] 2017 | EEG | 58,600 | CNN-BiLSTM | TensorFlow | 86.2 |
| Phan et al. [73] 2019 | EEG | − | Multi-task CNN | TensorFlow | 78.6 |
| Dong et al. [83] 2018 | EOG F4 | − | MNN RNN-LSTM | Theano | 85.9 |
| Dong et al. [83] 2018 | EOG Fp2 | − | MNN RNN-LSTM | Theano | 83.4 |
| Chambon et al. [84] 2018 | EEG/EOG + EMG | − | 2D-CNN | Keras | − |
| Phan et al. [85] 2019 | EEG + EOG + EMG | − | Hierarchical RNN | TensorFlow | 87.1 |
| Phan et al. [73] 2019 | EEG + EOG + EMG | − | Multi-task CNN | TensorFlow | 83.6 |
| Phan et al. [73] 2019 | EEG + EOG | − | Multi-task CNN | TensorFlow | 82.5 |

Table 4.
Summary of automated sleep stage classification approaches with DL applied to PSG recordings in Sleep Heart Health Study (SHHS) and Massachusetts Institute of Technology-Beth Israel Hospital (MIT-BIH) datasets.
Table 4.
Summary of automated sleep stage classification approaches with DL applied to PSG recordings in Sleep Heart Health Study (SHHS) and Massachusetts Institute of Technology-Beth Israel Hospital (MIT-BIH) datasets.
| Database | Author | Signals | Samples | Approach | Tools/Programming Languages | Accuracy (%) |
|---|---|---|---|---|---|---|
| MIT-BIH | Zhang et al. [86] 2020 | EEG | − | Orthogonal CNN | − | 87.6 |
| Zhang et al. [87] 2018 | EEG | − | CUCNN | MATLAB | 87.2 | |
| SHHS | Sors et al. [88] 2018 | EEG | 5793 | CNN | − | 87.0 |
| Seo et al. [70] 2020 | EEG | 5,421,338 | CRNN | PyTorch | 86.7 | |
| Fernández-Varela et al. [89] 2019 | EEG + EOG + EMG | 1,209,971 | 1D-CNN | − | 78.0 | |
| Zhang et al. [90] 2019 | EEG + EOG + EMG | 5793 | CNN-LSTM | − | − | |
| SHHS | Li et al. [60] 2018 | ECG HRV | 400,547 | CNN | MATLAB | 65.9 |
| MIT-BIH | Li et al. [60] 2018 | ECG HRV | 2829 | CNN | MATLAB | 75.4 |
| Tripathy et al. [61] 2018 | EEG + HRV | 7500 | DNN Autoencoder | MATLAB | 73.7 |

Table 5.
Summary of automated sleep stage classification approaches with DL applied to PSG recordings in ISRUC, Massachusetts General Hospital (MGH), and University College Dublin Sleep Apnea Database (UCD) datasets.
Table 5.
Summary of automated sleep stage classification approaches with DL applied to PSG recordings in ISRUC, Massachusetts General Hospital (MGH), and University College Dublin Sleep Apnea Database (UCD) datasets.
| Database | Author | Signals | Samples | Approach | Tools/Programming Languages | Accuracy (%) |
|---|---|---|---|---|---|---|
| ISRUC | Cui et al. [91] 2018 | EEG | − | CNN | − | 92.2 |
| Yang et al. [92] 2018 | EEG | − | CNN-LSTM | − | − | |
| UCD | Zhang et al. [86] 2020 | EEG | − | Orthogonal CNN | − | 88.4 |
| Zhang et al. [87] 2018 | EEG | − | CUCNN | MATLAB | 87.0 | |
| Yuan et al. [93] 2019 | Multivariate PSG signals | 287,840 | Hybrid CNN | PyTorch | 74.2 | |
| Private datasets | Zhang et al. [71] 2020 | EEG | 264,736 | CNN | − | 96.0 |
| Biswal et al. [94] 2018 | PSG signals | 10,000 | RCNN | PyTorch | 87.5 | |
| Biswal et al. [95] 2017 | EEG | 10,000 | RCNN | TensorFlow | 85.7 | |
| Class = 4 | ||||||
| Radha et al. [62] 2019 | ECG HRV | 541,214 | LSTM | − | 77.0 |
Figure 8 shows the number of times PSG recordings such as EEG, EOG, EMG, and ECG signals were used for sleep stage classification studies. It is not surprising that EEG signal was the most popular input for DL models. The characteristic waves and description of each sleep stages are often based on EEG characteristics (i.e., alpha waves, theta waves, delta waves, etc.); Figure 5.
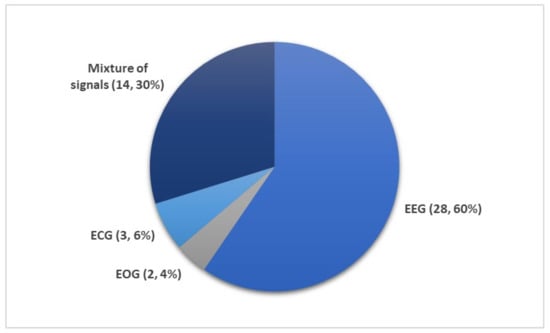
Figure 8.
Different subsets of PSG recordings used to train DL models for automated sleep stage classification as listed in Table 1, Table 2, Table 3, Table 4 and Table 5. Of the 36 studies, the mixture of signals (electrooculogram (EOG), electromyogram (EMG), and electroencephalography (EEG)) was employed 14 times while EEG signals were used 28 times. Only a small fraction (five studies) employed ECG or EOG time series. * Summary statistics: using EEG versus EEG + additional signals.
Nonetheless, other signals within the PSG recordings are indispensable, because they provide additional information on biological aspects of sleep that may not be manifested in EEG recordings. Since REM sleep is characterized by the movement of eyes and loss in muscle tone of the body core, EOG, and EMG signals may provide key information to separate the REM sleep stage from the other stages. It was shown that some of the REM sleep stages could be overlooked in single-channel EEG input [27]. Therefore, a combination of signals, comprising of EOG, EMG, and EEG, are second in terms of frequency of use after single-channel EEG inputs (Figure 8).
Although ECG is an important sleep parameter [96], it is not common to use raw ECG signals as a direct input for DL models. As seen in Table 4, heart rate variability (HRV) parameters derived from ECG signals, were used to train the DL models instead. There are only three studies that employed HRV parameters, and these studies classified sleep into four stages instead of five: wakefulness (W), light sleep (S1 and S2), deep sleep (S3 and S4), and REM sleep. Li et al. [60] proposed a 3-layer CNN model. They used a cardiorespiratory coupling (CRC) spectrogram, which was derived from ECG and HRV. Besides alternations in physiological signals, there are other changes in body system changes in some individuals such as cardiovascular [97], respiratory [98], or blood flow in the brain [99]. Hence, the CRC picks up the cardiovascular and respiratory changes. Their model achieved an overall accuracy of 65.9% and 75.4% for SHHS and MIT-BIH respectively, as seen in Table 4. Tripathy et al. [61] combined EEG and HRV features as input to an AE model. During testing, the model achieved an overall accuracy of 73.7%. Radha et al. [62] published the only study that was based on ECG signals from a private dataset that was collected as part of the European Union SIESTA project [100] as shown in Table 5. Likewise, they converted ECG signals into HRV and used the HRV features to train an LSTM model, which achieved an accuracy of 77.0%.
6. Discussion
Even though CNNs are primarily used in image classification, they can also be successfully applied to 1D PSG recordings. Most of the automated sleep stage classification studies rely on the CNN approach (Figure 9a). However, in order to convert 1D signals to 2D images, the input signals need to be reshaped so that the 2D convolutional layer is able to read the data. There are various 1D to 2D transformation methods, such as spectrogram [101], time-frequency representations, which can be established via Hilbert–Huang transform [86], or bispectrum algorithms [102]. To date, there are eight studies that included 2D-convolutional layers in their models [60,74,81,84,86,90,93,95]. However, the process of converting 1D signals to 2D signal representations should be carried out with caution due to potential loss of useful information during the conversion step [103].
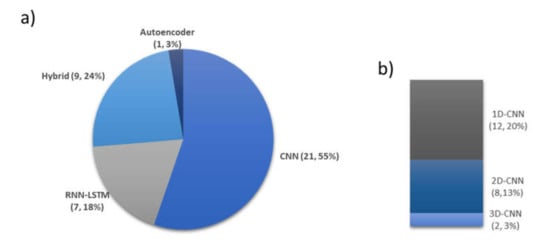
Figure 9.
Selection of various DL techniques for automated sleep stage classification based on 36 studies. (a) Number of times and percentage of DL models, (b) Various CNN-based models. * Summary statistics: using autoencoders versus hybrid versus LSTM versus CNN models (one-dimensional (1D), 2D, 3D).
One-dimensional (1D)-CNNs were specifically designed to process 1D signals [104]. Unlike traditional 2D-CNNs, which require the input data to be in a matrix format, and 1D-CNNs can run with a simple array, hence, significantly reducing the computational complexity. In addition, 2D-CNNs require deeper model architecture to learn 1D signals. In contrast, 1D-CNN can easily learn 1D signals with shallow model architecture. This means that training of 1D-CNN models with 1D signals is simpler, easier, faster and, therefore, more efficient. This also highlights the 1D-CNN models’ compatibility with near real-time processing and deployment in mobile applications, which can potentially be used to track and recognize sleep patterns at home [104]. The popularity of 1D-CNNs for the analysis of 1D PSG signals is demonstrated in this review. Almost all of the studies that proposed CNN-based models had employed 1D convolutional layers, as seen in Figure 9b. Furthermore, studies that proposed 1D-CNN models had higher performance compared to those with 2D-CNN. The highest accuracy score obtained by a 1D-CNN model was 96% [71]. Conversely, none of the eight studies that proposed 2D-CNN models surpassed an accuracy score of 90%.
Two studies included a 3D convolutional layer in their proposed CNN models. Phan et al. [73] used 3D filters to process a combination of signals, namely EEG, EOG, and EMG. Similar to signal pre-processing for a 2D-CNN, these three signals were converted to a 2D time-frequency image before arranging them as a 3D input. As a result, a higher accuracy was obtained with a combination of input signals when compared to a single channel input. Jadhav et al. [80] converted their EEG input into 2D-Continuous Wavelet Transform (CWT) images. The CWT images came with three main colors, red–green–blue (RGB), which provided the third-dimensional input. Hence, their convolutional layer in their proposed model had 3D filters to read the CWT images in terms of width, height, and color of the image.
From Table 4, it is evident that only Tripathy et al. [61] proposed AE models for automated sleep stage classification. In the study by Zhang et al. [87], AE was used solely as a dimensionality reduction tool to pre-process the EEG time-frequency distribution.
6.1. Proposed CNN-Based Models
From Figure 10, it can be observed that the performance of the CNN-based models had improved over the years. To date, the model by Zhang et al. [71] achieved the best overall accuracy (96%) for sleep stage classification. However, this accuracy was achieved using a private clinical dataset. When they evaluated the performance of their model using the Sleep-EDF dataset, an overall accuracy score of 86.4% was achieved. This was lower than the accuracy obtained by Zhu et al. [63] (93.7%) who used single channel EEG signals from the same database. The unique feature of the model proposed by Zhu et al. was the attention mechanism that they incorporated into the CNN’s learning framework. This attention mechanism improved the feature extraction performance of the model through intra- and inter-epoch feature learning. However, the same model achieved a lower accuracy of 82.8% when it was tested on EEG signals from the expanded Sleep-EDF database. Another unique CNN model was proposed by Cui et al. [91] wherein fine-grained methods were used to assist the CNN model to find the best time segment in the EEG signals. Fine-grained methods construct time series from the EEG signals. Basically, if the time window in fine-grained method is set to 3, every 3 time-steps along the EEG signals will be combined together as one-time segment, hence reducing the complexity of EEG signals. The layers in their proposed CNN were shallow-7 layers, including 2 convolutional layers. Yet, they were able to achieve a high overall accuracy of 92%.
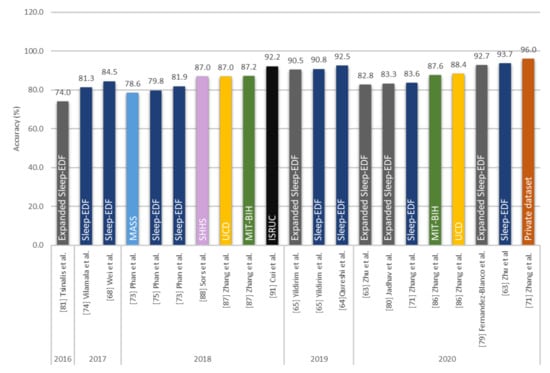
Figure 10.
Performance of CNN-based models analyzing only EEG signals. Sleep datasets are presented in different colors. * Summary statistics: Various sleep databases used to develop CNN models for sleep stage detection.
A more versatile and consistent CNN model was proposed by Yildirim et al. [65], which was a 19-layer 1D-CNN model with 10 convolutional layers. It achieved an accuracy higher than 90% in both Sleep-EDF dataset and its expanded version, as seen in Figure 10. The peak accuracy was achieved (91.2%) when a mixture of PSG signals was used as input (EEG and EOG), but when single channel EOG signals were used, the accuracy of the model decreased to below 90% (88.8% and 89.8%), (Figure 11).
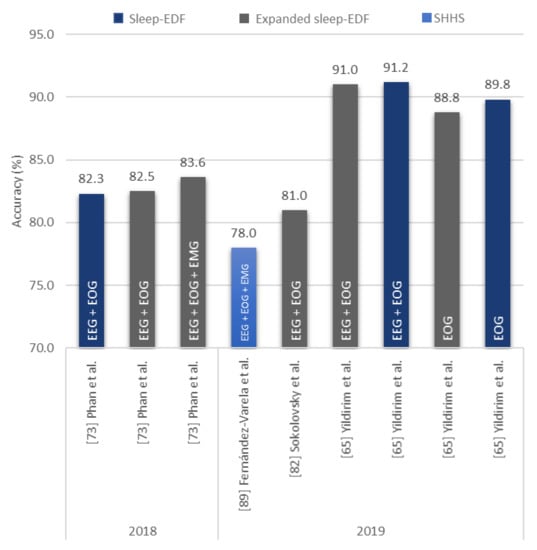
Figure 11.
Sleep stage classification accuracy of CNN-based models based on stand-alone EOG signals or a mixture of EEG, EOG, and/or EMG signals. Various sleep datasets are represented by different colors and the type of signals described in the bar chart. * Summary statistics: Using EEG+ other signals/EOG in 2018–2019.
Both Zhu et al. [63] and Cui et al. [91] showed that CNN models with a small number of layers can achieve a high classification performance of sleep stages by improving the feature extraction ability through additional tools, such as attention mechanism or fine-graining. On the other hand, Yildirim et al. [65] showed that a deeper CNN model can achieve high accuracy classifications across different inputs of PSG recordings (EEG, EOG, EEG + EOG).
6.2. Proposed RNN/LSTM-Based Models
Contrary to CNN models, very little automated sleep stage classification studies were done using RNN/LSTM models. The best performance was observed in a study by Hsu et al. [66] back in 2013, wherein a 4-layer RNN model was used. They had adopted the structure of an Elman network and had successfully classified various sleep stages with an overall accuracy score of 87.2%, as seen in Figure 12. A similar accuracy of 86.7% was achieved by Michielli et al. [67], where a cascaded RNN network was proposed, with 2 LSTM units. Two other studies explored a mixture of signals to train RNN-based models: Dong et al. [83] proposed a mixed neural network where a RNN model was substituted for a multi-layer perception and an LSTM model was exchanged for an RNN model. The proposed final model achieved accuracies of 85.9% and 83.4% using F4-EOG and Fp2-EOG inputs, respectively. Both F4 and Fp2 were single channel EEG signals recorded at different electrode placements. Hence, F4-EOG and Fp2-EOG inputs were considered a mixture of signals (EEG + EOG). Subsequently, Phan et al. [85] proposed an end-to-end hierarchical RNN model, known as SeqSleepNet, which consisted of an attention-based recurrent model and filter bank layers. Short-time Fourier transform was used to convert multiple PSG recordings (EEG, EOG, and EMG) into power spectra signals which were then fed to train the proposed model. The model achieved a high accuracy score of 87.1%, which was the highest amongst the RNN/LSTM-based models.
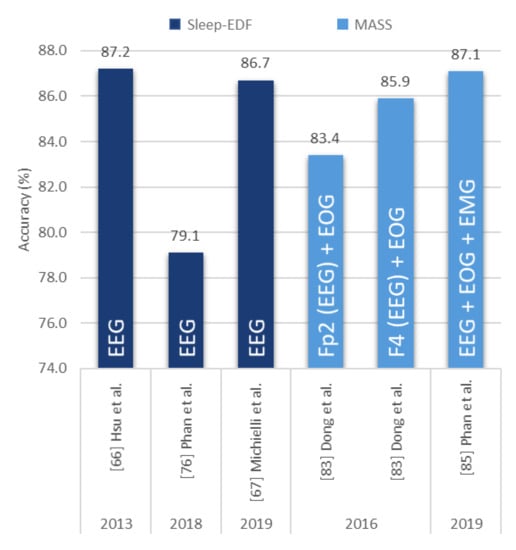
Figure 12.
Sleep stage classification accuracy of RNN/LSTM-based models based on stand-alone EEG signals or a mixture of EEG, EOG, and ECG signals. Various sleep datasets are represented by different colors and the type of signals described in the bar chart. * Summary statistics: Using RNN versus LSTM models for EEG/EEG + other signals from 2013–2019.
6.3. Proposed Hybrid Models
There are limited studies on the employment of hybrid models. The best performing hybrid model for EEG signals was proposed by Seo et al. [70]. Their IITNet model consisted of CNN layers and two bidirectional LSTM layers. The CNN layers were responsible for extracting representative features in each epoch and producing sequential feature maps, which were analyzed by the bidirectional LSTM layers to capture temporal sleep stage information [72]. Figure 13 shows that this model achieved an overall accuracy score of 86.7% (SHHS database), compared to 83.9% when applied to Sleep-EDF database. On the other hand, Mousavi et al. [69] proposed SleepEEGNet, a CNN-RNN model with bidirectional RNN units. The difference between this model and Seo et al.’s model [70] was in the architecture of bidirectional RNN units which resembled AEs in the former model. Comparing within the same Sleep-EDF databases, this model achieved a higher overall accuracy score of 84.3%, as seen in Figure 13. However, their proposed model achieved a lower accuracy score of 80.0%, with EEG signals from the Expanded Sleep-EDF database.
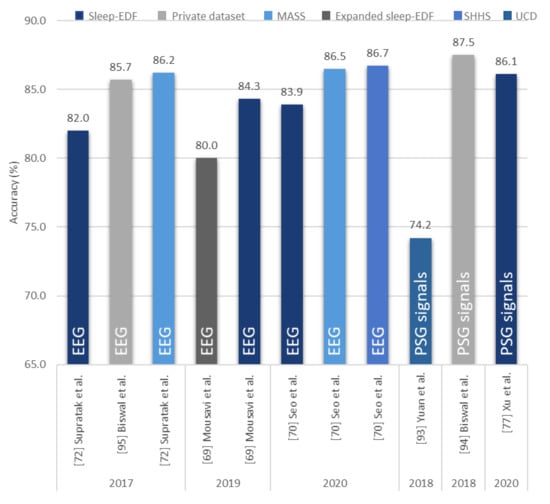
Figure 13.
Performance of proposed hybrid models using EEG signals or a mixture of signals. Various sleep datasets are represented by different colors and the type of signals are described in the bar chart. * Summary statistics: different datasets used to build hybrid models using EEG/PSG signals. * The accuracy scores in Figure 9, Figure 10, Figure 11 and Figure 12 are based on AASM guidelines and pertain to the five class classification [21].
When a mixture of PSG signals were taken into consideration, the CNN-RNN based model proposed by Biswal et al. [94] outperformed all other models. However, in their study they used a private sleep database with recordings from the Massachusetts General Hospital (MGH) sleep laboratory to train and test the model. They also trained their model with the MGH dataset and then tested it on the SHHS dataset. With that setup, they obtained an overall accuracy of 77.7%. This score is lower than that obtained by the CNN-based model proposed by Fernández-Varela et al. [89] who employed a mixture of signals from the SHHS database to train and test their model.
The number of studies employing DL methods for automated sleep stage classification has increased in the past few years (Figure 14), most of the studies incorporated CNN models. The CNN models became popular after the publication by Krizhevsky et al. [34] in 2015. However, the number of studies relying on this architecture started to decline after 2018. This implies that CNN-based approaches have perhaps reached their peak ability and peak performance in classifying sleep stages. This decline is also likely due to the fact that best CNN based architectures are now less competitive when compared to the other DL models. Conversely, the research on RNN/LSTM and hybrid models remained stagnant from 2018 to 2019, and no further studies were carried out using AE models after 2018. This suggests that more attention should be paid to improve the performances of these models, particularly to RNN/LSTM, and hybrid models in automated sleep stage classifications.
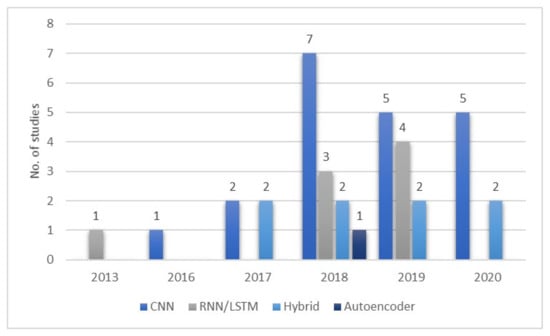
Figure 14.
Number of studies published between 2013 and 2020 describing implementation of various DL models for PSG recording analysis. * Summary statistics: year of the analyses for various deep learning models used in sleep classification.
When assessing polysomnographic recordings, clinical experts rely on a combination of EEG, EOG, and EMG signals before they determine the sleep stage for each sleep phase [63]. In order to be on par with the clinical experts, an ideal DL model should effectively classify sleep stages based on a mixture of signals. At present, the majority of automated sleep stage classification studies demonstrate a high performance with single-EEG channel, but only a small fraction (25%) evaluated the performance of their approaches on a mixture of signals (Figure 8).
In summary, research on a mixture of PSG signals should be the main focus in the automated detection of sleep stages. The RNN/LSTM and hybrid models have yet to reach their peak performance in classifying sleep stages. Further research on these models to evaluate their performance across different databases could decrease the bias in these methods and help to identify architectures capable of processing mixed PSG signals. A model with optimal architecture could be employed in various applications and platforms, such as mobile, point-of-care monitoring devices.
This review underscores the advantages (key points discussed) as follows:
- Numerous studies (15 from Figure 10) employed CNN models with EEG signals, and that CNN models are effective in recognizing characteristic features of sleep EEG.
- One-dimensional (1D)-CNN models were used more often than 2D- and 3D-CNN models. From Figure 9b, 12, 8, and 2 1D, 2D, and 3D models were used, respectively.
- Most studies (60% from Figure 8) used EEG signals and achieved high classification accuracy.
- EEG signals were mainly used in studies that explored a mixture of PSG signals. In other words, EEG could be a reference signal when considering mixture of PSG signals to train and evaluate newly proposed models.
The limitations of this review are as follows:
- It is difficult to compare various models and identify the best performing approach, because the majority of studies used data from only one sleep database to train and test the model.
- There is a lack of studies that utilized other PSG recordings, such as EOG, EMG, or ECG signals. Studies that used these PSG recordings also did not perform equally well as those that used only EEG signals. Hence, this limits the implementation of these PSG recordings in real world applications for automated sleep stages classification.
7. Future Work
We anticipate that more public databases with large data records will become available to conduct studies on sleep stage classification and the development of more accurate approaches. When constructing a DL model, the ability to implement it in cloud-based processing systems should be considered, (Figure 15). Extracted EEG or PSG signals could then be sent to the model, for processing in the cloud and results of the analysis returned to the clinician. The analysis could be performed on-line or off-line with the goal to reduce expert workload who otherwise would have to review hours of recordings manually. Subsequently, verified results could be sent to the patient’s mobile phone. With the DL model deployed in the cloud, any device with access to internet, such as mobile applications, could use it anytime and anywhere. For example, Patanaik et al. [105] developed a cloud-based framework that could classify sleep stages using real-time data with 90% accuracy. Their model performed better than expert scorers (82%). This type of framework is suitable for wearable sleep devices, such as the Kokoon and Dreem headband that reads real-time data [105].
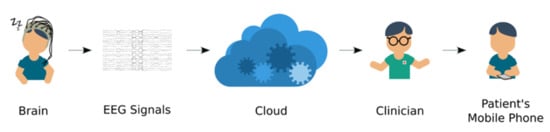
Figure 15.
Block diagram of cloud-based sleep stage classification system using EEG signals.
Furthermore, future studies should focus on using various EEG signals to detect sleep stages. Currently, the most common electrode placing to record EEG signals are Fpz-Cz, Pz-Oz, C4-A1, and C3-A2.
Another area of development and research is the microstructure of sleep, such as Cyclic Alternating Pattern (CAP) and arousal. Their duration is shorter than a half-minute epoch, hence, often undetected by humans in conventional sleep stage scoring [6]. Since quantifying microstructures by humans has poor reproducibility and is prone to errors, it would be desirable to develop automated approaches to address this deficiency [6]. However, CAP sleep database (CAPSLPDB) is currently the only database that provide PSG recordings for development of tools for CAP detection [106]. Hence, more public databases designed for microstructure of sleep assessment should be available to allow advancement of PDT in the detection of CAP.
8. Conclusions
Sleep disorders are a pressing global issue and the most dangerous sleep disorder is the obstructive sleep apnea, which can lead to cardiovascular diseases, if left untreated. Hence, efficient and accurate diagnostic tools are required for early interventions. In this work, we reviewed 36 studies that employed programmed diagnostic tools with the DL models as the backbone, analyzing overnight polysomnogram recordings to classify sleep stages. Presently, CNN models can offer higher performance in classifying sleep stages, especially with EEG signals. Hence, they are consistently and favorably used by researchers to classify sleep stages as compared to the other machine learning models and physiological signals. Moreover, employing 1D-CNN models is advantageous, because they yield high classification results on EEG signals. However, EEG signals alone may not be sufficient to achieve robust classifications. To achieve robustness and high accuracy one could develop a system that takes advantage of an automated processing and human experts in the interpretation of EEG, EOG, and EMG signals together when classifying sleep stages. Therefore, in this review, we highlighted that future studies should focus on classifying sleep stages using all or a combination of these signals. Furthermore, other DL models, such as RNN/LSTM and hybrid models, should also be explored as their full potential has yet to be realized. Future studies could focus on the compatibility and applicability of the DL models in mobile and real time applications. Lastly, more research in developing DL models to detect sleep microstructures is required, as these are often undetected in sleep stage scoring.
Author Contributions
All authors contributed to this article. The idea for the article was provided by H.W.L. and U.R.A.; H.W.L. drafted the paper. S.L.O. provided some of the diagrams. C.P.O., O.F., A.G., J.V., and U.R.A. edited the paper and provided suggestions to improve the quality of the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Laposky, A.; Bass, J.; Kohsaka, A.; Turek, F.W. Sleep and circadian rhythms: Key components in the regulation of energy metabolism. FEBS Lett. 2007, 582, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.W.; Duffy, J.F. Sleep, sleep disorders, and sexual dysfunction. World J. Men’s Health 2019, 37, 261–275. [Google Scholar] [CrossRef] [PubMed]
- Institute of Medicine (US), Committee on Sleep Medicine and Research. Sleep Disorders and Sleep Deprivation: An Unmet Public Health Problem; Colten, H.R., Altevogt, B.M., Eds.; National Academies Press: Washington, DC, USA, 2006. [Google Scholar]
- Stranges, S.; Tigbe, W.; Gómez-Olivé, F.X.; Thorogood, M.; Kandala, N.-B. Sleep problems: An emerging global epidemic? Findings from the INDEPTH WHO-SAGE study among more than 40,000 older adults from 8 countries across Africa and Asia. Sleep 2012, 35, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Schulz, H. Rethinking sleep analysis. J. Clin. Sleep Med. 2008, 4, 99–103. [Google Scholar] [CrossRef]
- Spriggs, W.H. Essentials of Polysomnography; Jones & Bartlett Learning: Burlington, MA, USA, 2014. [Google Scholar]
- Silber, M.H.; Ancoli-Israel, S.; Bonnet, M.H.; Chokroverty, S.; Grigg-Damberger, M.M.; Hirshkowitz, M.; Kapen, S.; A Keenan, S.; Kryger, M.H.; Penzel, T.; et al. The visual scoring of sleep in adults. J. Clin. Sleep Med. 2007, 3, 121–131. [Google Scholar] [CrossRef]
- Corral, J.; Pepin, J.-L.; Barbé, F. Ambulatory monitoring in the diagnosis and management of obstructive sleep apnoea syndrome. Eur. Respir. Rev. 2013, 22, 312–324. [Google Scholar] [CrossRef]
- Jung, R.; Kuhlo, W. Neurophysiological studies of abnormal night sleep and the Pickwickian syndrome. Prog. Brain Res. 1965, 18, 140–159. [Google Scholar] [CrossRef]
- Bahammam, A. Obstructive sleep apnea: From simple upper airway obstruction to systemic inflammation. Ann. Saudi Med. 2011, 31, 1–2. [Google Scholar] [CrossRef][Green Version]
- Marshall, N.S.; Wong, K.K.H.; Liu, P.Y.; Cullen, S.R.J.; Knuiman, M.; Grunstein, R.R. Sleep apnea as an independent risk factor for all-cause mortality: The Busselton health study. Sleep 2008, 31, 1079–1085. [Google Scholar] [CrossRef]
- Hirotsu, C.; Tufik, S.; Andersen, M.L. Interactions between sleep, stress, and metabolism: From physiological to pathological conditions. Sleep Sci. 2015, 8, 143–152. [Google Scholar] [CrossRef]
- Schilling, C.; Schredl, M.; Strobl, P.; Deuschle, M. Restless legs syndrome: Evidence for nocturnal hypothalamic-pituitary-adrenal system activation. Mov. Disord. 2010, 25, 1047–1052. [Google Scholar] [CrossRef] [PubMed]
- Hungin, A.P.S.; Close, H. Sleep disturbances and health problems: Sleep matters. Br. J. Gen. Pract. 2010, 60, 319–320. [Google Scholar] [CrossRef]
- Hudgel, D.W. The role of upper airway anatomy and physiology in obstructive sleep. Clin. Chest Med. 1992, 13, 383–398. [Google Scholar] [PubMed]
- Shahar, E.; Whitney, C.W.; Redline, S.; Lee, E.T.; Newman, A.B.; Nieto, F.J.; O’Connor, G.; Boland, L.L.; Schwartz, J.E.; Samet, J.M. Sleep-disordered Breathing and Cardiovascular Disease. Am. J. Respir. Crit. Care Med. 2001, 163, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Mohsenin, V. Obstructive sleep apnea and hypertension: A critical review. Curr. Hypertens. Rep. 2014, 16, 482. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, J.S.; Patel, S.R. Obstructive sleep apnea. Ann. Intern. Med. 2014, 161. [Google Scholar] [CrossRef]
- Williamson, A.; Lombardi, D.A.; Folkard, S.; Stutts, J.; Courtney, T.K.; Connor, J.L. The link between fatigue and safety. Accid. Anal. Prev. 2011, 43, 498–515. [Google Scholar] [CrossRef]
- Léger, D.; Guilleminault, C.; Bader, G.; Lévy, E.; Paillard, M. Medical and socio-professional impact of insomnia. Sleep 2002, 25, 625–629. [Google Scholar] [CrossRef]
- Iber, C.; Ancoli-Israel, S.; Chesson, A.L.; Quan, S.F. The AASM Manual for the Scoring of Sleep and Associated Events: Rules, Terminology and Technical Specification; American Academy of Sleep Medicine: Darien, IL, USA, 2007. [Google Scholar]
- Svetnik, V.; Ma, J.; Soper, K.A.; Doran, S.; Renger, J.J.; Deacon, S.; Koblan, K.S. Evaluation of automated and semi-automated scoring of polysomnographic recordings from a clinical trial using zolpidem in the treatment of insomnia. Sleep 2007, 30, 1562–1574. [Google Scholar] [CrossRef][Green Version]
- Pittman, M.S.D.; Macdonald, R.M.M.; Fogel, R.B.; Malhotra, A.; Todros, K.; Levy, B.; Geva, D.A.B.; White, D.P. Assessment of automated scoring of polysomnographic recordings in a population with suspected sleep-disordered breathing. Sleep 2004, 27, 1394–1403. [Google Scholar] [CrossRef]
- Anderer, P.; Gruber, G.; Parapatics, S.; Woertz, M.; Miazhynskaia, T.; Klösch, G.; Saletu, B.; Zeitlhofer, J.; Barbanoj, M.J.; Danker-Hopfe, H.; et al. An E-health solution for automatic sleep classification according to Rechtschaffen and Kales: Validation study of the Somnolyzer 24 × 7 utilizing the siesta database. Neuropsychobiology 2005, 51, 115–133. [Google Scholar] [CrossRef] [PubMed]
- Acharya, U.R.; Bhat, S.; Faust, O.; Adeli, H.; Chua, E.C.-P.; Lim, W.J.E.; Koh, J.E.W. Nonlinear dynamics measures for automated EEG-based sleep stage detection. Eur. Neurol. 2015, 74, 268–287. [Google Scholar] [CrossRef] [PubMed]
- Mirza, B.; Wang, W.; Wang, J.; Choi, H.; Chung, N.C.; Ping, P. Machine learning and integrative analysis of biomedical big data. Genes 2019, 10, 87. [Google Scholar] [CrossRef] [PubMed]
- Faust, O.; Razaghi, H.; Barika, R.; Ciaccio, E.J.; Acharya, U.R. A review of automated sleep stage scoring based on physiological signals for the new millennia. Comput. Methods Programs Biomed. 2019, 176, 81–91. [Google Scholar] [CrossRef]
- Shoeibi, A.; Ghassemi, N.; Khodatars, M.; Jafari, M.; Hussain, S.; Alizadehsani, R.; Moridian, P.; Khosravi, A.; Hosseini-Nejad, H.; Rouhani, M.; et al. Epileptic Seizure Detection Using Deep Learning Techniques: A Review. arXiv 2020, arXiv:2007.01276. [Google Scholar]
- Faust, O.; Hagiwara, Y.; Hong, T.J.; Lih, O.S.; Acharya, U.R. Deep learning for healthcare applications based on physiological signals: A review. Comput. Methods Programs Biomed. 2018, 161, 1–13. [Google Scholar] [CrossRef]
- Silva, D.B.; Cruz, P.P.; Molina, A.; Molina, A.M. Are the long–short term memory and convolution neural networks really based on biological systems? ICT Express 2018, 4, 100–106. [Google Scholar] [CrossRef]
- Hubel, D.H.; Wiesel, T.N. Receptive fields, binocular interaction and functional architecture in the cat’s visual cortex. J. Physiol. 1962, 160, 106–154. [Google Scholar] [CrossRef]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef]
- Tabian, I.; Fu, H.; Khodaei, Z.S. A convolutional neural network for impact detection and characterization of complex composite structures. Sensors 2019, 19, 4933. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Goehring, T.; Keshavarzi, M.; Carlyon, R.P.; Moore, B.C.J. Using recurrent neural networks to improve the perception of speech in non-stationary noise by people with cochlear implants. J. Acoust. Soc. Am. 2019, 146, 705. [Google Scholar] [CrossRef] [PubMed]
- Coto-Jiménez, M. Improving post-filtering of artificial speech using pre-trained LSTM neural networks. Biomimetics 2019, 4, 39. [Google Scholar] [CrossRef] [PubMed]
- Lyu, C.; Chen, B.; Ren, Y.; Ji, D. Long short-term memory RNN for biomedical named entity recognition. BMC Bioinform. 2017, 18, 462. [Google Scholar] [CrossRef] [PubMed]
- Graves, A.; Liwicki, M.; Fernández, S.; Bertolami, R.; Bunke, H.; Schmidhuber, J. A Novel connectionist system for unconstrained handwriting recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 31, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Sharma, A.; Tsunoda, T. Brain wave classification using long short-term memory network based OPTICAL predictor. Sci. Rep. 2019, 9, 1–13. [Google Scholar] [CrossRef]
- Kim, B.-H.; Pyun, J.-Y. ECG identification for personal authentication using LSTM-based deep recurrent neural networks. Sensors 2020, 20, 3069. [Google Scholar] [CrossRef]
- Yu, Y.; Si, X.; Hu, C.; Zhang, J.-X. A Review of recurrent neural networks: LSTM cells and network architectures. Neural Comput. 2019, 31, 1235–1270. [Google Scholar] [CrossRef]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Gers, F.A.; Schmidhuber, J.; Cummins, F. Learning to forget: Continual prediction with LSTM. In Proceedings of the 9th International Conference on Artificial Neural Networks—ICANN’99, Edinburgh, UK, 7–10 September 1999. [Google Scholar]
- Masuko, T. Computational cost reduction of long short-term memory based on simultaneous compression of input and hidden state. In Proceedings of the 2017 IEEE Automatic Speech Recognition and Understanding Workshop, Okinawa, Japan, 16–20 December 2017; pp. 126–133. [Google Scholar] [CrossRef]
- Dash, S.; Acharya, B.R.; Mittal, M.; Abraham, A.; Kelemen, A. (Eds.) Deep Learning Techniques for Biomedical and Health Informatics; Springer International Publishing: Cham, Switzerland, 2020. [Google Scholar]
- Rumelhart, D.E.; McClelland, J.L. Learning internal representations by error propagation. In Parallel Distributed Processing: Explorations in the Microstructure of Cognition: Foundations; MIT Press: Cambridge, MA, USA, 1987; pp. 318–362. [Google Scholar]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep learning for computer vision: A brief review. Comput. Intell. Neurosci. 2018, 2018, 1–13. [Google Scholar] [CrossRef]
- Testolin, A.; Diamant, R. Combining denoising autoencoders and dynamic programming for acoustic detection and tracking of underwater moving targets. Sensors 2020, 20, 2945. [Google Scholar] [CrossRef] [PubMed]
- Trabelsi, A.; Chaabane, M.; Ben-Hur, A. Comprehensive evaluation of deep learning architectures for prediction of DNA/RNA sequence binding specificities. Bioinformatics 2019, 35, i269–i277. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Liao, B.; Xu, X.; Yang, J. A hybrid deep learning model for predicting protein hydroxylation sites. Int. J. Mol. Sci. 2018, 19, 2817. [Google Scholar] [CrossRef] [PubMed]
- Hori, T.; Sugita, Y.; Koga, E.; Shirakawa, S.; Inoue, K.; Uchida, S.; Kuwahara, H.; Kousaka, M.; Kobayashi, T.; Tsuji, Y.; et al. Proposed sments and amendments to ‘A Manual of Standardized Terminology, Techniques and Scoring System for Sleep Stages of Human Subjects’, the Rechtschaffen & Kales (1968) standard. Psychiatry Clin. Neurosci. 2001, 55, 305–310. [Google Scholar] [CrossRef]
- Carley, D.W.; Farabi, S.S. Physiology of sleep. Diabetes Spectr. 2016, 29, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.-K.; Stanley, H.E. PhysioBank, PhysioToolkit, and PhysioNet. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef]
- Kemp, B.; Zwinderman, A.; Tuk, B.; Kamphuisen, H.; Oberye, J. Analysis of a sleep-dependent neuronal feedback loop: The slow-wave microcontinuity of the EEG. IEEE Trans. Biomed. Eng. 2000, 47, 1185–1194. [Google Scholar] [CrossRef]
- Zhang, G.-Q.; Cui, L.; Mueller, R.; Tao, S.; Kim, M.; Rueschman, M.; Mariani, S.; Mobley, D.R.; Redline, S. The national sleep research resource: Towards a sleep data commons. J. Am. Med. Inform. Assoc. 2018, 25, 1351–1358. [Google Scholar] [CrossRef]
- Quan, S.F.; Howard, B.V.; Iber, C.; Kiley, J.P.; Nieto, F.J.; O’Connor, G.; Rapoport, D.M.; Redline, S.; Robbins, J.; Samet, J.M.; et al. The sleep heart health study: Design, rationale, and methods. Sleep 1997, 20, 1077–1085. [Google Scholar] [CrossRef]
- Ichimaru, Y.; Moody, G. Development of the polysomnographic database on CD-ROM. Psychiatry Clin. Neurosci. 1999, 53, 175–177. [Google Scholar] [CrossRef]
- Khalighi, S.; Sousa, T.; Santos, J.M.; Nunes, U. ISRUC-Sleep: A comprehensive public dataset for sleep researchers. Comput. Methods Programs Biomed. 2016, 124, 180–192. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, C.; Gosselin, N.; Carrier, J.; Nielsen, T. Montreal archive of sleep studies: An open-access resource for instrument benchmarking and exploratory research. J. Sleep Res. 2014, 23, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, Q.C.; Liu, C.; Shashikumar, S.P.; Nemati, S.; Clifford, G.D. Deep learning in the cross-time frequency domain for sleep staging from a single-lead electrocardiogram. Physiol. Meas. 2018, 39, 124005. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, R.; Acharya, U.R. Use of features from RR-time series and EEG signals for automated classification of sleep stages in deep neural network framework. Biocybern. Biomed. Eng. 2018, 38, 890–902. [Google Scholar] [CrossRef]
- Radha, M.; Fonseca, P.; Moreau, A.; Ross, M.; Cerny, A.; Anderer, P.; Long, X.; Aarts, R.M. Sleep stage classification from heart-rate variability using long short-term memory neural networks. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Zhu, T.; Luo, W.; Yu, F. Convolution-and attention-based neural network for automated sleep stage classification. Int. J. Environ. Res. Public Health 2020, 17, 4152. [Google Scholar] [CrossRef]
- Qureshi, S.; Karrila, S.; Vanichayobon, S. GACNN SleepTuneNet: A genetic algorithm designing the convolutional neuralnetwork architecture for optimal classification of sleep stages from a single EEG channel. Turk. J. Electr. Eng. Comput. Sci. 2019, 27, 4203–4219. [Google Scholar] [CrossRef]
- Yıldırım, Ö.; Baloglu, U.B.; Acharya, U.R. A deep learning model for automated sleep stages classification using PSG signals. Int. J. Environ. Res. Public Health 2019, 16, 599. [Google Scholar] [CrossRef]
- Hsu, Y.-L.; Yang, Y.-T.; Wang, J.-S.; Hsu, C.-Y. Automatic sleep stage recurrent neural classifier using energy features of EEG signals. Neurocomputing 2013, 104, 105–114. [Google Scholar] [CrossRef]
- Michielli, N.; Acharya, U.R.; Molinari, F. Cascaded LSTM recurrent neural network for automated sleep stage classification using single-channel EEG signals. Comput. Biol. Med. 2019, 106, 71–81. [Google Scholar] [CrossRef]
- Wei, L.; Lin, Y.; Wang, J.; Ma, Y. Time-Frequency Convolutional Neural Network for Automatic Sleep Stage Classification Based on Single-Channel EEG. In Proceedings of the 2017 IEEE 29th International Conference on Tools with Artificial Intelligence, Boston, MA, USA, 6–8 November 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 88–95. [Google Scholar]
- Mousavi, S.; Afghah, F.; Acharya, U.R. SleepEEGNet: Automated sleep stage scoring with sequence to sequence deep learning approach. PLoS ONE 2019, 14, e0216456. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.; Back, S.; Lee, S.; Park, D.; Kim, T.; Lee, K. Intra- and inter-epoch temporal context network (IITNet) using sub-epoch features for automatic sleep scoring on raw single-channel EEG. Biomed. Signal Process. Control 2020, 61, 102037. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, M.; Li, Y.; Su, M.; Xu, Z.; Wang, C.; Kang, D.; Li, H.; Mu, X.; Ding, X.; et al. Automated multi-model deep neural network for sleep stage scoring with unfiltered clinical data. Sleep Breath. 2020, 24, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Supratak, A.; Dong, H.; Wu, C.; Guo, Y. DeepSleepNet: A model for automatic sleep stage scoring based on raw single-channel EEG. IEEE Trans. Neural Syst. Rehabil. Eng. 2017, 25, 1998–2008. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.; Andreotti, F.; Cooray, N.; Chén, O.Y.; de Vos, M. Joint classification and prediction CNN framework for automatic sleep stage classification. IEEE Trans. Biomed. Eng. 2019, 66, 1285–1296. [Google Scholar] [CrossRef]
- Vilamala, A.; Madsen, K.H.; Hansen, L.K. Deep convolutional neural networks for interpretable analysis of EEG sleep stage scoring. In Proceedings of the 2017 IEEE 27th International Workshop on Machine Learning for Signal Processing (MLSP), Tokyo, Japan, 25–28 September 2017; pp. 1–6. [Google Scholar] [CrossRef]
- Phan, H.; Andreotti, F.; Cooray, N.; Chen, O.Y.; de Vos, M. DNN filter bank improves 1-max pooling CNN for single-channel EEG automatic sleep stage classification. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018; pp. 453–456. [Google Scholar] [CrossRef]
- Phan, H.; Andreotti, F.; Cooray, N.; Chen, O.Y.; De Vos, M. Automatic Sleep Stage Classification Using Single-Channel EEG: Learning Sequential Features with Attention-Based Recurrent Neural Networks. In Proceedings of the 2018 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Honolulu, HI, USA, 18–21 July 2018. [Google Scholar]
- Xu, M.; Wang, X.; Zhangt, X.; Bin, G.; Jia, Z.; Chen, K. Computation-Efficient Multi-Model Deep Neural Network for Sleep Stage Classification. In Proceedings of the ASSE ’20: 2020 Asia Service Sciences and Software Engineering Conference, Nagoya, Japan, 13–15 May 2020; Association for Computing Machinery: New York, NY, USA, 2020; pp. 1–8. [Google Scholar]
- Wang, Y.; Wu, D. Deep Learning for Sleep Stage Classification. In Proceedings of the 2018 Chinese Automation Congress (CAC), Xi’an, China, 30 November–2 December 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 3833–3838. [Google Scholar]
- Fernandez-Blanco, E.; Rivero, D.; Pazos, A. Convolutional neural networks for sleep stage scoring on a two-channel EEG signal. Soft Comput. 2019, 24, 4067–4079. [Google Scholar] [CrossRef]
- Jadhav, P.; Rajguru, G.; Datta, D.; Mukhopadhyay, S. Automatic sleep stage classification using time-frequency images of CWT and transfer learning using convolution neural network. Biocybern. Biomed. Eng. 2020, 40, 494–504. [Google Scholar] [CrossRef]
- Tsinalis, O.; Matthews, P.M.; Guo, Y.; Zafeiriou, S. Automatic Sleep Stage Scoring with Single-Channel EEG Using Convolutional Neural Networks; Imperial College London: London, UK, 2016. [Google Scholar]
- Sokolovsky, M.; Guerrero, F.; Paisarnsrisomsuk, S.; Ruiz, C.; Alvarez, S.A. Deep learning for automated feature discovery and classification of sleep stages. IEEE/ACM Trans. Comput. Biol. Bioinform. 2019, 17. [Google Scholar] [CrossRef]
- Dong, H.; Supratak, A.; Pan, W.; Wu, C.; Matthews, P.M.; Guo, Y. Mixed neural network approach for temporal sleep stage classification. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 324–333. [Google Scholar] [CrossRef]
- Chambon, S.; Galtier, M.N.; Arnal, P.J.; Wainrib, G.; Gramfort, A. A deep learning architecture for temporal sleep stage classification using multivariate and multimodal time series. IEEE Trans. Neural Syst. Rehabil. Eng. 2018, 26, 758–769. [Google Scholar] [CrossRef]
- Phan, H.; Andreotti, F.; Cooray, N.; Chén, O.Y.; de Vos, M. SeqSleepNet: End-to-end hierarchical recurrent neural network for sequence-to-sequence automatic sleep staging. IEEE Trans. Neural Syst. Rehabil. Eng. 2019, 27, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yao, R.; Ge, W.; Gao, J. Orthogonal convolutional neural networks for automatic sleep stage classification based on single-channel EEG. Comput. Methods Programs Biomed. 2020, 183, 105089. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wu, Y. Complex-valued unsupervised convolutional neural networks for sleep stage classification. Comput. Methods Programs Biomed. 2018, 164, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Sors, A.; Bonnet, S.; Mirek, S.; Vercueil, L.; Payen, J.-F. A convolutional neural network for sleep stage scoring from raw single-channel EEG. Biomed. Signal Process. Control 2018, 42, 107–114. [Google Scholar] [CrossRef]
- Fernández-Varela, I.; Hernández-Pereira, E.; Alvarez-Estevez, D.; Moret-Bonillo, V. A Convolutional Network for Sleep Stages Classification. arXiv 2019, arXiv:1902.05748v1. [Google Scholar] [CrossRef]
- Zhang, L.; Fabbri, D.; Upender, R.; Kent, D.T. Automated sleep stage scoring of the Sleep Heart Health Study using deep neural networks. Sleep 2019, 42. [Google Scholar] [CrossRef]
- Cui, Z.; Zheng, X.; Shao, X.; Cui, L. Automatic sleep stage classification based on convolutional neural network and fine-grained segments. Complexity 2018, 2018, 1–13. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, X.; Yuan, F. A Study on Automatic Sleep Stage Classification Based on CNN-LSTM. In Proceedings of the ICCSE’18: The 3rd International Conference on Crowd Science and Engineering, Singapore, 28–31 July 2018; Association for Computing Machinery: New York, NY, USA, 2018; pp. 1–5. [Google Scholar]
- Yuan, Y.; Jia, K.; Ma, F.; Xun, G.; Wang, Y.; Su, L.; Zhang, A. A hybrid self-attention deep learning framework for multivariate sleep stage classification. BMC Bioinform. 2019, 20, 1–10. [Google Scholar] [CrossRef]
- Biswal, S.; Sun, H.; Goparaju, B.; Westover, M.B.; Sun, J.; Bianchi, M.T. Expert-level sleep scoring with deep neural networks. J. Am. Med. Inform. Assoc. 2018, 25, 1643–1650. [Google Scholar] [CrossRef]
- Biswal, S.; Kulas, J.; Sun, H.; Goparaju, B.; Westover, M.B.; Bianchi, M.T.; Sun, J. SLEEPNET: Automated Sleep Staging System via Deep Learning. arXiv 2017, arXiv:1707.08262. [Google Scholar]
- Hoshide, S.; Kario, K. Sleep Duration as a risk factor for cardiovascular disease—A review of the recent literature. Curr. Cardiol. Rev. 2010, 6, 54–61. [Google Scholar] [CrossRef]
- Woods, S.L.; Froelicher, E.S.S.; Motzer, S.U.; Bridges, S.J. Cardiac Nursing, 5th ed.; Lippincott Williams and Wilkins: London, UK, 2005. [Google Scholar]
- Krieger, J. Breathing during sleep in normal subjects. Clin. Chest Med. 1985, 6, 577–594. [Google Scholar] [PubMed]
- Madsen, P.L.; Schmidt, J.F.; Wildschiodtz, G.; Friberg, L.; Holm, S.; Vorstrup, S.; Lassen, N.A. Cerebral O2 metabolism and cerebral blood flow in humans during deep and rapid-eye-movement sleep. J. Appl. Physiol. 1991, 70, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Klosh, G.; Kemp, B.; Penzel, T.; Schlogl, A.; Rappelsberger, P.; Trenker, E.; Gruber, G.; Zeithofer, J.; Saletu, B.; Herrmann, W.; et al. The SIESTA project polygraphic and clinical database. IEEE Eng. Med. Boil. Mag. 2001, 20, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Yıldırım, Ö.; Talo, M.; Ay, B.; Baloglu, U.B.; Aydin, G.; Acharya, U.R. Automated detection of diabetic subject using pre-trained 2D-CNN models with frequency spectrum images extracted from heart rate signals. Comput. Biol. Med. 2019, 113, 103387. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.-H.; Vicnesh, J.; Koh, J.E.; Oh, S.L.; Arunkumar, N.; Abdulhay, E.; Ciaccio, E.J.; Acharya, U.R. Autism spectrum disorder diagnostic system using HOS bispectrum with EEG Signals. Int. J. Environ. Res. Public Health 2020, 17, 971. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Kim, J.-M. Automated bearing fault diagnosis using 2D analysis of vibration acceleration signals under variable speed conditions. Shock. Vib. 2016, 2016, 1–11. [Google Scholar] [CrossRef]
- Kiranyaz, S.; Avci, O.; Abdeljaber, O.; Ince, T.; Gabbouj, M.; Inman, D.J. 1D convolutional neural networks and applications: A survey. Mech. Syst. Signal Process. 2021, 151. [Google Scholar] [CrossRef]
- Patanaik, A.; Ong, J.L.; Gooley, J.J.; Ancoli-Israel, S.; Chee, M.W.L. An end-to-end framework for real-time automatic sleep stage classification. Sleep 2018, 41. [Google Scholar] [CrossRef]
- Terzano, M.G.; Parrino, L.; Sherieri, A.; Chervin, R.; Chokroverty, S.; Guilleminault, C.; Hirshkowitz, M.; Mahowald, M.; Moldofsky, H.; Rosa, A.; et al. Atlas, rules, and recording techniques for the scoring of cyclic alternating pattern (CAP) in human sleep. Sleep Med. 2001, 2, 537–553. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).