An Efficient Lightweight CNN and Ensemble Machine Learning Classification of Prostate Tissue Using Multilevel Feature Analysis
Abstract
1. Introduction
2. Related Work
3. Tissue Staining and Data Collection
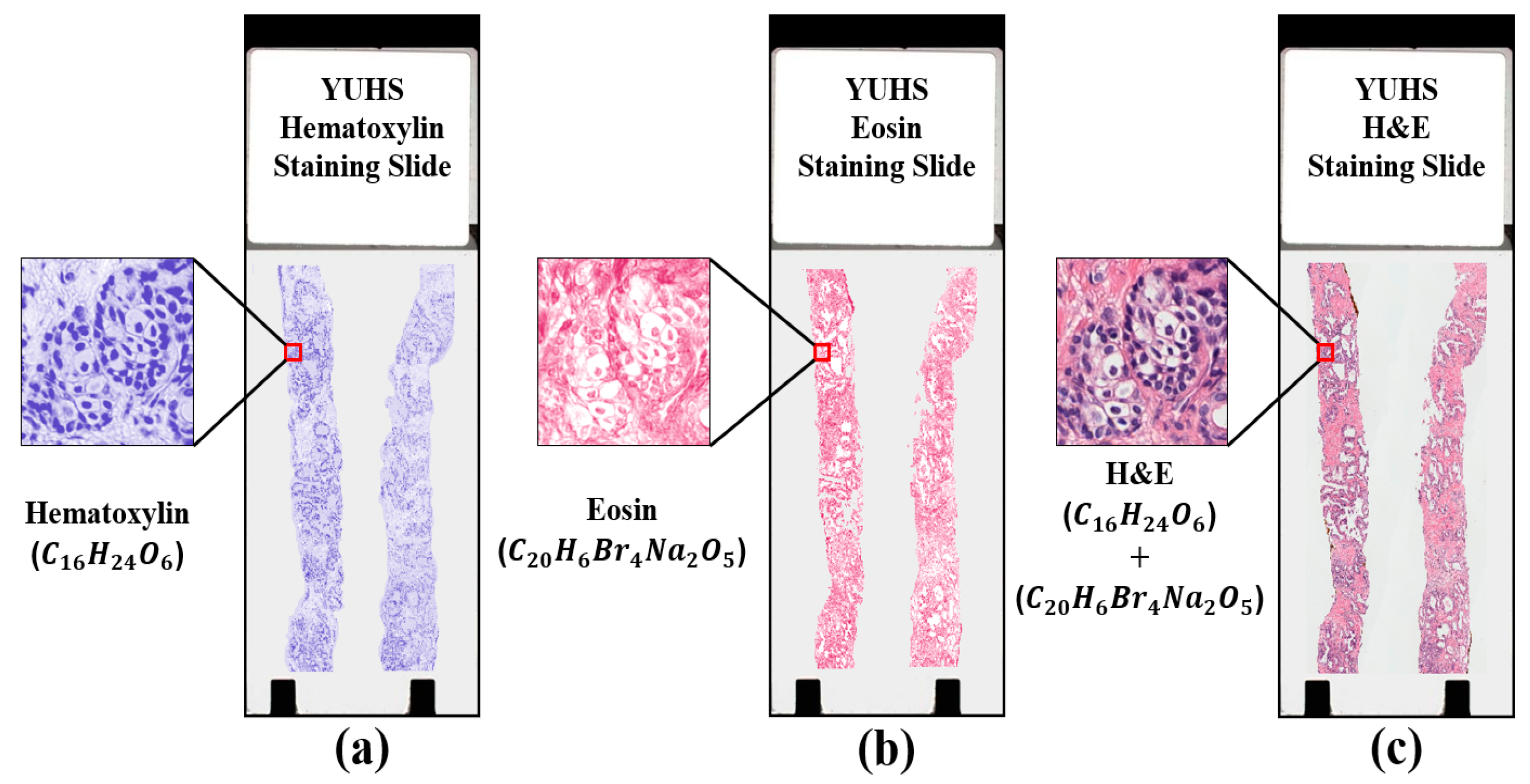
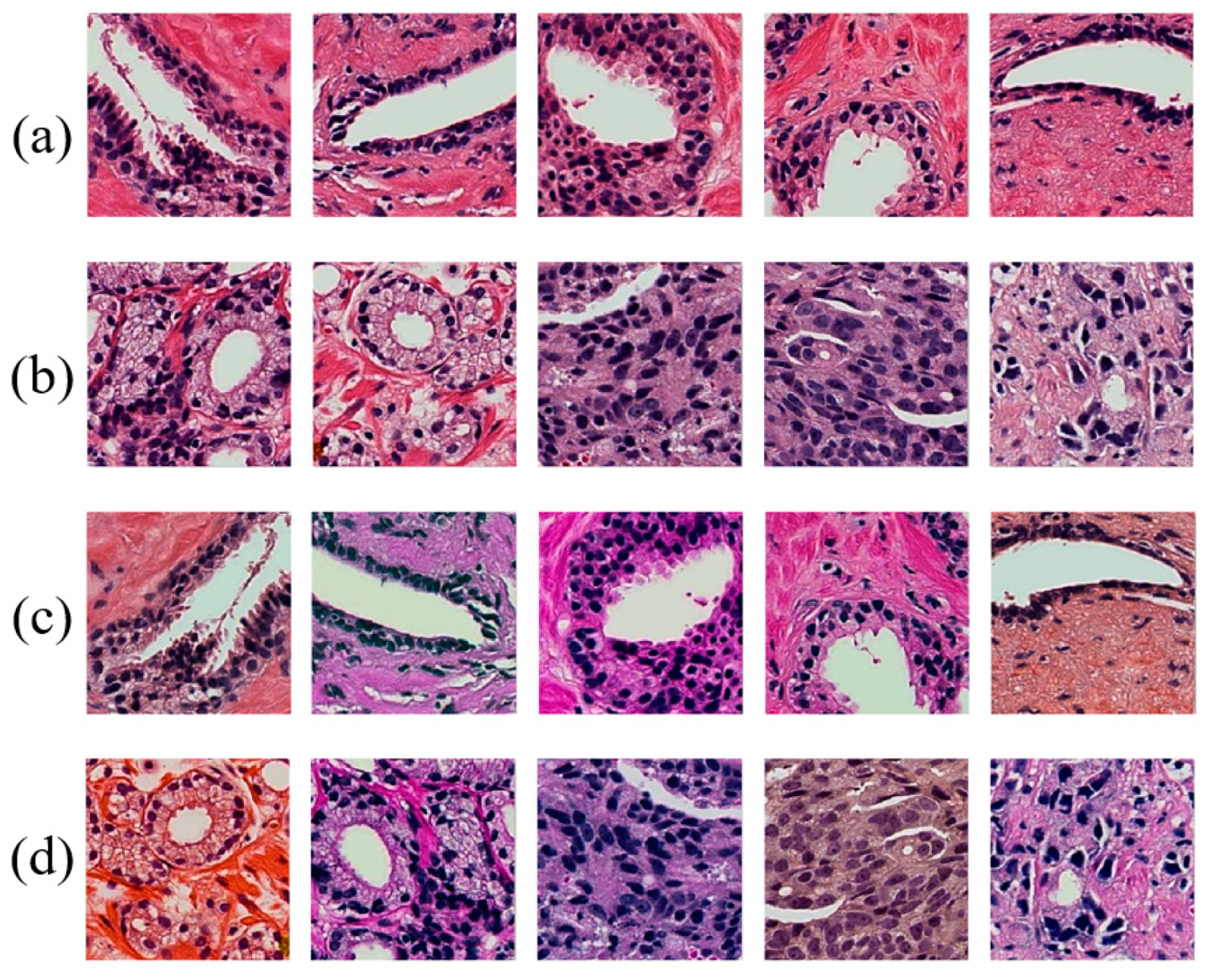
3.1. Tissue Staining
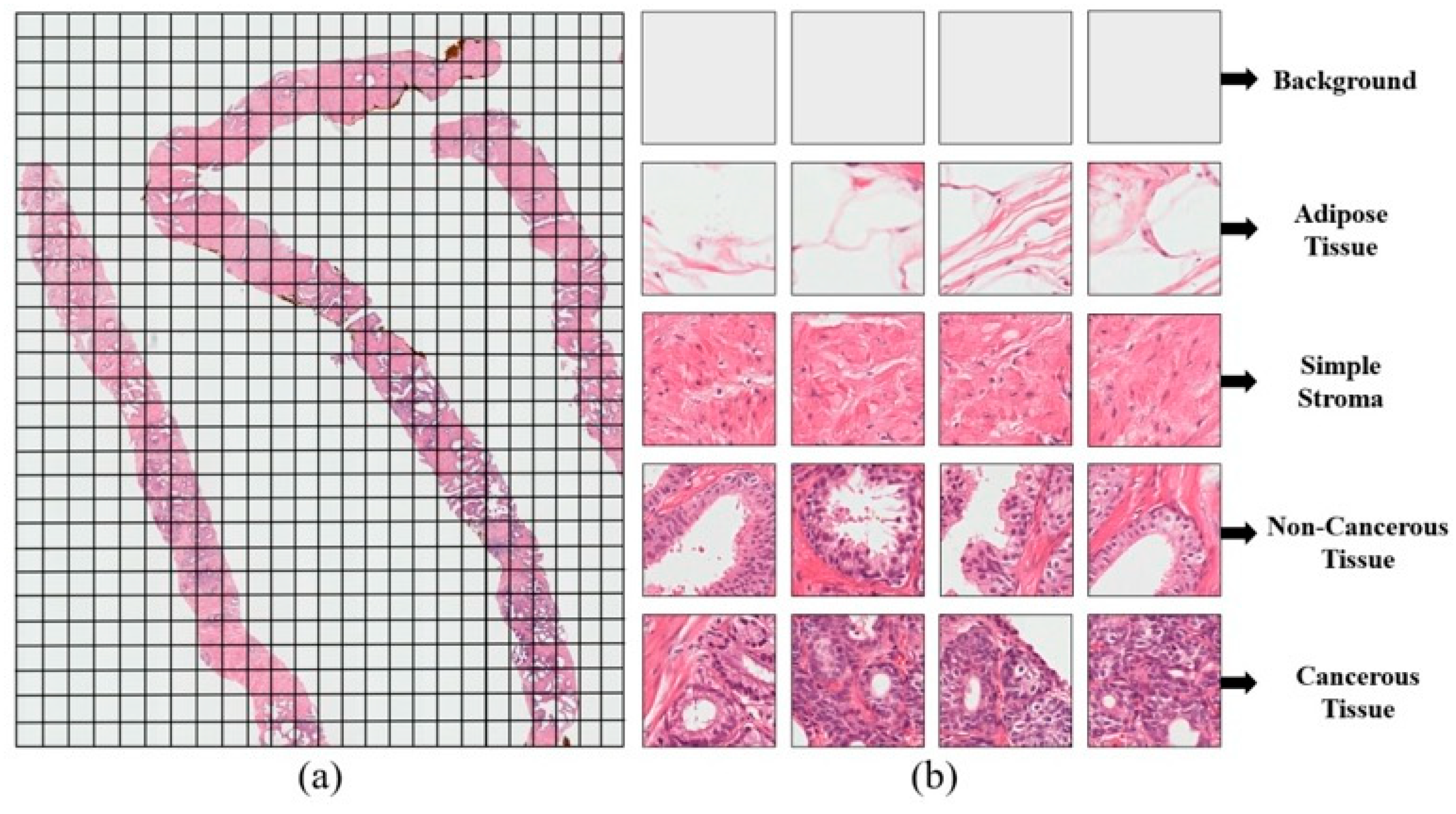
3.2. Data Collection
4. Materials and Methods
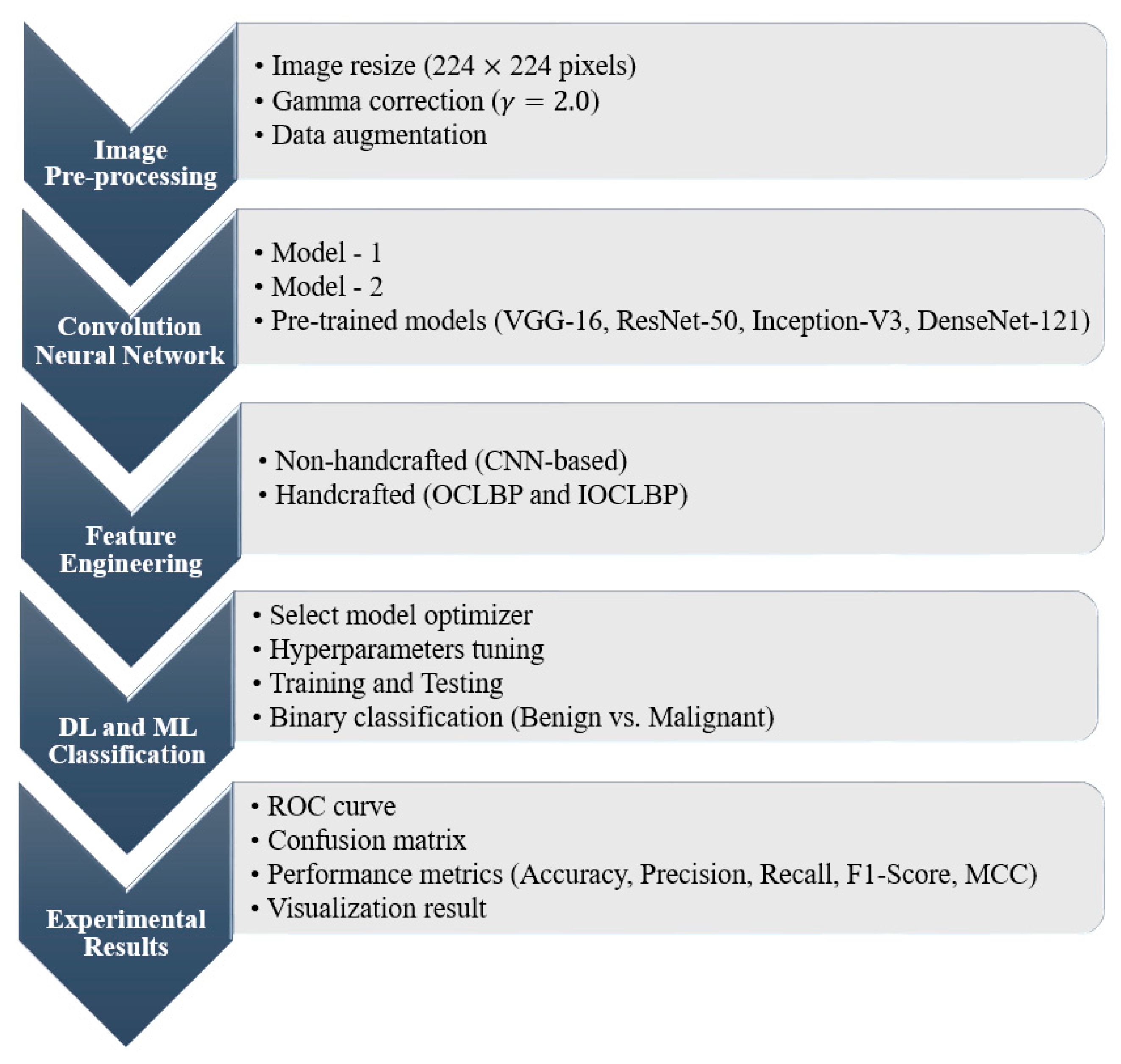
4.1. Proposed Pipeline
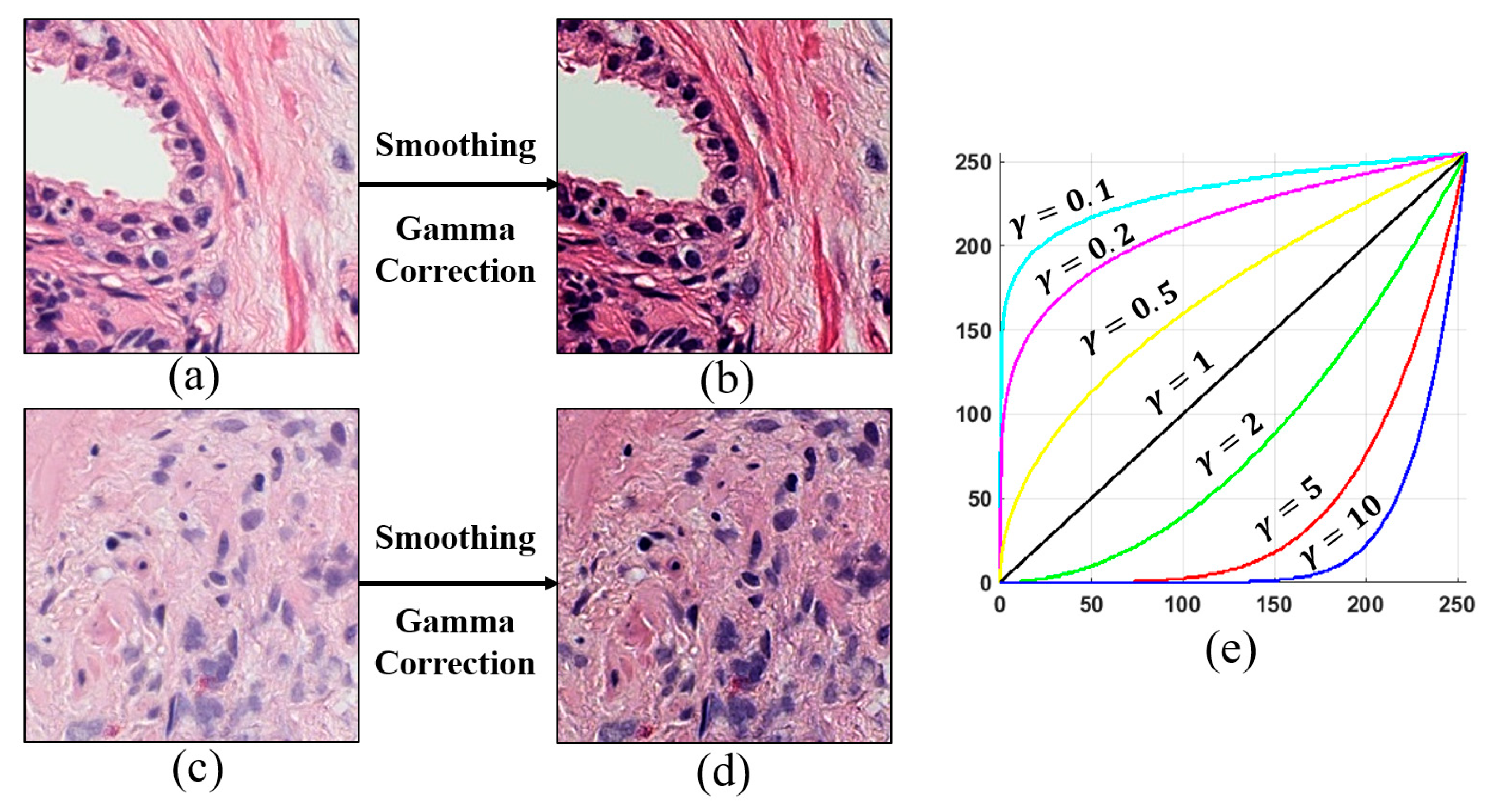
4.2. Image Preprocessing
4.3. Convolution Neural Network
4.4. Feature Engineering
4.5. DL and ML Classification
5. Experimental Results
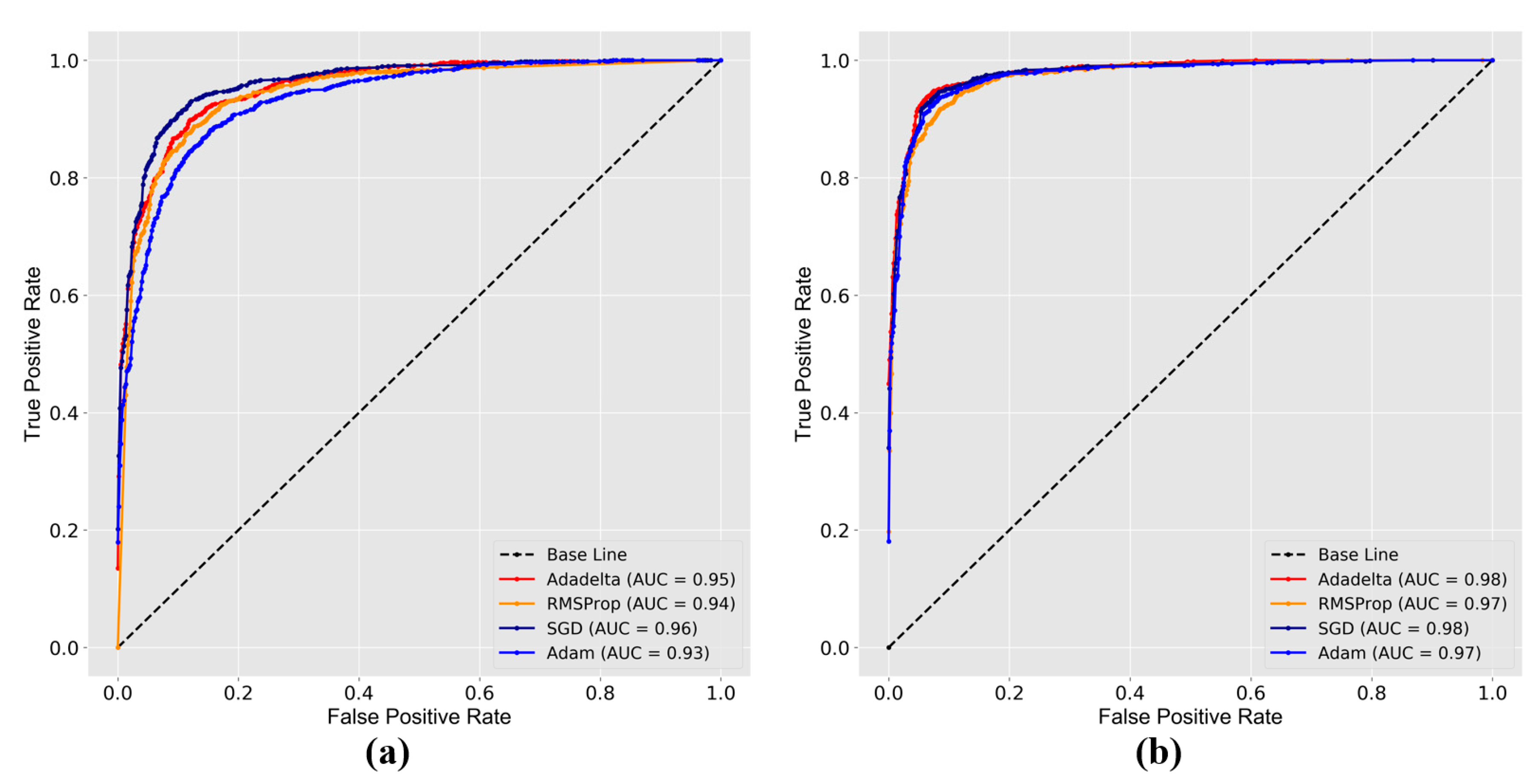
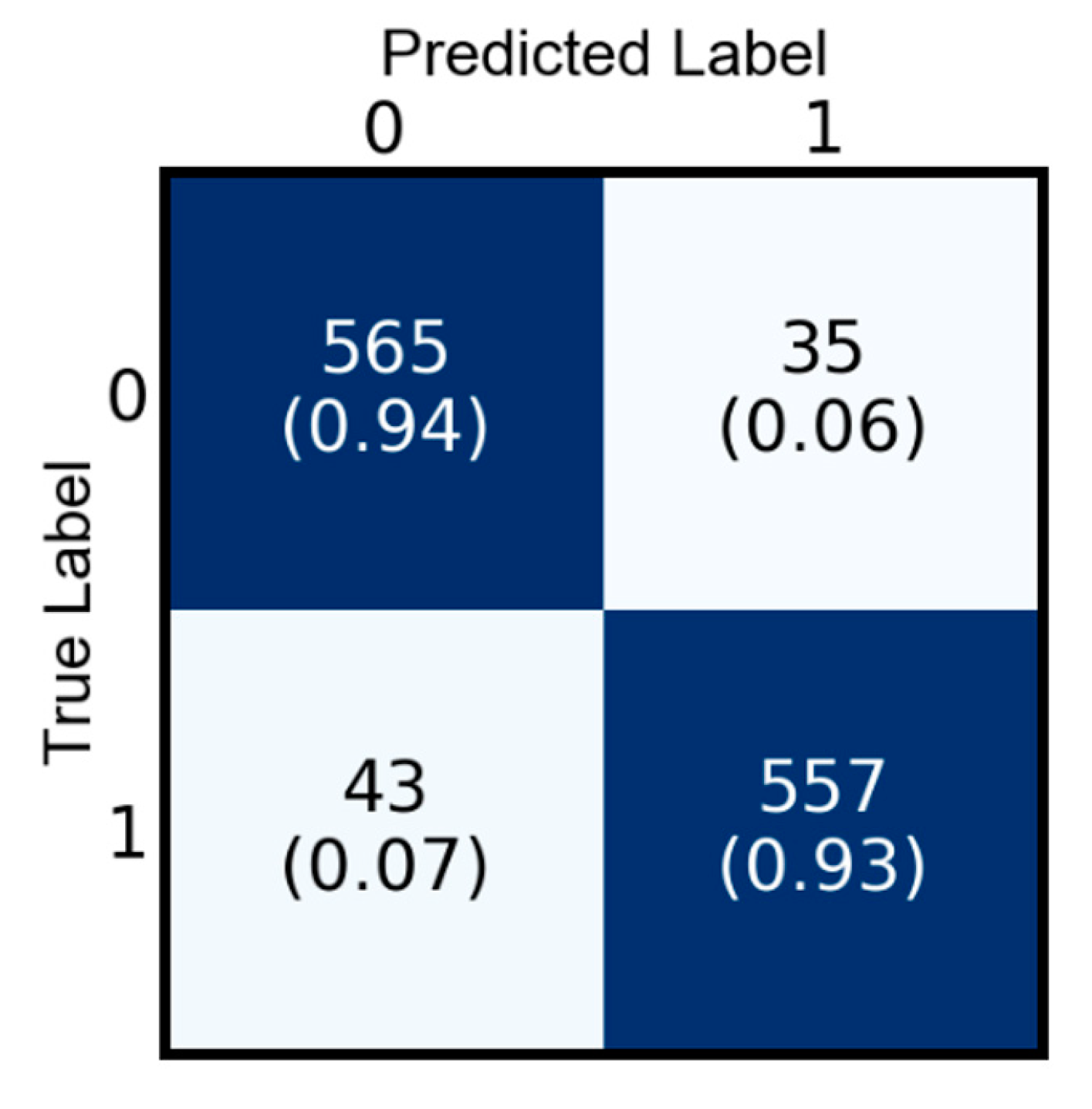
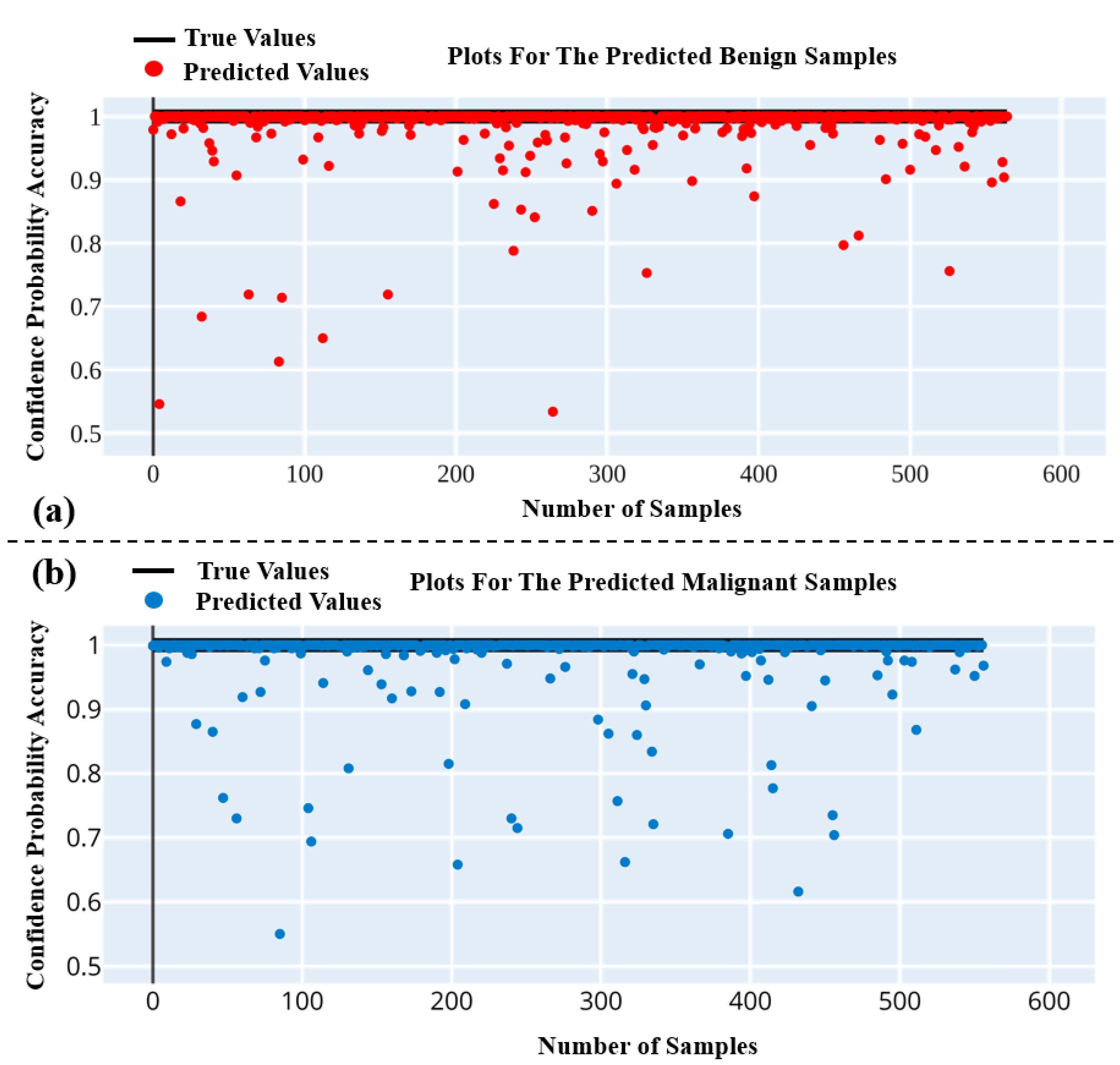
5.1. Performance Analysis
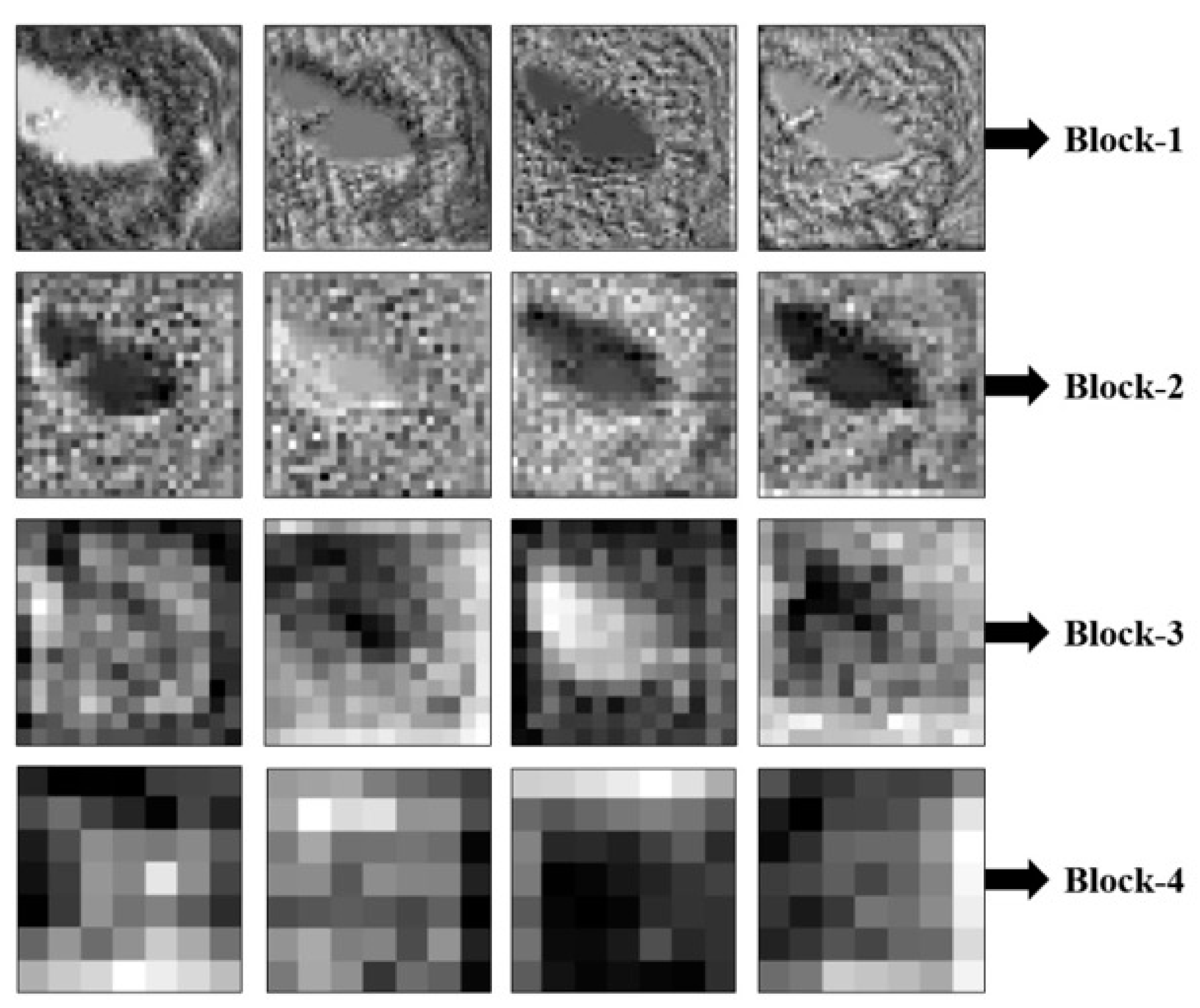
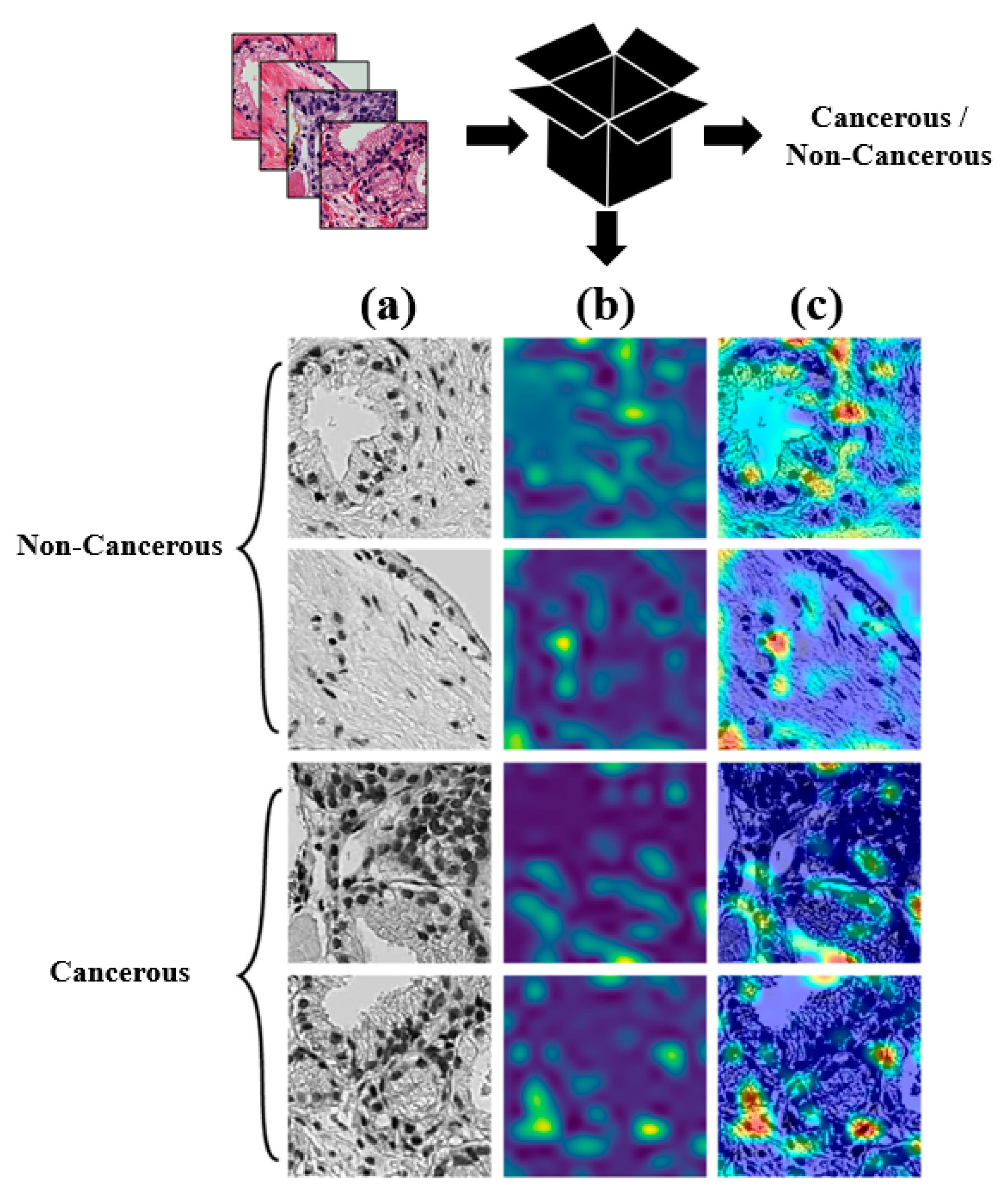
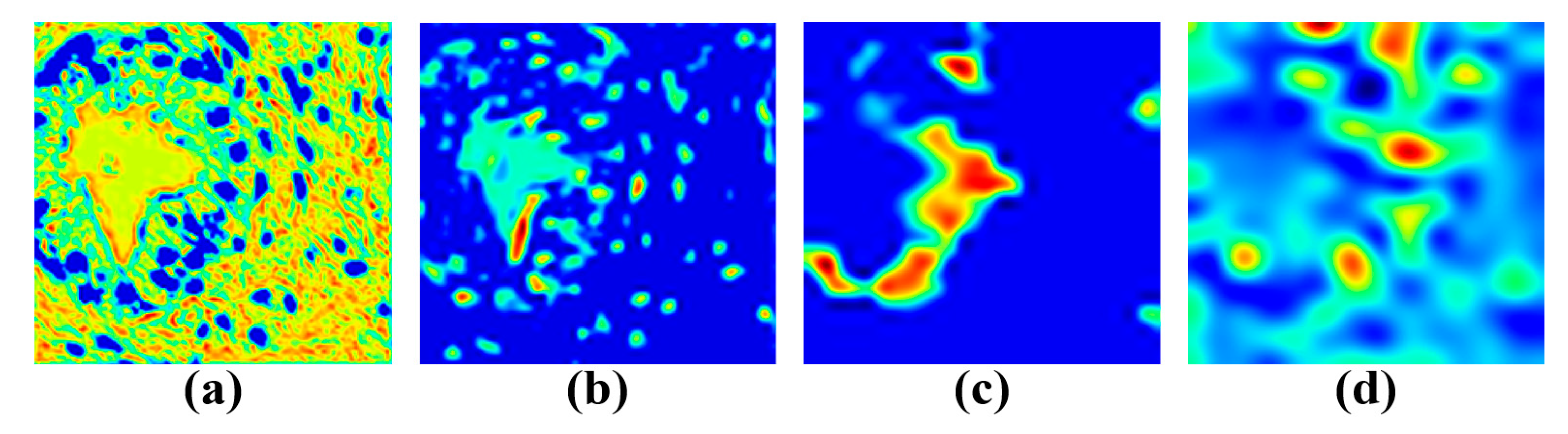
5.2. Visualization Results
6. Discussion
7. Conclusions
Author Contributions
Funding
Ethical Approval
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2015. CA Cancer J. Clin. 2015, 65, 5–29. [Google Scholar] [CrossRef]
- Chung, M.S.; Shim, M.; Cho, J.S.; Bang, W.; Kim, S.I.; Cho, S.Y.; Rha, K.H.; Hong, S.J.; Hong, C.-H.; Lee, K.S.; et al. Pathological Characteristics of Prostate Cancer in Men Aged <50 Years Treated with Radical Prostatectomy: A Multi-Centre Study in Korea. J. Korean Med. Sci. 2019, 34, 78. [Google Scholar] [CrossRef]
- Yoo, S.; Gujrathi, I.; Haider, M.A.; Khalvati, F. Prostate Cancer Detection using Deep Convolutional Neural Networks. Sci. Rep. 2019, 9, 19518. [Google Scholar] [CrossRef]
- Humphrey, P.A. Diagnosis of adenocarcinoma in prostate needle biopsy tissue. J. Clin. Pathol. 2007, 60, 35–42. [Google Scholar] [CrossRef]
- Van Der Kwast, T.H.; Lopes, C.; Santonja, C.; Pihl, C.-G.; Neetens, I.; Martikainen, P.; Di Lollo, S.; Bubendorf, L.; Hoedemaeker, R.F. Guidelines for processing and reporting of prostatic needle biopsies. J. Clin. Pathol. 2003, 56, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Andriole, G.L. Improved biopsy efficiency with MR/ultrasound fusion-guided prostate biopsy. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef] [PubMed]
- Heidenreich, A.; Bastian, P.J.; Bellmunt, J.; Bolla, M.; Joniau, S.; Van Der Kwast, T.; Mason, M.; Matveev, V.; Wiegel, T.; Zattoni, F.; et al. EAU Guidelines on Prostate Cancer. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent—Update 2013. Eur. Urol. 2014, 65, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, P.A. Gleason grading and prognostic factors in carcinoma of the prostate. Mod. Pathol. 2004, 17, 292–306. [Google Scholar] [CrossRef]
- Nagpal, K.; Foote, D.; Liu, Y.; Chen, P.-H.C.; Wulczyn, E.; Tan, F.; Olson, N.; Smith, M.C.; Mohtashamian, A.; Wren, J.H.; et al. Development and validation of a deep learning algorithm for improving Gleason scoring of prostate cancer. NPJ Digit. Med. 2019, 2, 48. [Google Scholar] [CrossRef]
- Alqahtani, S.; Wei, C.; Zhang, Y.; Szewczyk-Bieda, M.; Wilson, J.; Huang, Z.; Nabi, G. Prediction of prostate cancer Gleason score upgrading from biopsy to radical prostatectomy using pre-biopsy multiparametric MRI PIRADS scoring system. Sci. Rep. 2020, 10, 7722. [Google Scholar] [CrossRef]
- Zhu, Y.; Freedland, S.J.; Ye, D. Prostate Cancer and Prostatic Diseases Best of Asia, 2019: Challenges and opportunities. Prostate Cancer Prostatic Dis. 2019, 23, 197–198. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Srivastava, R.; Srivastava, S.K. Detection and Classification of Cancer from Microscopic Biopsy Images Using Clinically Significant and Biologically Interpretable Features. J. Med. Eng. 2015, 2015, 457906. [Google Scholar] [CrossRef] [PubMed]
- Cahill, L.C.; Fujimoto, J.G.; Giacomelli, M.G.; Yoshitake, T.; Wu, Y.; Lin, D.I.; Ye, H.; Carrasco-Zevallos, O.M.; Wagner, A.A.; Rosen, S. Comparing histologic evaluation of prostate tissue using nonlinear microscopy and paraffin H&E: A pilot study. Mod. Pathol. 2019, 32, 1158–1167. [Google Scholar] [CrossRef]
- Otali, D.; Fredenburgh, J.; Oelschlager, D.K.; Grizzle, W.E. A standard tissue as a control for histochemical and immunohistochemical staining. Biotech. Histochem. 2016, 91, 309–326. [Google Scholar] [CrossRef] [PubMed]
- Alturkistani, H.A.; Tashkandi, F.M.; Mohammedsaleh, Z.M. Histological Stains: A Literature Review and Case Study. Glob. J. Health Sci. 2015, 8, 72. [Google Scholar] [CrossRef] [PubMed]
- Zarella, M.D.; Yeoh, C.; Breen, D.E.; Garcia, F.U. An alternative reference space for H&E color normalization. PLoS ONE 2017, 12, 0174489. [Google Scholar]
- Lahiani, A.; Klaiman, E.; Grimm, O. Enabling histopathological annotations on immunofluorescent images through virtualization of hematoxylin and eosin. J. Pathol. Inform. 2018, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Gavrilovic, M.; Azar, J.C.; Lindblad, J.; Wählby, C.; Bengtsson, E.; Busch, C.; Carlbom, I.B. Blind Color Decomposition of Histological Images. IEEE Trans. Med. Imaging 2013, 32, 983–994. [Google Scholar] [CrossRef]
- Bautista, P.A.; Yagi, Y. Staining Correction in Digital Pathology by Utilizing a Dye Amount Table. J. Digit. Imaging 2015, 28, 283–294. [Google Scholar] [CrossRef]
- Bianconi, F.; Kather, J.N.; Reyes-Aldasoro, C.C. Evaluation of Colour Pre-Processing on Patch-Based Classification of H&E-Stained Images. In Digital Pathology. ECDP; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2019; Volume 11435, pp. 56–64. [Google Scholar] [CrossRef]
- Diamant, A.; Chatterjee, A.; Vallières, M.; Shenouda, G.; Seuntjens, J. Deep learning in head & neck cancer outcome prediction. Sci. Rep. 2019, 9, 2764. [Google Scholar]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef] [PubMed]
- Sahiner, B.; Pezeshk, A.; Hadjiiski, L.; Wang, X.; Drukker, K.; Cha, K.H.; Summers, R.M.; Giger, M.L. Deep learning in medical imaging and radiation therapy. Med. Phys. 2019, 46, e1–e36. [Google Scholar] [CrossRef] [PubMed]
- Nanni, L.; Ghidoni, S.; Brahnam, S. Handcrafted vs. non-handcrafted features for computer vision classification. Pattern Recognit. 2017, 71, 158–172. [Google Scholar] [CrossRef]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-G.; Jun, S.; Cho, Y.-W.; Lee, H.; Kim, G.B.; Seo, J.B.; Kim, N. Deep Learning in Medical Imaging: General Overview. Korean J. Radiol. 2017, 18, 570–584. [Google Scholar] [CrossRef] [PubMed]
- Bi, W.L.; Hosny, A.; Schabath, M.B.; Giger, M.L.; Birkbak, N.J.; Mehrtash, A.; Allison, T.; Arnaout, O.; Abbosh, C.; Dunn, I.F.; et al. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J. Clin. 2019, 69, 127–157. [Google Scholar] [CrossRef]
- Jha, S.; Topol, E.J. Adapting to Artificial Intelligence. JAMA 2016, 316, 2353–2354. [Google Scholar] [CrossRef] [PubMed]
- Badejo, J.A.; Adetiba, E.; Akinrinmade, A.; Akanle, M.B. Medical Image Classification with Hand-Designed or Machine-Designed Texture Descriptors: A Performance Evaluation. In Internatioanl Conference on Bioinformatics and Biomedical Engineering; Springer: Cham, Switzerland, 2018; pp. 266–275. [Google Scholar] [CrossRef]
- Bianconi, F.; Bello-Cerezo, R.; Napoletano, P. Improved opponent color local binary patterns: An effective local image descriptor for color texture classification. J. Electron. Imaging 2017, 27, 011002. [Google Scholar] [CrossRef]
- Kather, J.N.; Bello-Cerezo, R.; Di Maria, F.; Van Pelt, G.W.; Mesker, W.E.; Halama, N.; Bianconi, F. Classification of Tissue Regions in Histopathological Images: Comparison Between Pre-Trained Convolutional Neural Networks and Local Binary Patterns Variants. In Intelligent Systems Reference Library; Springer: Cham, Switzerland, 2020; pp. 95–115. [Google Scholar] [CrossRef]
- Khairunnahar, L.; Hasib, M.A.; Bin Rezanur, R.H.; Islam, M.R.; Hosain, K. Classification of malignant and benign tissue with logistic regression. Inform. Med. Unlocked 2019, 16, 100189. [Google Scholar] [CrossRef]
- Guidotti, R.; Monreale, A.; Ruggieri, S.; Turini, F.; Giannotti, F.; Pedreschi, D. A Survey of Methods for Explaining Black Box Models. ACM Comput. Surv. 2019, 51, 93. [Google Scholar] [CrossRef]
- Hayashi, Y. New unified insights on deep learning in radiological and pathological images: Beyond quantitative performances to qualitative interpretation. Inform. Med. Unlocked 2020, 19, 100329. [Google Scholar] [CrossRef]
- Lo, S.-C.; Lou, S.-L.; Lin, J.-S.; Freedman, M.; Chien, M.; Mun, S. Artificial convolution neural network techniques and applications for lung nodule detection. IEEE Trans. Med. Imaging 1995, 14, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.-C.B.; Chan, H.-P.; Lin, J.-S.; Li, H.; Freedman, M.T.; Mun, S.K. Artificial convolution neural network for medical image pattern recognition. Neural Netw. 1995, 8, 1201–1214. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef]
- Liu, S.; Zheng, H.; Feng, Y.; Li, W. Prostate cancer diagnosis using deep learning with 3D multiparametric MRI. In Medical Imaging 2017: Computer-Aided Diagnosis; SPIE 10134; International Society for Optics and Photonics: Orlando, FL, USA, 2017; p. 1013428. [Google Scholar]
- Han, Z.; Wei, B.; Zheng, Y.; Yin, Y.; Li, K.; Li, S. Breast Cancer Multi-classification from Histopathological Images with Structured Deep Learning Model. Sci. Rep. 2017, 7, 4172. [Google Scholar] [CrossRef]
- Abraham, B.; Nair, M.S. Automated grading of prostate cancer using convolutional neural network and ordinal class classifier. Inform. Med. Unlocked 2019, 17, 100256. [Google Scholar] [CrossRef]
- Truki, T. An Empirical Study of Machine Learning Algorithms for Cancer Identification. In Proceedings of the 2018 IEEE 15th International Conference on Networking, Sensing and Control (ICNSC), Zhuhai, China, 27–29 March 2018; pp. 1–5. [Google Scholar]
- Veta, M.M.; Pluim, J.P.W.; Van Diest, P.J.; Viergever, M.A. Breast Cancer Histopathology Image Analysis: A Review. IEEE Trans. Biomed. Eng. 2014, 61, 1400–1411. [Google Scholar] [CrossRef]
- Moradi, M.; Mousavi, P.; Abolmaesumi, P. Computer-Aided Diagnosis of Prostate Cancer with Emphasis on Ultrasound-Based Approaches: A Review. Ultrasound Med. Biol. 2007, 33, 1010–1028. [Google Scholar] [CrossRef]
- Alom, Z.; Yakopcic, C.; Nasrin, M.S.; Taha, T.M.; Asari, V.K. Breast Cancer Classification from Histopathological Images with Inception Recurrent Residual Convolutional Neural Network. J. Digit. Imaging 2019, 32, 605–617. [Google Scholar] [CrossRef]
- Wang, C.; Shi, J.; Zhang, Q.; Ying, S. Histopathological image classification with bilinear convolutional neural networks. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Seogwipo, Korea, 15–16 July 2017; Volume 2017, pp. 4050–4053. [Google Scholar]
- Smith, S.A.; Newman, S.J.; Coleman, M.P.; Alex, C. Characterization of the histologic appearance of normal gill tissue using special staining techniques. J. Vet. Diagn. Investig. 2018, 30, 688–698. [Google Scholar] [CrossRef]
- Vodyanoy, V.; Pustovyy, O.; Globa, L.; Sorokulova, I. Primo-Vascular System as Presented by Bong Han Kim. Evid. Based Complement. Altern. Med. 2015, 2015, 361974. [Google Scholar] [CrossRef] [PubMed]
- Larson, K.; Ho, H.H.; Anumolu, P.L.; Chen, M.T. Hematoxylin and Eosin Tissue Stain in Mohs Micrographic Surgery: A Review. Dermatol. Surg. 2011, 37, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-C.; Cheng, F.-C.; Chiu, Y.-S. Efficient Contrast Enhancement Using Adaptive Gamma Correction With Weighting Distribution. IEEE Trans. Image Process. 2012, 22, 1032–1041. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.; Rahman, M.; Abdullah-Al-Wadud, M.; Al-Quaderi, G.D.; Shoyaib, M. An adaptive gamma correction for image enhancement. EURASIP J. Image Video Process. 2016, 2016, 35. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 60. [Google Scholar] [CrossRef]
- Lecun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Kieffer, B.; Babaie, M.; Kalra, S.; Tizhoosh, H.R. Convolutional neural networks for histopathology image classification: Training vs. Using pre-trained networks. In Proceedings of the 2017 Seventh International Conference on Image Processing Theory, Tools and Applications (IPTA), Montreal, QC, Canada, 28 November–1 December 2017; pp. 1–6. [Google Scholar]
- Mourgias-Alexandris, G.; Tsakyridis, A.; Passalis, N.; Tefas, A.; Vyrsokinos, K.; Pleros, N. An all-optical neuron with sigmoid activation function. Opt. Express 2019, 27, 9620–9630. [Google Scholar] [CrossRef]
- Elfwing, S.; Uchibe, E.; Doya, K. Sigmoid-weighted linear units for neural network function approximation in reinforcement learning. Neural Netw. 2018, 107, 3–11. [Google Scholar] [CrossRef]
- Kouretas, I.; Paliouras, V. Simplified Hardware Implementation of the Softmax Activation Function. In Proceedings of the 2019 8th International Conference on Modern Circuits and Systems Technologies (MOCAST), Thessaloniki, Greece, 13–15 May 2019; pp. 1–4. [Google Scholar]
- Zhu, Q.; He, Z.; Zhang, T.; Cui, W. Improving Classification Performance of Softmax Loss Function Based on Scalable Batch-Normalization. Appl. Sci. 2020, 10, 2950. [Google Scholar] [CrossRef]
- Dietterich, T.G. Ensemble Methods in Machine Learning. In International Workshop on Multiple Classifier System; Springer: Berlin, Heidelberg, 2000; pp. 1–15. [Google Scholar] [CrossRef]
- Dikaios, N.; Alkalbani, J.; Sidhu, H.S.; Fujiwara, T.; Abd-Alazeez, M.; Kirkham, A.; Allen, C.; Ahmed, H.; Emberton, M.; Freeman, A.; et al. Logistic regression model for diagnosis of transition zone prostate cancer on multi-parametric MRI. Eur. Radiol. 2015, 25, 523–532. [Google Scholar] [CrossRef]
- Nguyen, C.; Wang, Y.; Nguyen, H.N. Random forest classifier combined with feature selection for breast cancer diagnosis and prognostic. J. Biomed. Sci. Eng. 2013, 6, 551–560. [Google Scholar] [CrossRef]
- Cruz, J.A.; Wishart, D.S. Applications of Machine Learning in Cancer Prediction and Prognosis. Cancer Inform. 2006, 2, 59–77. [Google Scholar] [CrossRef]
- Tang, T.T.; Zawaski, J.A.; Francis, K.N.; Qutub, A.A.; Gaber, M.W. Image-based Classification of Tumor Type and Growth Rate using Machine Learning: A preclinical study. Sci. Rep. 2019, 9, 12529. [Google Scholar] [CrossRef] [PubMed]
- Madabhushi, A.; Lee, G. Image analysis and machine learning in digital pathology: Challenges and opportunities. Med. Image Anal. 2016, 33, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Huang, H.; Zhang, Z.; Chen, X.; Huang, K.; Zhang, S. Towards Rich Feature Discovery With Class Activation Maps Augmentation for Person Re-Identification. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 1389–1398. [Google Scholar]
- Hou, X.; Gong, Y.; Liu, B.; Sun, K.; Liu, J.; Xu, B.; Duan, J.; Qiu, G. Learning Based Image Transformation Using Convolutional Neural Networks. IEEE Access 2018, 6, 49779–49792. [Google Scholar] [CrossRef]
- Chai, X.; Gu, H.; Li, F.; Duan, H.; Hu, X.; Lin, K. Deep learning for irregularly and regularly missing data reconstruction. Sci. Rep. 2020, 10, 3302. [Google Scholar] [CrossRef]



| Dataset | Benign (0) | Malignant (1) | Total |
|---|---|---|---|
| Training | 1800 | 1800 | 3600 |
| Validation | 600 | 600 | 1200 |
| Testing | 600 | 600 | 1200 |
| Total | 3000 | 3000 | 6000 |
| Layer Type | Filters | Output Shape | Kernel Size/Strides | |
|---|---|---|---|---|
| Model-1 Specification | ||||
| Input | Image | 1 | 224 × 224 × 3 | - |
| Block-1 | 2× convolutional + ReLU + BN | 32 | 56 × 56 × 32 | 3 × 3/2 |
| Block-2 | 2× convolutional + ReLU + BN | 64 | 56 × 56 × 64 | 3 × 3/1 |
| - | Max pooling + dropout (0.25) | 64 | 28 × 28 × 64 | 2 × 2/2 |
| Block-3 | 3× convolutional + ReLU + BN | 128 | 28 × 28 × 128 | 3 × 3/1 |
| - | Max pooling + dropout (0.25) | 128 | 14 × 14 × 128 | 2 × 2/2 |
| Block-4 | 3× convolutional + ReLU + BN | 256 | 14 × 14 × 256 | 3 × 3/1 |
| - | Max pooling + dropout (0.25) | 256 | 7 × 7 × 256 | 2 × 2/2 |
| - | Flatten | - | 12,544 | - |
| - | Dense-1 + ReLU + BN | 1024 | 1024 | - |
| - | Dense-2 + ReLU + BN | 1024 | 1024 | - |
| - | Dropout (0.5) | 1024 | 1024 | - |
| Output | Sigmoid | 2 | 2 | - |
| Model-2 specification | ||||
| Input | Image | 1 | 224 × 224 × 3 | - |
| Block-1 | 2× convolutional + ReLU + BN | 92 | 56 × 56 × 92 | 5 × 5/2 |
| Block-2 | 2× convolutional + ReLU + BN | 192 | 56 × 56 × 192 | 3 × 3/1 |
| - | Max pooling | 192 | 28 × 28 × 192 | 2 × 2/2 |
| Block-3 | 3× convolutional + ReLU + BN | 384 | 28 × 28 × 384 | 3 × 3/1 |
| - | Max pooling + dropout (0.25) | 384 | 14 × 14 × 384 | 2 × 2/2 |
| - | GAP | - | 384 | 2 × 2/2 |
| - | Dense-1 + ReLU + BN | 64 | 64 | - |
| - | Dense-2 + ReLU + BN | 32 | 32 | - |
| - | Dropout (0.5) | 32 | 32 | - |
| Output | Softmax | 2 | 2 | - |
| Models | Specification |
|---|---|
| Model-1, VGG-16, ResNet-50, Inception-V3, DenseNet-121 | loss = binary_crossentropy; learning rate = start:1.0—auto reduce on plateau fraction: 0.8 after 10 consecutive non-declines of validation loss; classifier = sigmoid; epochs = 300 |
| Model-2 | loss = binary_crossentropy; learning rate = start:1.0—auto reduce on plateau fraction: 0.8 after 10 consecutive non-declines of validation loss; classifier = softmax; epochs = 300, kernel initializer = glorot_uniform |
| LR | C = 100, max_iter = 500, tol = 0.001, method = isotonic, penalty = l2 |
| RF | n_estimators = 500, criterion = gini, max_depth = 9, min_samples_split = 5, min_samples_leaf = 4, method = isotonic |
| Model-1 | Model-2 | |||
|---|---|---|---|---|
| Optimizers | Test Loss (%) | Test Accuracy (%) | Test Loss (%) | Test Accuracy (%) |
| SGD | 0.51 | 85.7 | 0.25 | 93.3 |
| RMSProp | 1.00 | 85.5 | 0.62 | 89.3 |
| Adam | 0.45 | 84.4 | 0.28 | 91.1 |
| Adadelta | 0.54 | 89.1 | 0.25 | 94.0 |
| Deep Learning | ||||||
|---|---|---|---|---|---|---|
| Model-1 | Model-2 | VGG-16 | ResNet-50 | Inception-V3 | DenseNet-121 | |
| Accuracy | 89.1% | 94.0% | 92.0% | 93.0% | 94.6% | 95.0% |
| Precision | 89.2% | 94.2% | 92.2% | 95.0% | 96.5% | 96.2% |
| Recall | 89.1% | 92.9% | 91.9% | 90.6% | 93.2% | 94.6% |
| F1-Score | 89.0% | 93.5% | 92.0% | 92.8% | 94.8% | 95.4% |
| MCC | 78.3% | 87.0% | 84.0% | 85.3% | 89.5% | 90.7% |
| Ensemble Machine Learning | ||||
|---|---|---|---|---|
| CNN-Based | OCLBP | IOCLBP | OCLBP + IOCLBP | |
| Accuracy | 92.0% | 69.3% | 83.6% | 85.0% |
| Precision | 92.7% | 66.0% | 83.2% | 85.5% |
| Recall | 91.0% | 70.6% | 83.9% | 84.5% |
| F1-Score | 91.8% | 68.2% | 83.5% | 85.0% |
| MCC | 83.5% | 38.6% | 67.2% | 69.8% |
| Models | Parameter (Trainable) | Model Memory Usage | Time to Solution (Minutes) | |
|---|---|---|---|---|
| Train | Test | |||
| VGG-16 | 27,823,938 | 60.9 MB | 570 | <1 |
| ResNet-50 | 23,538,690 | 148.4 MB | 660 | <1 |
| Inception-V3 | 22,852,898 | 66.9 MB | 600 | <1 |
| DenseNet-121 | 7,479,682 | 199.7 MB | 700 | <1 |
| Model-2 (LWCNN) | 5,386,638 | 44.7 MB | 190 | <1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhattacharjee, S.; Kim, C.-H.; Prakash, D.; Park, H.-G.; Cho, N.-H.; Choi, H.-K. An Efficient Lightweight CNN and Ensemble Machine Learning Classification of Prostate Tissue Using Multilevel Feature Analysis. Appl. Sci. 2020, 10, 8013. https://doi.org/10.3390/app10228013
Bhattacharjee S, Kim C-H, Prakash D, Park H-G, Cho N-H, Choi H-K. An Efficient Lightweight CNN and Ensemble Machine Learning Classification of Prostate Tissue Using Multilevel Feature Analysis. Applied Sciences. 2020; 10(22):8013. https://doi.org/10.3390/app10228013
Chicago/Turabian StyleBhattacharjee, Subrata, Cho-Hee Kim, Deekshitha Prakash, Hyeon-Gyun Park, Nam-Hoon Cho, and Heung-Kook Choi. 2020. "An Efficient Lightweight CNN and Ensemble Machine Learning Classification of Prostate Tissue Using Multilevel Feature Analysis" Applied Sciences 10, no. 22: 8013. https://doi.org/10.3390/app10228013
APA StyleBhattacharjee, S., Kim, C.-H., Prakash, D., Park, H.-G., Cho, N.-H., & Choi, H.-K. (2020). An Efficient Lightweight CNN and Ensemble Machine Learning Classification of Prostate Tissue Using Multilevel Feature Analysis. Applied Sciences, 10(22), 8013. https://doi.org/10.3390/app10228013






