Figure 1 shows that the proposed fusion method contains three steps: inversion, fusion, and selector. Firstly, a morphology image is inverted into a series of inverted images using different averages. The max rule is then used to fuse the inverted images and acoustic image into the pre-fused images sequences. Finally, a selector is used to choose the expected image from the pre-fused images sequence as the fusion images’ output.
2.1.1. Inversion
Our proposed method’s first step is inverting the morphology image into a series of inverted images whose average intensities are different from each other. Our grayscale inversion approach is a simple linear approach, which is defined as below:
where
is the original morphology image.
is the
inverted morphology image,
is the largest gray level of
.
is the collection of inverted morphology images, and
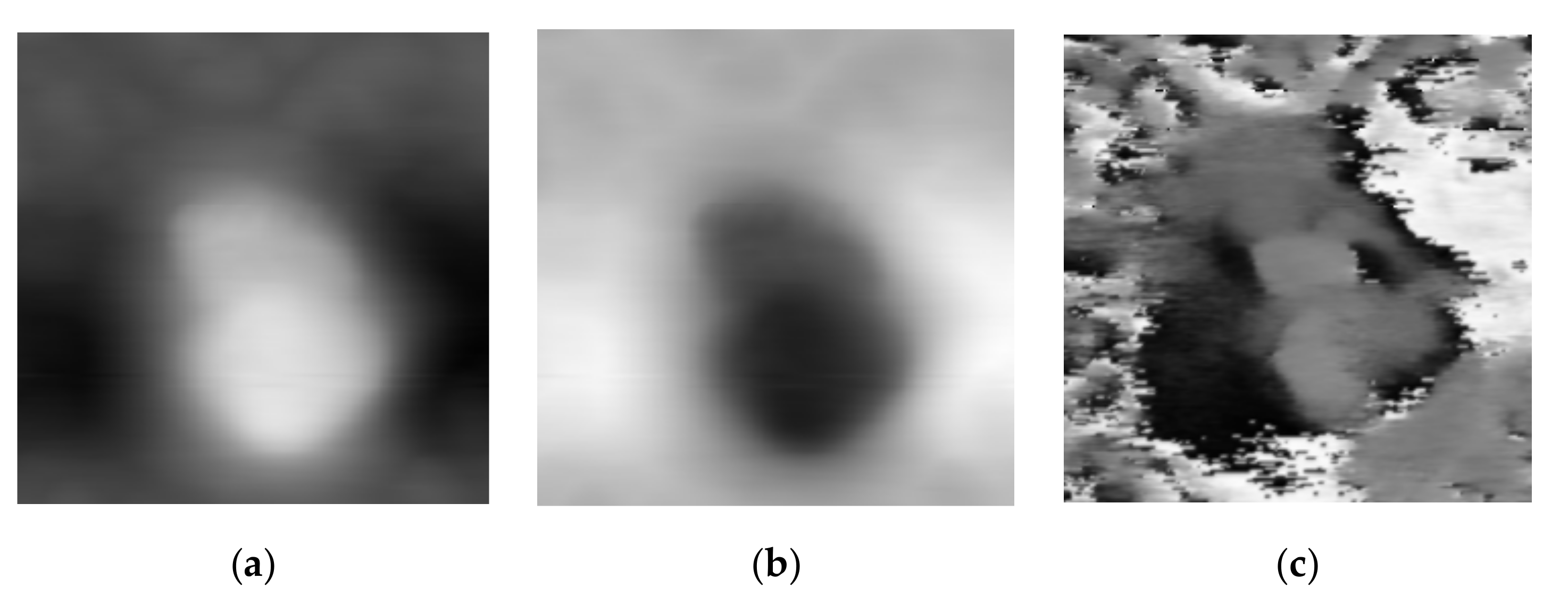
is the maximum gray value of the acoustic image. We apply greyscale inversion to match the gray level diffusion of morphology and acoustic images. In
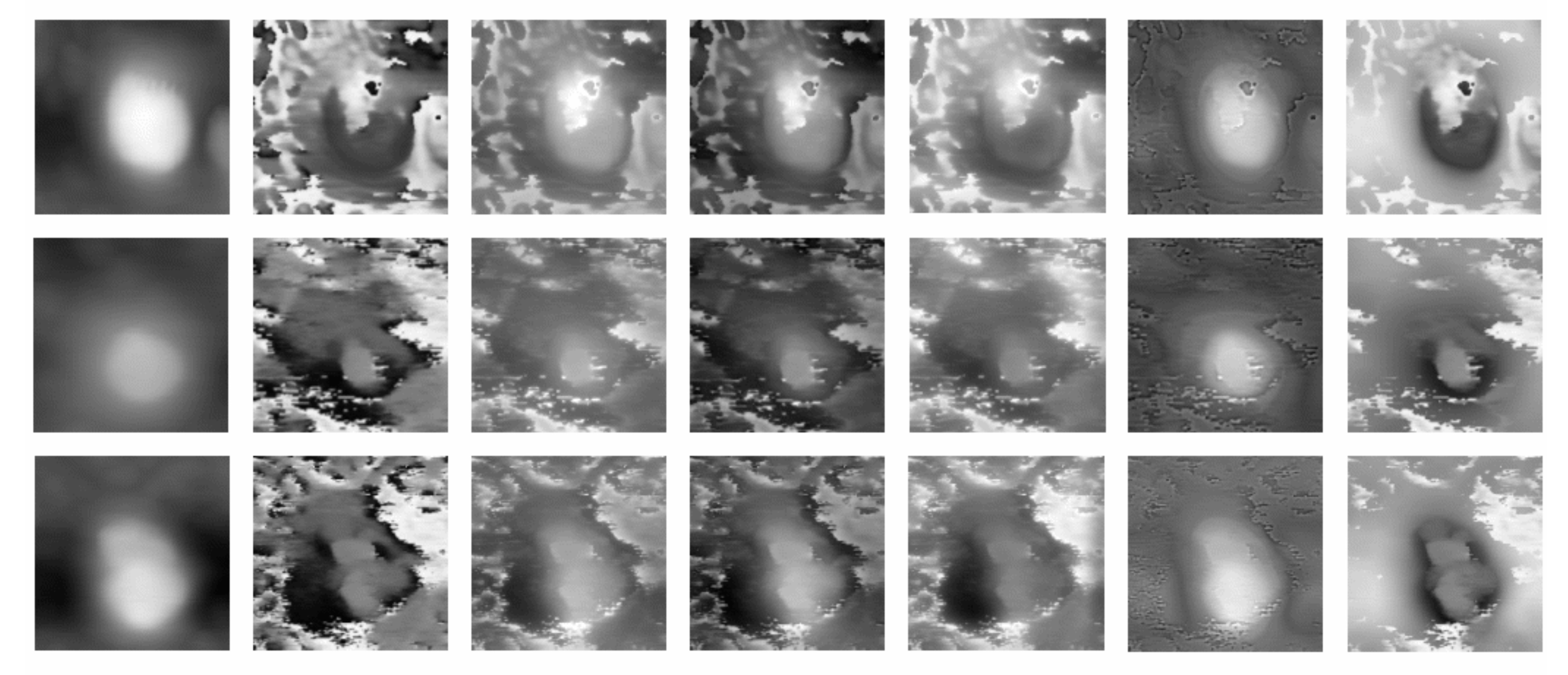
Figure 2, we show the morphology, inverted, and acoustic image. The morphology image’s intensity relates to the sample’s height, so the bright area is the cell region, and the dark area is the background, respectively. As for acoustic images, many high-intensity noises surround the cell region, and the acoustic details in the cell are darker than the same region in the morphology image. Grayscale inversion aims to produce a high-intensity background and low-intensity cell region. The high-intensity background will reduce the contract of the noise and even cover it. Simultaneously, the low-intensity cell region makes acoustic details easier to be presented. The bright region, which is considered as background, may be beyond the bounds of the gray level. To present the cell’s shape and gradient in the morphology image, the dark areas, which is considered the cell region, should take priority of the fusion work. As a trade-off, we truncate the image gray level to the bounds of the morphological image gray level, i.e., the pixels whose intensity is larger than 255 will be truncated to 255 for the 8-bit image.
2.1.2. Max Rule
The fusion rule of inverted and acoustic images is the max rule. At the pixel (i,j), it compares the intensity of the
inverted image and acoustic image, and selects the larger one as the output. It is defined as follows:
where
is the
pre-fused image,
is the acoustic image. Now, we fuse the information from both morphology and acoustic image. In the background, because the inversed image has a higher average gray level, the pre-fused image has smooth and high-intensity areas. In the boundary of the cell, the gradient from the bright region to dark region indicates the cell’s shape. In the cell region, acoustic details are preserved because they have higher intensity than the inversed image. In
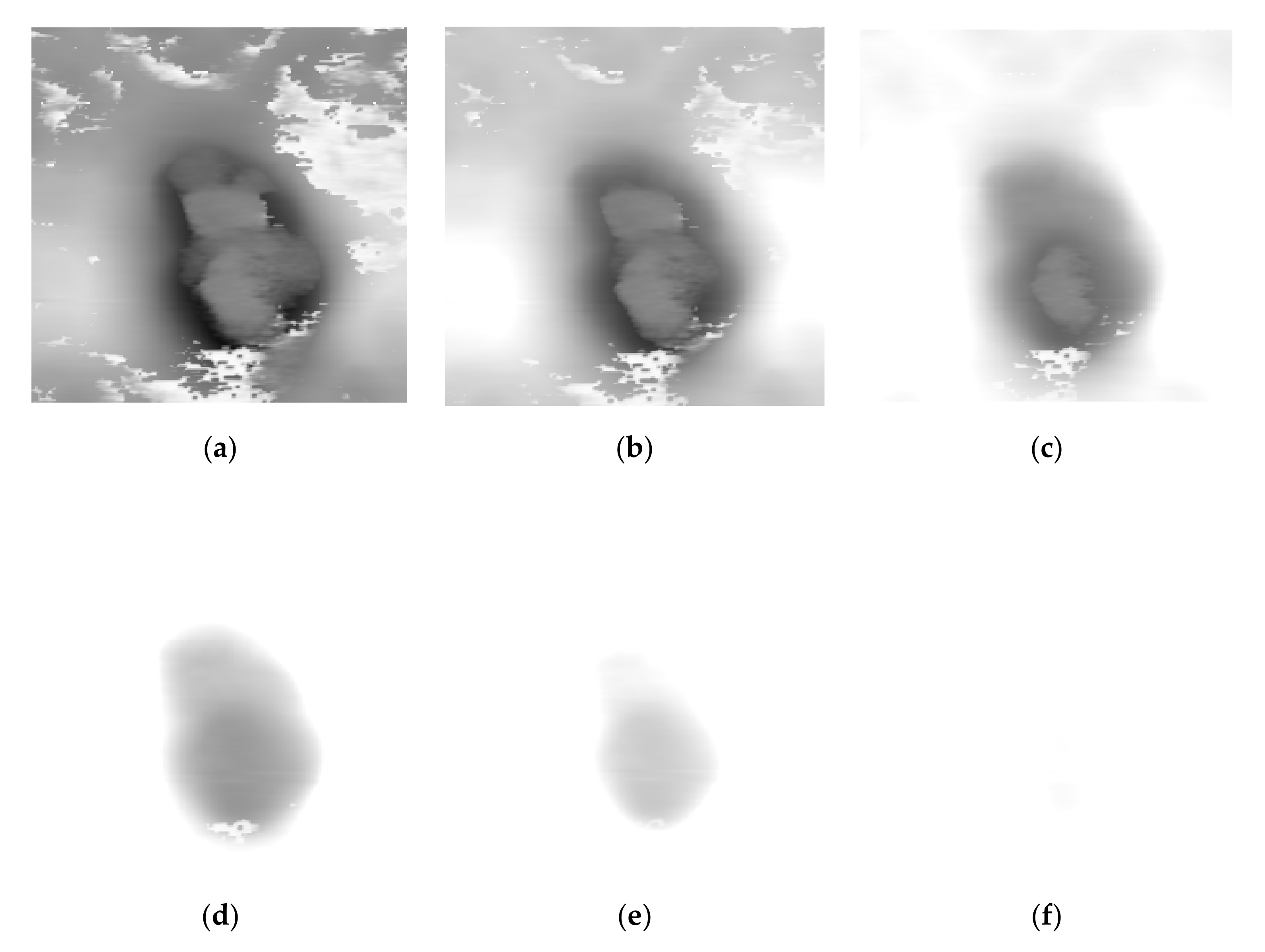
Figure 3, as the increasing value of k, the pre-fused image provides more gradient information of the inverted morphology image but loses details of the acoustic image. Thus, the next step is to choose the image that preserves both morphology and acoustic information. We name this ideal image as the best-fit intensity image.
2.1.3. Selector
To choose the best-fit intensity image, a selector is designed to measure the information loss of the pre-fused images to select the best-fit intensity image as the final output of the fused image.
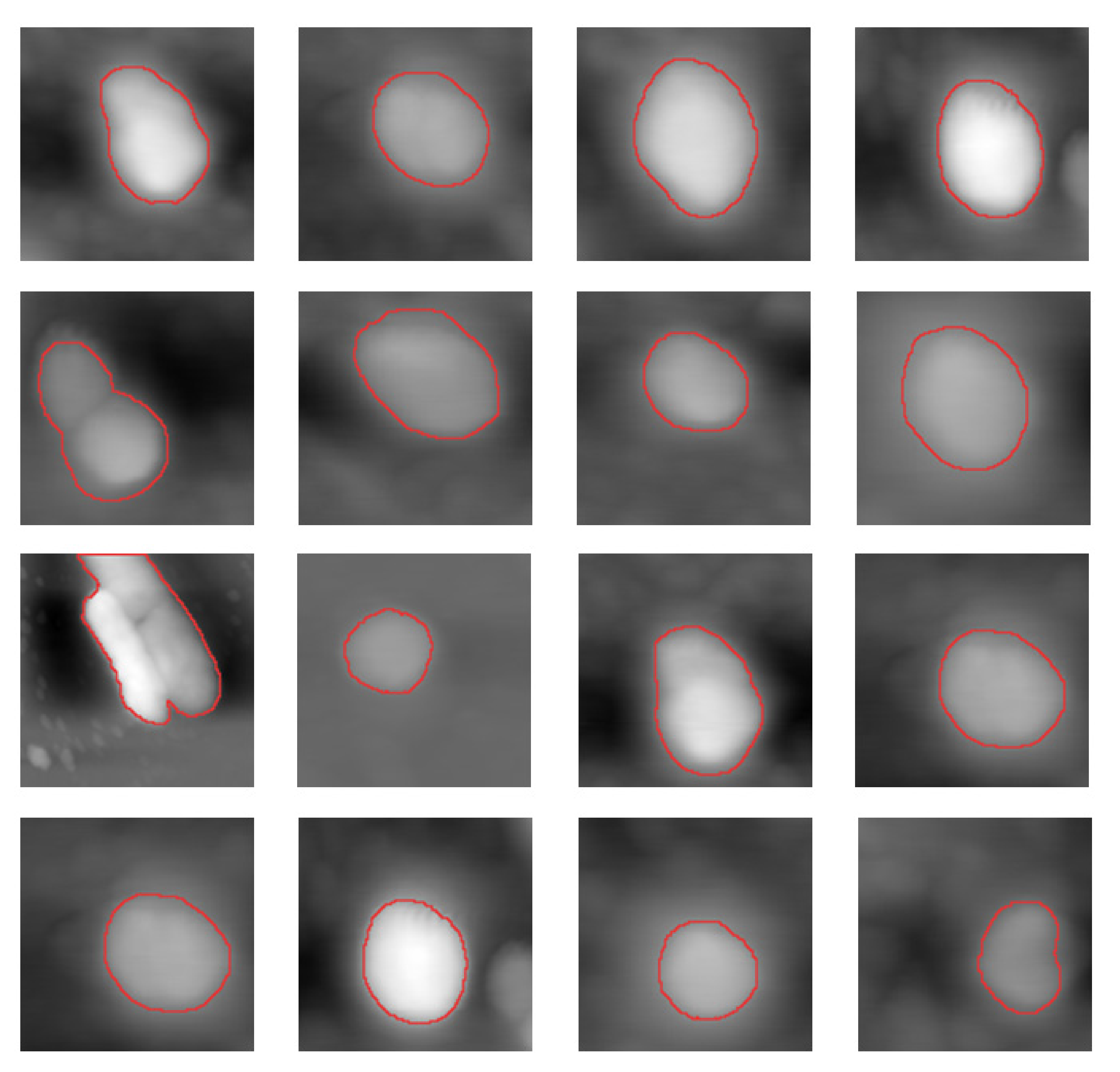
Because AFAM fusion aims is to combine the information of the cell, the regions of the cells should be extracted, and the fusion problem could be simplified to the fusion problem in the regions of cells. To acquire the position and shape of the cell, a cell extraction algorithm based on Otsu’s approach [
14] was developed. Erosion and dilation steps are used to reduce the influence of the noise and smoothen the boundary of the cell region. Our cell extraction method is a triple-step process. The first step is detection. Otsu’s threshold is used to segment the morphology image into
and
at gray level
, which is defined as follows:
where
is the threshold of classifying background and the cell. To clarifiy, Otsu’s threshold is not an accurate segmentation method because it does not perform well in segmenting smooth boundary. Our method is not sensible to the segmentation accuracy.
However, the small object, which is not the region of interest in the view of content but has the same gray level section, does harm to the fusion performance. To clear them, we apply the erosion step. We assume that the main part of the morphology image is the cell, and noise occupies small and separated regions. To clear these areas, the erosion step is defined as follows:
where
is a disk-shaped structuring element,
is the size of
, and
is the multiple coefficient that determines the narrowing of the original segmentation of the cell. The function of
is to clear the noise regions which have a similar intensity feature of the cells, and the proper value of
is to clear all the noise regions but still preserve the region of the cells. According to our hypothesis, the noise regions are removed during the erosion process but the cell region is narrowed either. Therefore, the dilation step, which is the third step, is used to recover the cell region. It is defined as follows:
where
is a disk-shaped structuring element,
is the size of
, and
is a multiple coefficient that determines the extension of
. The function of
is to recover the region of cells and make the boundary of the cells smooth. The value of
is determined by
, and it is defined as follows:
At last, we obtain
, which represents the region of cell, and
, representing the background. The relationship of morphology image,
and
is defined as follows:
Next, to set the standard of the best-fit intensity image among the pre-fused images, we define a multi-regional information loss (MRIL) to measure the information loss. It is a double-stage algorithm, which would calculate the total information loss of the inner and outer cells. To define the set of the criterion of the pre-fused images, characteristic of inverted morphology and acoustic image should be taken into consideration. As it is shown in
Figure 2, inverted morphology and acoustic images show different information in different regions. In the cell region, because of grayscale inversion, the intensities of inverted morphology are weaker than those in the background and provide little information about the inner structures. By contrast, the acoustic image provides much richer details in the region of the cell. Thus, the fused image should preserve as many details of the acoustic image as we can. In terms of background, inverted morphology image is used to show the cell’s shape and position, which is not clear in acoustic image. To measure the total information loss, MRIL is defined as follows:
where
is the loss of cell and
is the loss of background. They are given by:
where
is the
pre-fused image,
is the acoustic image,
has a similar definition of the ratio of SF error [
15]. The difference is that our rSFe method measures the space-frequency error between the pre-fused image and inverted morphology image in the region of
instead of fused image and source images. The reason why we choose rSFe to measure the loss in the background for the following two reasons. Firstly, the space-frequency remains the same no matter what the value of k is. Secondly, rSFe is a suitable regulation that makes the background as bright as possible to cover the acoustic noise, and it is not sensible to the segmentation accuracy of the cell either. Given that the acoustic image is characterized by their pixel intensities, MRIL is sensitive to among the pixels preserved from the acoustic image in the region of the cell.
According to the definition of information loss, the expected image is the image with the lowest
. Thus, the selected image is defined as follows: