An Adaptive Generative Adversarial Network for Cardiac Segmentation from X-ray Chest Radiographs
Abstract
1. Introduction
- Quick initial diagnosis
- Joint diagnosis
- Guidance for clinical surgery
- The first stage
- The second stage
- The third stage
2. Basic Theories and Methods Related to the Proposed AGAN
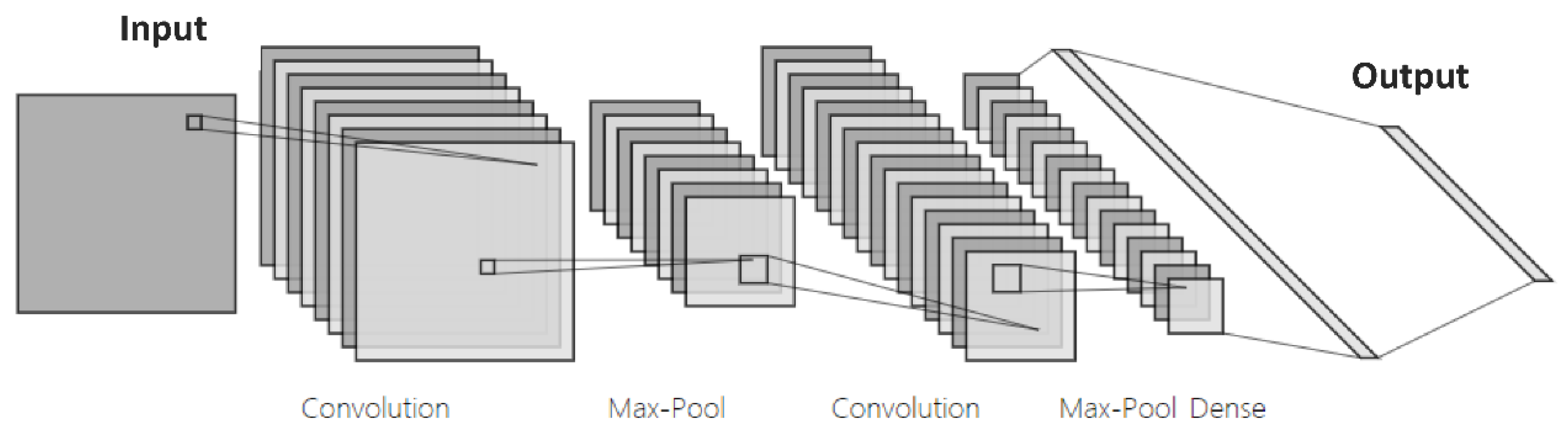
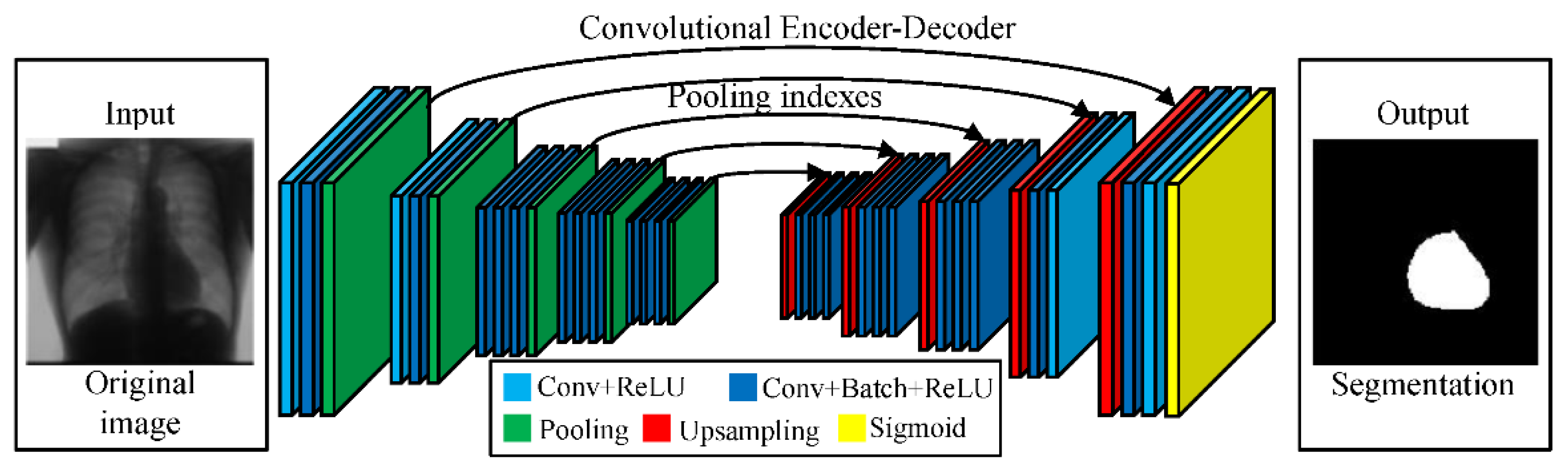
2.1. Segmentation of Images Using Neural Networks
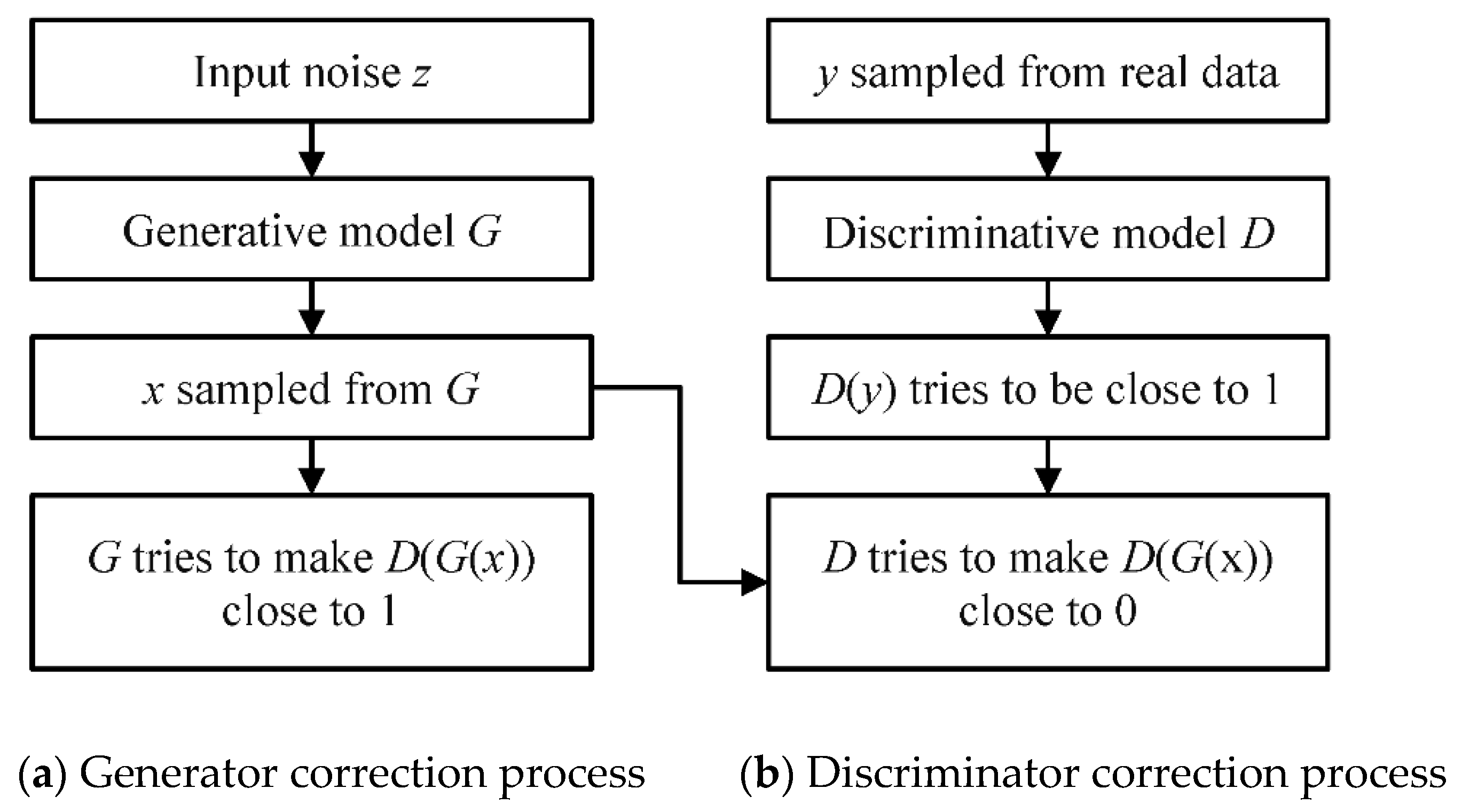
2.2. Generate Adversarial Network Model
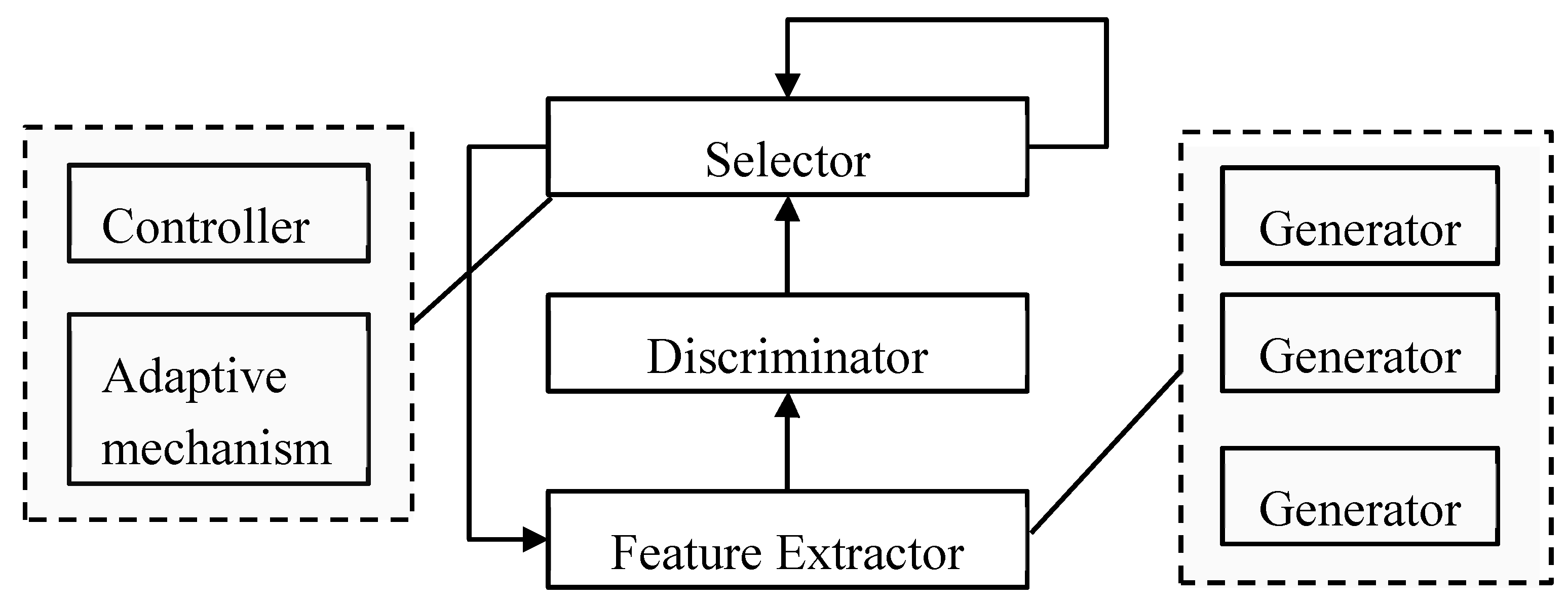
2.3. Adaptive Model
3. Image Segmentation Based on an AGAN
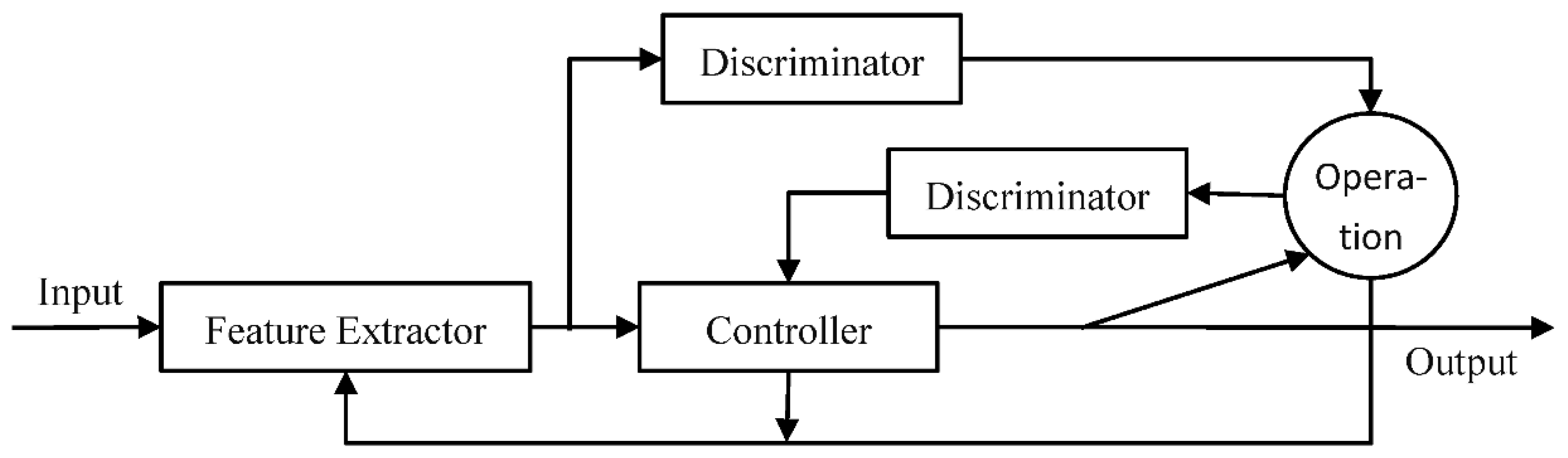
3.1. Agan Framework
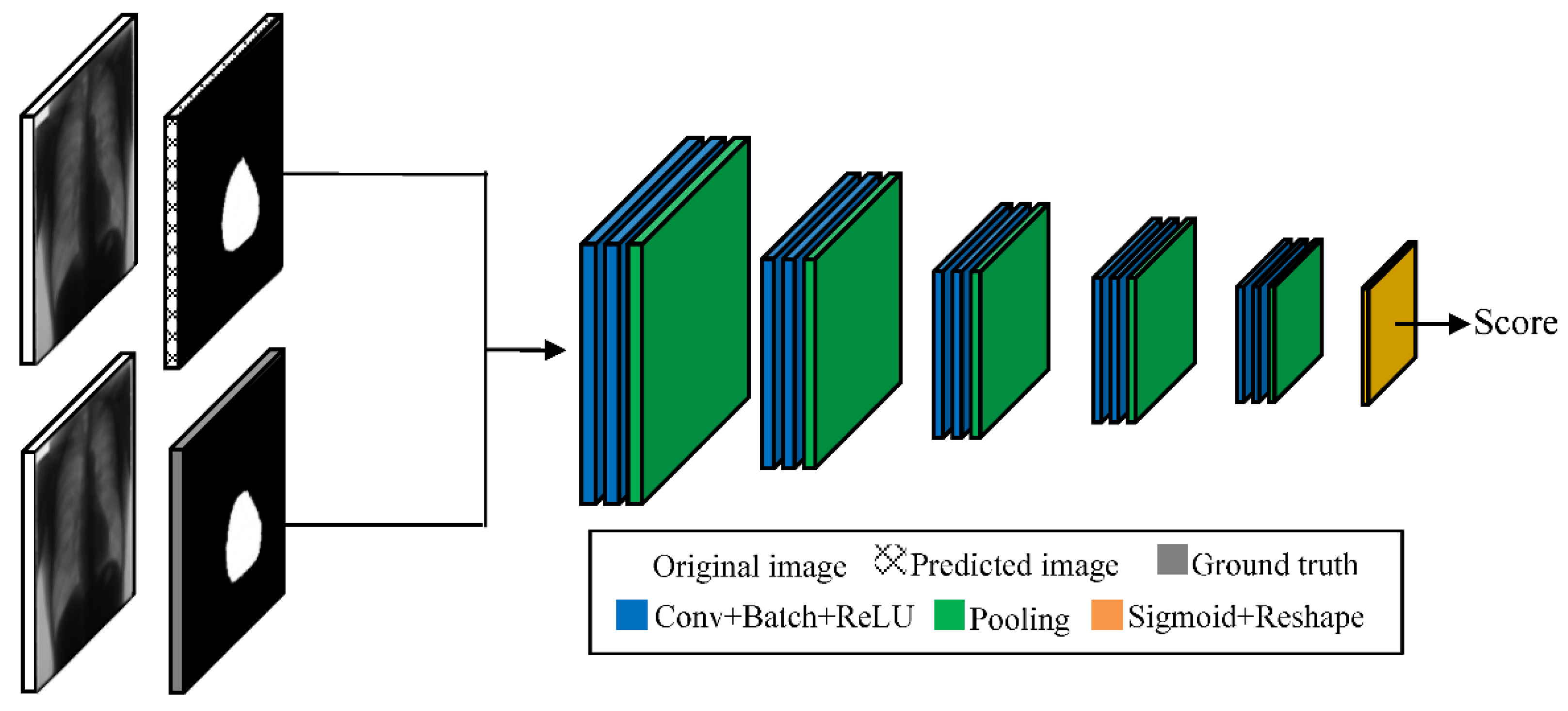
3.2. Feature Extractor
3.3. Discriminator
3.4. Selector
3.5. Training and Testing Process of the Agan
- (1).
- First, image features are extracted by the feature extractor.
- (2).
- Second, the features of different dimensions extracted by the two generators in the feature extractor are scored by the discriminator.
- (3).
- Finally, the selector adjusts the system through feedback and adaptive adjustment.
- (4).
- The above three steps are repeated until the network has a good representation ability; that is, the first loop of the adaptive adjustment process in the selector automatically terminates, and then the result of the calculation in the third loop is output.
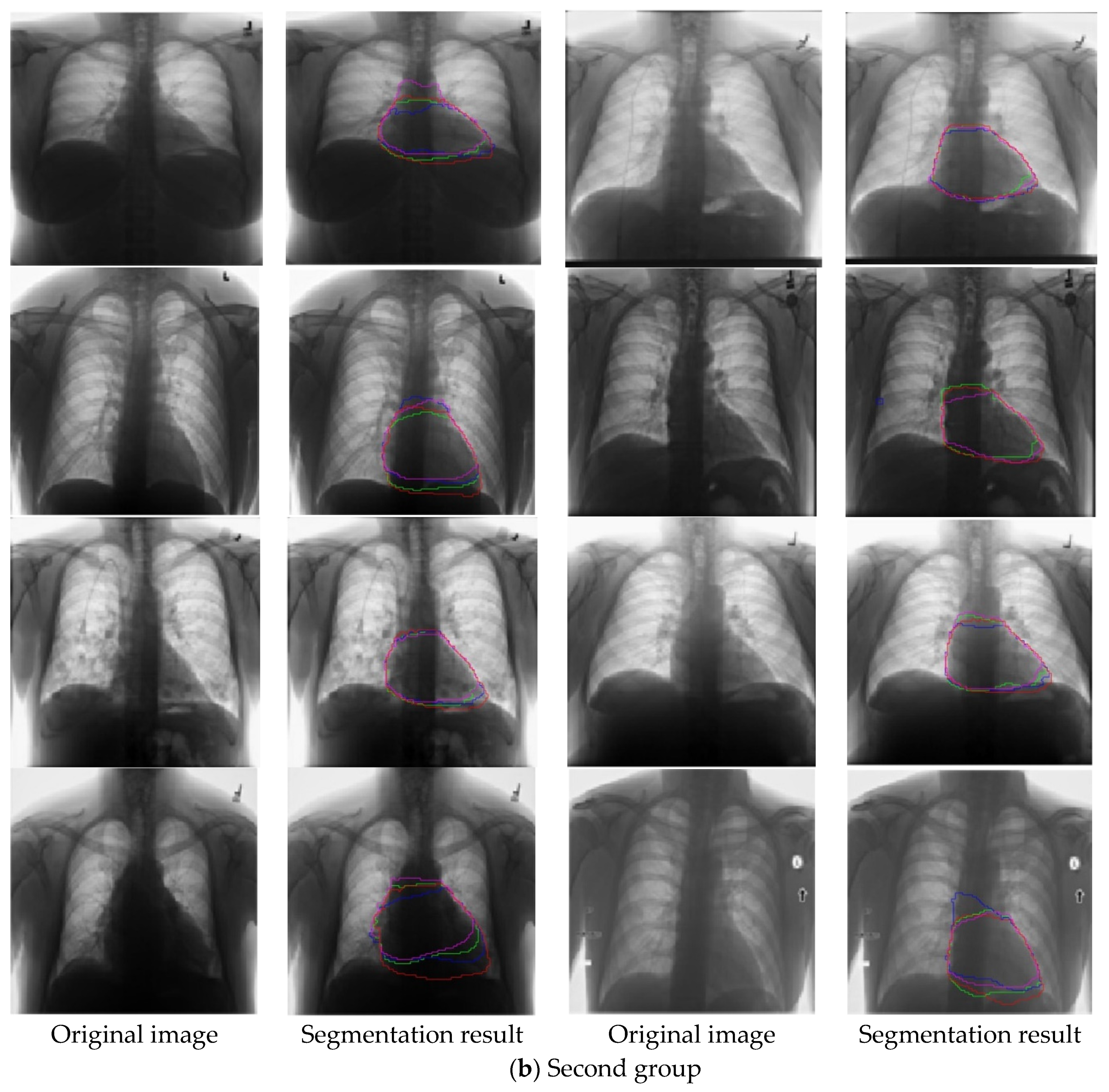
4. Experiments
4.1. Image Database
4.2. Evaluation Criteria
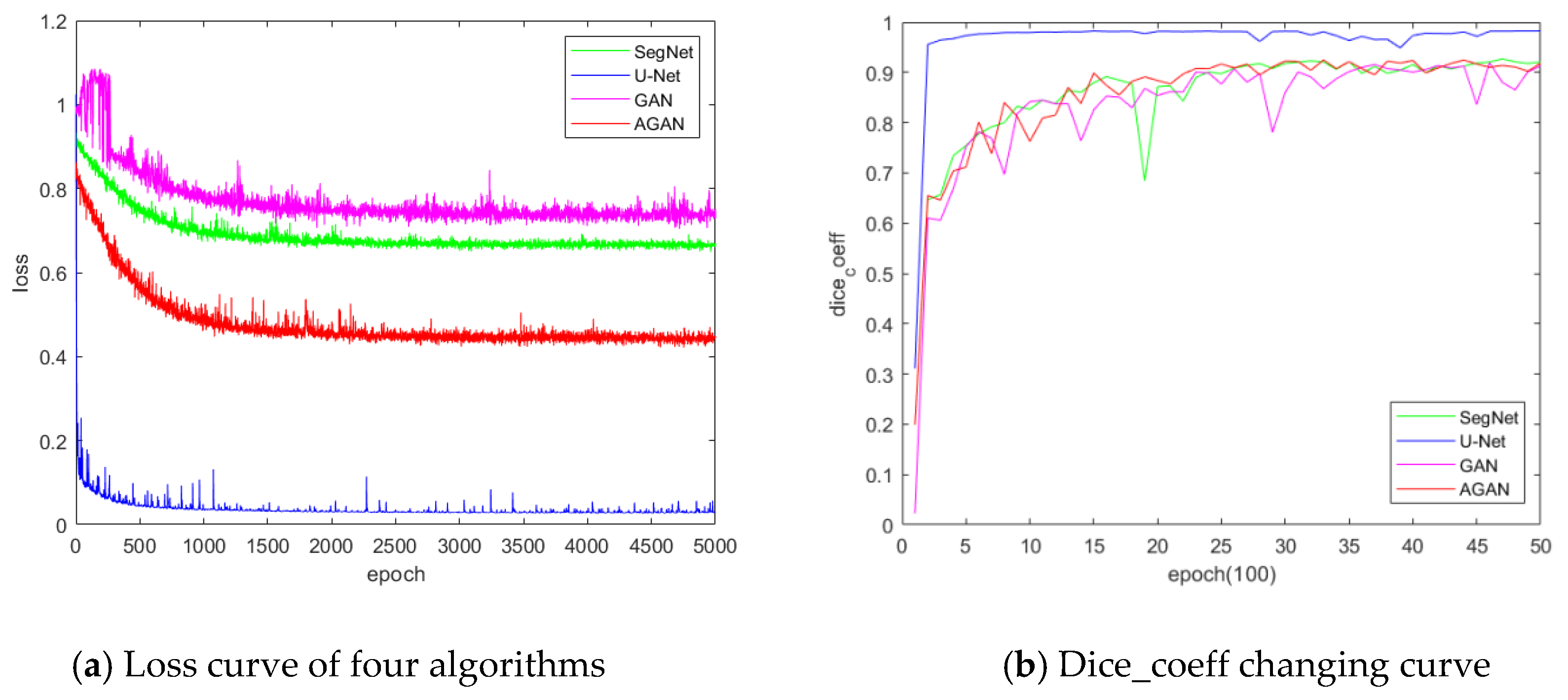
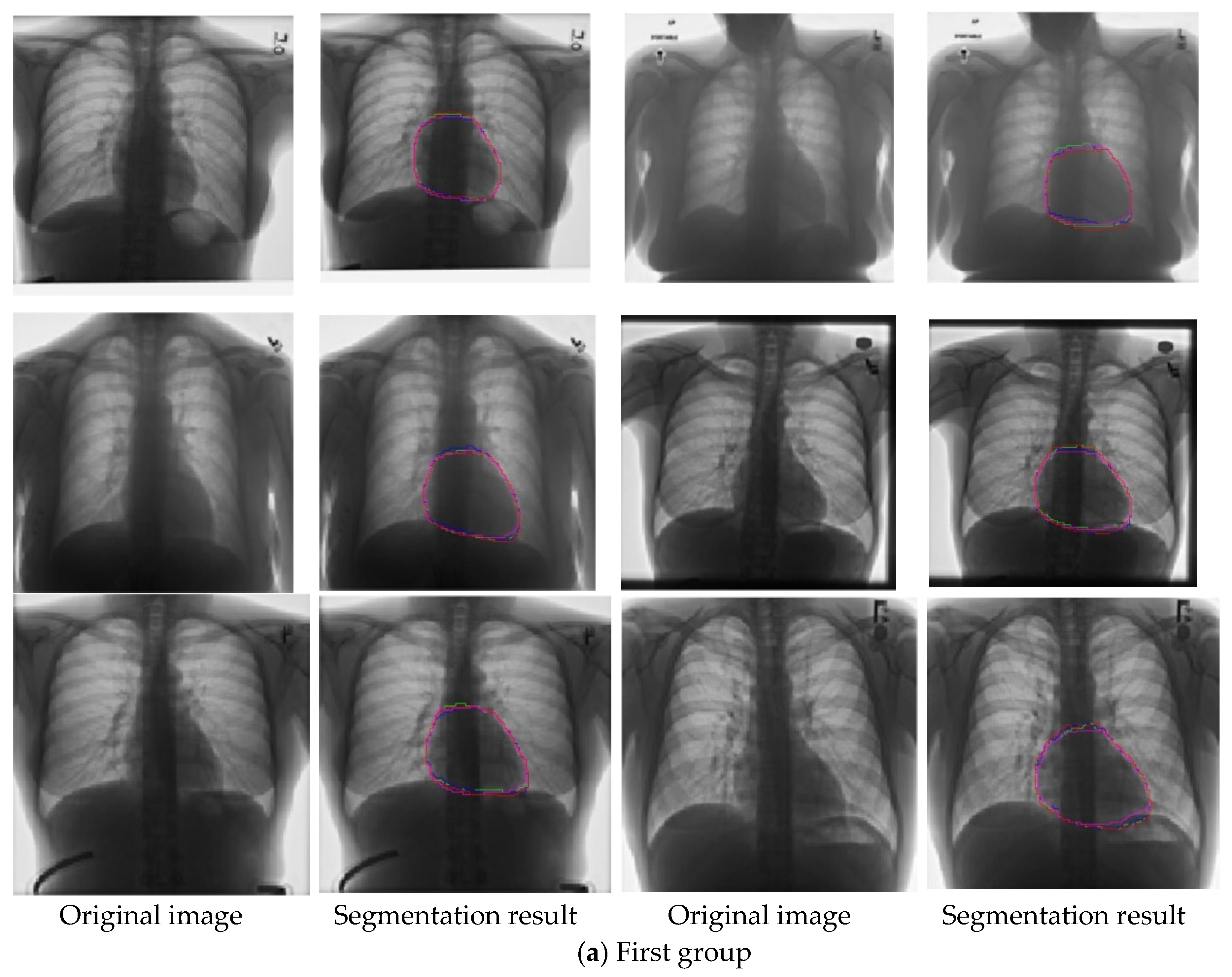
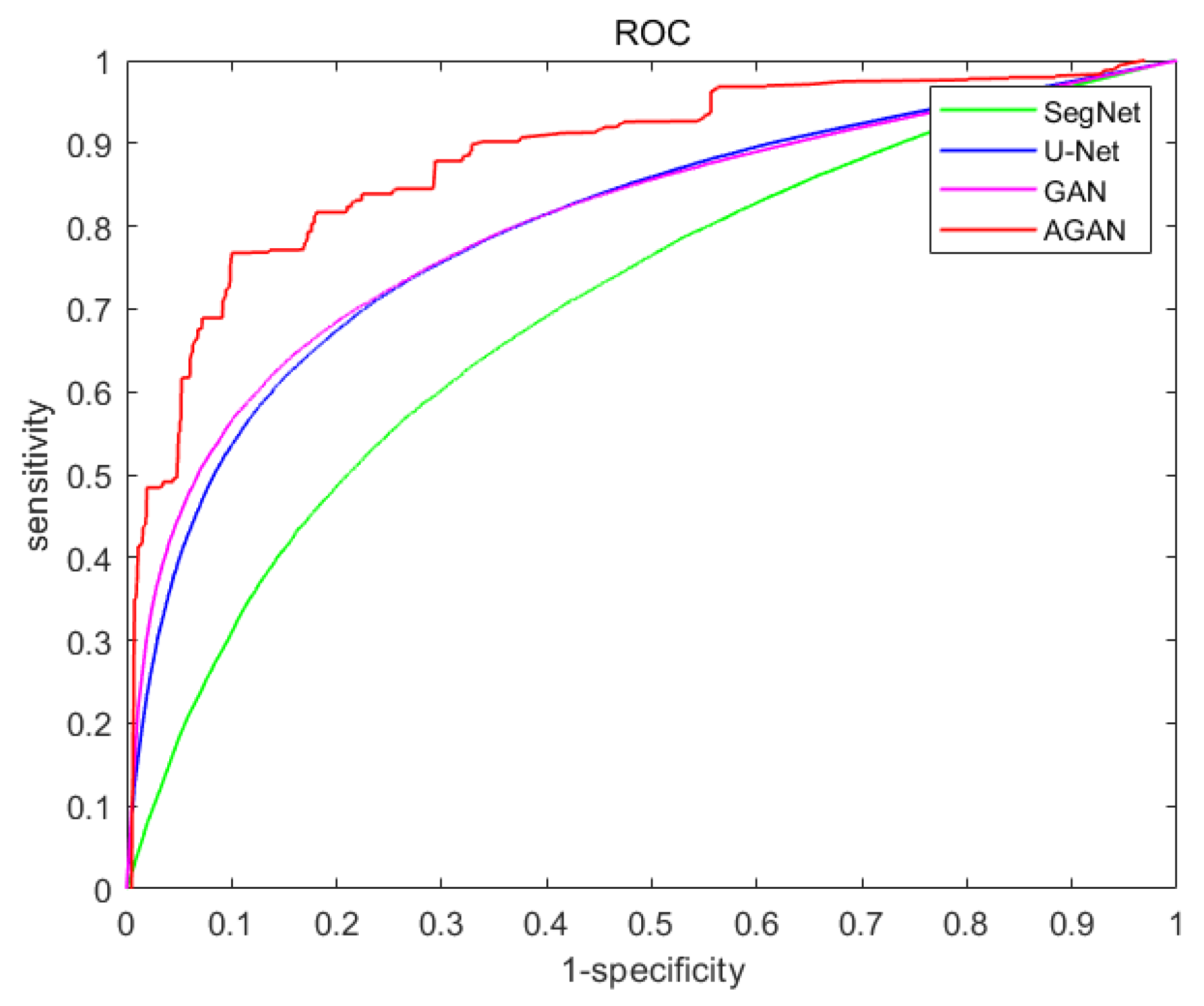
4.3. Experimental Performance Comparison and Analysis
- Accuracy
- Reliability
- Stability
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Lesage, D.; Angelini, E.D.; Bloch, I.; Funka-Lea, G. A review of 3d vessel lumen segmentation techniques: Models, features and extraction schemes. Med. Image Anal. 2009, 13, 819–845. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Shuang, Q.T.; Bin, C.Z. An Automated Calculation of Cardiothoracic Ratio on Chest Radiography. Chin. J. Biomed. Eng. 2009, 28, 149–152. [Google Scholar]
- Toth, D.; Panayiotou, M.; Brost, A.; Behar, J.M.; Rinaldi, C.A.; Rhode, K.S.; Mountney, P. 3D/2D Registration with Superabundant Vessel Reconstruction for Cardiac Resynchronization Therapy. Med. Image Anal. 2017, 42, 160. [Google Scholar] [CrossRef] [PubMed]
- Hatt, C.R.; Speidel, M.A.; Raval, A.N. Real-Time Pose Estimation of Devices from X-ray Images: Application to X-ray/Echo Registration for Cardiac Interventions. Med Image Anal. 2016, 34, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Yan, P.; Zhang, W.; Turkbey, B.; Choyke, P.L.; Li, X. Global structure constrained local shape prior estimation for medical image segmentation. Comput. Vis. Image Underst. 2013, 117, 1017–1026. [Google Scholar] [CrossRef]
- Yu, H.; He, F.; Pan, Y. A novel segmentation model for medical images with intensity inhomogeneity based on adaptive perturbation. Multimed. Tools Appl. 2019, 78, 11779–11798. [Google Scholar] [CrossRef]
- Iakovidis, D.K. Versatile approximation of the lung field boundaries in chest radiographs in the presence of bacterial pulmonary infections. In Proceedings of the IEEE International Conference on Bioinformatics & Bioengineering IEEE 2008, Athens, Greece, 8–10 October 2008. [Google Scholar]
- Umehara, K.; Ota, J.; Ishimaru, N.; Ohno, S.; Okamoto, K.; Suzuki, T.; Shirai, N.; Ishida, T. Super-resolution convolutional neural network for the improvement of the image quality of magnified images in chest radiographs. In Proceedings of the Conference on Medical Imaging—Image Processing, Orlando, FL, USA, 12–14 February 2017. [Google Scholar]
- Becker, H.C.; Nettleton, W.J.; Meyers, P.H.; Sweeney, J.W.; Nice, C.M. Digital Computer Determination of a Medical Diagnostic Index Directly from Chest X-Ray Images. IEEE Trans. Biomed. Eng. 1964, BME-11, 67–72. [Google Scholar] [CrossRef]
- Hall, D.L.; Lodwick, G.S.; Kruger, R.; Dwyer, S.J. Computer diagnosis of heart disease. Radiol. Clin. N. Am. 1972, 9, 533–541. [Google Scholar]
- Nakamori, N.; Doi, K.; Sabeti, V.; MacMahon, H. Image feature analysis and computer-aided diagnosis in digital radiography: Automated analysis of sizes of heart and lung in chest images. Med. Phys. 1990, 17, 342. [Google Scholar] [CrossRef]
- Viergever, M.A.; Romeny, B.H.; van Goudoever, J.B. Computer-aided diagnosis in chest radiography: A survey. IEEE Trans. Med. Imaging 2001, 20, 228–241. [Google Scholar]
- James, S.D.; Nicholas, A. Medical image analysis: Progress over two decades and the challenges ahead. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 85–106. [Google Scholar]
- Kass, M.; Witkin, A.; Terzopoulos, D. Snakes: Active Contour Models. Int. J. Comput. Vis. 1988, 1, 321–331. [Google Scholar] [CrossRef]
- Behiels, G.; Vandermeulen, D.; Maes, F.; Suetens, P.; Dewaele, P. Active Shape Model-Based Segmentation of Digital X-ray Images. Lect. Notes Comput. Sci. 1999, 1679, 128–137. [Google Scholar]
- Cootes, F.T.; Edwards, G.J.; Taylor, C.J. Active appearance models. IEEE Trans. Pattern Anal. Mach. Intell. 2001, 23, 681–685. [Google Scholar] [CrossRef]
- Van Ginneken, B.; Frangi, A.F.; Staal, J.J.; ter Haar Romeny, B.M.; Viergever, M.A. Active shape model segmentation with optimal features. IEEE Trans. Med. Imaging 2002, 21, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Alex, K.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. Available online: http://www.cs.toronto.edu/~hinton/absps/imagenet.pdf (accessed on 22 June 2020).
- Olaf, R.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the 18th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Goodfellow, I.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative adversarial nets. In Proceedings of the 2014 Conference on Advances in Neural Information Processing Systems 27; Curran Associates, Inc.: Montreal, QC, Canada, 2014; pp. 2672–2680. [Google Scholar]
- Dan, P.; Longfei, J.; An, Z.; Xiaowei, S. Applications of Generative Adversarial Networks in medical image processing. J. Biomed. Eng. 2018, 6, 970–976. [Google Scholar]
- Chen, C.; Dou, Q.; Chen, H.; Heng, P.A. Semantic-Aware Generative Adversarial Nets for Unsupervised Domain Adaptation in Chest X-Ray Segmentation. In Machine Learning in Medical Imaging Workshop with MICCAI 2018; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Li, X.; Wang, W.; Sun, W. Multi-model Adaptive Control. Control. Decis. 2000, 15, 390–394. [Google Scholar]
- Lv, Y.; Na, J.; Yang, Q.; Wu, X.; Guo, Y. Online adaptive optimal control for continuous-time nonlinear systems with completely unknown dynamics. Int. J. Control. 2016, 89, 99–112. [Google Scholar] [CrossRef]
- Affonso, C.; Rossi, A.L.D.; Vieira, F.H.A.; de Leon Ferreira de Carvalhob, A.C.P. Deep learning for biological image classification. Expert Syst. Appl. 2017, 85, 114–122. [Google Scholar] [CrossRef]
- Ma, Z.; Wu, X.; Wang, X.; Song, Q.; Yin, Y.; Cao, K.; Wang, Y.; Zhou, J. An iterative multi-path fully convolutional neural network for automatic cardiac segmentation in cine MR images. Med. Phys. 2019, 46, 12. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Li, X.; Huang, H.; Guo, N.; Li, Q. Deep Learning-based Image Segmentation on Multi-modal Medical Imaging. IEEE Trans. Radiat. Plasma Med. Sci. 2019, 3, 162–169. [Google Scholar] [CrossRef]
- Xia, K.; Yin, H.; Qian, P.; Jiang, Y.; Wang, S. Liver Semantic Segmentation Algorithm Based on Improved Deep Adversarial Networks in combination of Weighted Loss Function on Abdominal CT Images. IEEE Access 2019, 7, 96349–96358. [Google Scholar] [CrossRef]



| Network | Dice_Coeff | Acc | Sensitivity | Specificity |
|---|---|---|---|---|
| SegNet | 92.41% | 98.65% | 88.86% | 99.64% |
| U-Net | 92.09% | 98.59% | 88.31% | 99.64% |
| GAN | 93.16% | 98.75% | 91.58% | 99.48% |
| AGAN | 94.06% | 98.92% | 92.25% | 99.60% |
| Evaluation Index | SegNet | U-Net | GAN | AGAN |
|---|---|---|---|---|
| STR | 1.95 | 1.96 | 1.93 | 1.91 |
| STE | 1.93 | 1.91 | 1.90 | 1.91 |
| BR | 97.58% | 97.65% | 97.38% | 98.37% |
| WR | 69.18% | 60.02% | 70.18% | 72.60% |
| N | 3 | 4 | 4 | 1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, X.; Tian, X. An Adaptive Generative Adversarial Network for Cardiac Segmentation from X-ray Chest Radiographs. Appl. Sci. 2020, 10, 5032. https://doi.org/10.3390/app10155032
Wu X, Tian X. An Adaptive Generative Adversarial Network for Cardiac Segmentation from X-ray Chest Radiographs. Applied Sciences. 2020; 10(15):5032. https://doi.org/10.3390/app10155032
Chicago/Turabian StyleWu, Xiaochang, and Xiaolin Tian. 2020. "An Adaptive Generative Adversarial Network for Cardiac Segmentation from X-ray Chest Radiographs" Applied Sciences 10, no. 15: 5032. https://doi.org/10.3390/app10155032
APA StyleWu, X., & Tian, X. (2020). An Adaptive Generative Adversarial Network for Cardiac Segmentation from X-ray Chest Radiographs. Applied Sciences, 10(15), 5032. https://doi.org/10.3390/app10155032




